Abstract
Myelin/oligodendrocyte glycoprotein (MOG) is a target antigen for myelin-destructive Abs in autoimmune central nervous system demyelinating disorders. Little is known about the molecular and structural basis of these pathogenic Ab responses. Here, we have characterized anti-MOG Ab specificities in the marmoset model of experimental allergic encephalomyelitis, by means of a combinatorial IgG-Fab library. We found that a diverse population of Ig genes encodes for auto-Abs that exclusively recognize conformation-dependent antigenic targets on MOG. These antigenic domains correspond to exposed epitopes in vivo, as the Fab fragments recognize native MOG in situ in marmoset brain tissue. The Ab fragments described here represent Ab specificities that are common constituents of the humoral immune repertoire against MOG in outbred populations, as demonstrated by their ability to displace native anti-MOG Abs present in sera from MOG-immune marmosets and patients with multiple sclerosis. Furthermore, neuropathological analysis and characterization of Ab epitope specificities in animals immunized with MOG or MOG-derived peptides revealed that only conformation-dependent Abs are associated with demyelinating activity, suggesting that epitope recognition is an important factor for Ab pathogenicity. Our findings provide novel and unexpected knowledge on the diversity of anti-MOG Ab responses in nonhuman primates and humans, and will permit the dissection of pathogenic auto-Ab properties in multiple sclerosis.
Multiple sclerosis (MS)† is a chronic, demyelinating disease of the central nervous system (CNS) that is thought to be mediated by autoaggressive immune responses against myelin antigens (reviewed in ref. 1). Extensive investigations have addressed the respective roles of T and B cell responses against myelin antigens in experimental allergic encephalomyelitis (EAE), a disease model for MS. It is now recognized that, whereas myelin-reactive T cell responses are essential to disease pathogenesis, auto-Abs may play a major role as effectors of tissue damage (1–4). Myelin/oligodendrocyte glycoprotein (MOG) is a surface-exposed protein of myelin that has been identified as a prime target for demyelinating auto-Abs in several species (5–7). Anti-MOG auto-Abs mediate a characteristic vesicular transformation of compact myelin in acutely demyelinating lesions, a neuropathological feature which has also been documented in human MS (8).
Despite these advances, the significance of polyclonal Ab responses against MOG measured in humans remains unclear. Anti-MOG Abs seem to be equally prevalent in the peripheral blood of affected patients and healthy controls (9, 10), and precise definition of the disease-relevant Ab epitopes of MOG is lacking. Similarly, the pathogenic significance of humoral responses directed against MOG has not been established with certainty for all EAE models (11). Indeed, these findings raise the possibility that the MOG-specific humoral response may be heterogeneous in terms of their potential to mediate demyelination. Analyses of the fine specificities of anti-MOG Abs in EAE and MS have mainly been conducted with short peptides derived from the amino acid sequence of MOG (12–14). This approach cannot provide an understanding of the full complexity of anti-MOG humoral responses, because it does not account for epitopes that depend on the tertiary structure of the folded protein. Similarly, whereas molecular studies have independently established that CNS-specific clonal expansion of B cells occurs in MS (15–18), the antigenic specificities of these responses have not been identified. The use of systems that permit analysis of gene usage and individual Ab specificities should facilitate characterization of humoral responses against myelin autoantigens.
Here, we used a combinatorial Ab library of Fab fragments to characterize the humoral immune response against MOG in the common marmoset, an outbred primate species that develop an MS-like, Ab-mediated form of EAE after immunization with MOG (19). We have observed that the recombinant MOG-specific Ab fragments use a limited repertoire of heavy (H)- and light (L)-chain genes and identify epitopes of MOG with specificities that are strictly conformation-dependent. The conformational epitopes of MOG defined by these Fab fragments are consistently targeted by the humoral repertoire in all outbred marmosets studied to date. Furthermore, we show that MOG-immune marmosets do not develop demyelinating EAE unless their humoral repertoire includes conformation-dependent Abs, a finding that underscores the relevance of this Ab subgroup in disease pathogenesis.
Materials and Methods
Animals and Induction of EAE.
All Callithrix jacchus marmosets used in this study were maintained in a primate colony at the University of California, San Francisco, and were cared for in accordance with all guidelines of the local Institutional Animal Care and Usage Committee. EAE was induced by active immunization with either 50 μg of recombinant protein corresponding to the extracellular domain of rat MOGaa1–125 (rMOG) expressed in Escherichia coli and purified to homogeneity following published procedures (20) or a mixture of 100 μg each of overlapping synthetic 20-mer peptides corresponding to the sequence of MOGaa1–120 (Research Genetics, Huntsville, AL). Peptides were purified >95% by HPLC, and purity was confirmed by mass spectrometry. Antigens were dissolved in 200 μl of PBS, emulsified with an equal volume of complete Freund's adjuvant, and injected intradermally as described (5). Animals were killed under deep phenobarbital anesthesia 4–70 days after the onset of clinical signs of EAE. Brain and spinal cord were dissected and fixed in 4% para-formaldehyde, and serial sections of the entire neuraxis were processed for routine histology.
Construction of a Combinatorial IgGκ-Fab Library from a MOG-Immunized C. jacchus Marmoset.
The system used to generate the combinatorial library involved the phage display vector pCOMB3H (provided by C. F. Barbas III, The Scripps Research Institute, La Jolla, CA). This system permits the construction of a cloning product containing L and H chains flanked by SfiI restriction sites for directional cloning (21).
Bone marrow and spleen cells were obtained from an rMOG-immunized C. jacchus that was killed after onset of clinical EAE. RNA was extracted with the Trizol reagent (Invitrogen) and first strand synthesis was performed with Superscript II reverse transcriptase (Invitrogen). In brief, three steps of PCR reactions were necessary to generate cloning inserts containing the Fab portions of C. jacchus IgGκ. For a detailed description of these PCR steps, see ref. 21. First, all known marmoset H-chain variable region (IGHV) and L-chain variable region (IGKV) genes, as well as H-chain IgG CH1-domain and κL chain C-region (IGKC) genes, were amplified in separate reactions (for primer sequences, see http://itsa.ucsf.edu/∼claudeg/Primers.htm and supporting information published on the PNAS web site, www.pnas.org). The template for IGHC and IGKC was a marmoset Fab library previously constructed in pCOMB3H, to include an SfiI restriction site on the 3′ end of IGHC and the pelB leader sequence with IGKC. In the second step, IGHV was joined with IGHC (H-chain assembly), and IGKV with IGKC (κL-chain assembly). Third, the κL chain (IGKV-IGKC-pelB) was joined with the H chain (IGHV-IGHC-SfiI) to yield a ∼1,460-bp cloning product containing SfiI-κL chain-pelB-H chain-SfiI. Finally, the cloning product and pCOMB3H were digested with SfiI (Roche Molecular Biochemicals) and purified. Equal amounts of pCOMB3H and C. jacchus κL/H DNA were ligated with T4 ligase (Roche Molecular Biochemicals) and electroporated into electrocompetent XL1-Blue cells (Stratagene) with a Bio-Rad GenepulserII (2.5 kV, 200 ohms, 25 μF). The complexity of the obtained C. jacchus IgGκ-pCOMB3H library was ≈1 × 107 recombinants. Infective phagemid particles were generated by rescue with the helper phage VCSM13 (Stratagene).
Screening of the C. jacchus IgGκ-Fab Library with rMOG.
Approximately 1012 Fab-expressing phagemids were incubated (37°C, 1 h) in ELISA wells coated with rMOG (1 μg per well). In the first round of the selection process (panning), wells were washed 5 times with PBS containing 0.05% Tween20 (PBS-T), bound phagemid eluted with trypsin (500 μg per well), and eluted phagemid used to infect XL1-Blue cells. After incubation at 37°C overnight, phagemids were precipitated and resuspended in PBS containing 1% BSA and submitted to the panning process 3 more times with increasing washing stringency (second round, 10 times; third round, 15 times; fourth round, 15 times). Enrichment of rMOG-specific Fab fragments was confirmed by measuring bound phagemid from each panning round in rMOG-coated ELISA wells with an anti-M13, horseradish peroxidase-conjugated Ab (Amersham Pharmacia Biotech).
DNA Sequence Analysis.
Phagemid DNA was extracted with the Qiagen (Valencia, CA) MaxiFilter kit and digested with SpeI and NheI for removal of the gIII protein gene, which permitted expression of soluble Fab fragments. SpeI/NheI-digested DNA was gel-purified, religated with T4 ligase, and transformed into XL1-Blue cells. Sixty randomly picked, Fab-expressing clones from the last panning round were grown in Superbroth containing 100 μg/ml of carbenicillin for minipreps, plasmid DNA was extracted with the Qiagen MiniPrep kit, and DNA was sequenced at the University of California, San Francisco, Genomics Core Facility by automated fluorescent chain termination sequencing. Sequences of both H and κL chains were aligned with megalign (DNAstar, Madison, WI).
Expression of Soluble Fab Fragments.
Fab-expressing clones representing all IGHV/IGKV combinations and H chain complementarity determining region (CDR) 3 motifs (Table 1) were grown in 3 liters of Superbroth until OD600 = 1.2, and expression was induced with 1 mM IPTG. After overnight incubation at 30°C, bacteria were lysed by sonication in 30 ml of PBS and Fabs were purified from the soluble fraction over a protein L column (Pierce) following the manufacturer's protocol. Where desired, purified Fab fragments were biotinylated with a sulfo-N-hydroxysuccinimide (NHS) biotinylation reagent (Pierce) following the manufacturer's instruction. Unreacted sulfo-NHS biotin was removed by extensive dialysis against PBS.
Table 1.
MOG-Fab IGHV and IGKV subgroup usage and H/κL-CDR3 motifs
| IGHV | H chain CDR3 | GenBank accession no. | IGKV | κL chain CDR3 | GenBank accession no. | % of total clones | Clone ID |
|---|---|---|---|---|---|---|---|
| 1 | CARDVNFGNYFDY | AF393235 | 3 | CQQYSSWPPTF | AF393236 | 73 | M26 |
| 1 | CARDRGMGNYFDY | AF393237 | 3 | CQQYSSWPLTF | AF393238 | 13 | M38 |
| 1 | CARDATRILADVLDY | AF393239 | 3 | CQQYSSWYTF | AF393240 | 8 | M45 |
| 3 | CARAWRLSARAGYFDY | AF393229 | 1 | CQQHYSTPLTF | AF393230 | 2 | M3-8 |
| 3 | CILSDTGAFDV | AF393231 | 3 | CQQYSSWYTF | AF393232 | 2 | M3-24 |
| 3 | CTGAGPTYYFDY | AF393233 | 1 | CQQGYTTPVTF | AF393234 | 2 | M3-31 |
Amino acid sequences of CDR3 motifs are deduced from cDNA. For complete H- and L-chain sequences please refer to GenBank.
Purification of Serum Anti-MOG Abs and Fractionation of Ab Specificities.
rMOG-reactive fractions of serum Abs were purified on 1-ml prepacked N-hydroxysuccinimide (NHS)-Sepharose columns reacted with 200 μg of rMOG, following the manufacturer's instructions (Amersham Pharmacia Biotech). rMOG-Sepharose columns were loaded with C. jacchus immune sera, diluted 1:5 in PBS, extensively washed with PBS, and bound Abs were eluted in 0.1 M glycine buffer, pH 2.2. For human sera, the protein G-reactive fraction (IgG) was extracted before purification by rMOG-affinity chromatography. To separate the Ab fractions binding to linear peptides from those binding to structural determinants of rMOG, MOG-peptide-reactive Ab fractions were removed from serum by repeated passes (n = 5) on 1-ml PepMOG (mixture of overlapping 20-mer peptides spanning the entire sequence of rMOG) columns, which were synthesized by reacting 5.5 mg of PepMOG (500 μg per peptide) with NHS-Sepharose. Where desired, purified Abs were biotinylated as described previously.
Epitope Specificities of MOG-Reactive Ab and Recombinant Fab Fragments.
Maleic anhydride-activated ELISA plates (Pierce) were coated with 1 μg per well of rMOG or PepMOG (1 μg per peptide), blocked with PBS-T/3% BSA, and washed with PBS-T. Samples were added in PBS-T/3% BSA as follows: rMOG- or PepMOG-immune C. jacchus serum, or fractions thereof (after removal of peptide-specific Abs), 1:200; monoclonal Fab fragments, 1 μg per well. After incubation at 37°C for 1 h, wells were washed with PBS-T, and appropriate secondary Ab added in PBS-T/3% BSA [serum and serum fractions: anti-monkey IgG-horseradish peroxidase (HRP) 1:6,000, Sigma; Fab fragments: protein L-HRP 1:5,000, Pierce] for 1 h at 37°C. After a final wash with PBS-T, plates were developed with tetramethylbenzidine (Pierce) and read at 450 nm.
Competition Assays.
Competition experiments were designed to examine the ability of Fab fragments to compete against each other and against native C. jacchus anti-MOG Abs for binding to rMOG. First, the amount of biotinylated Ab or Fab necessary to achieve 50% saturation of rMOG (50–100 ng per well) adsorbed on Ni-coated ELISA plates (Pierce) with biotinylated anti-MOG Abs or MOG-specific Fab was determined.
To study competitive displacement, nonbiotinylated Fabs or native Abs were added to MOG-coated wells at increasing concentrations (∼10−12 to 10−5 M) in the presence of the 50% saturation concentrations of the biotinylated reagent. After overnight incubation at 4°C, wells were washed and incubated with a streptavidin-peroxidase conjugate (Invitrogen; 1:1,000 in PBS-T/3% BSA, 20 min at room temperature). Bound Abs were detected with tetramethylbenzidine.
Competition experiments with human anti-MOG Abs were performed with a similar protocol, in Immunosorp ELISA wells (Nunc) coated with rMOG (500 ng per well) in PBS. A constant concentration of unlabeled, MOG-affinity-purified Abs was incubated in the presence of increasing concentrations of the M26 and M3-8 Fab fragments overnight at 4°C. Bound human anti-MOG Abs were detected with an Fc-specific, alkaline phosphatase-conjugated anti-human IgG (Sigma, 1:5,000; this Ab was not cross-reactive with Fab fragments), and plates were developed with para-nitrophenol phosphate and read at 405 nm. Displacement was quantitated as the ratio of OD in the presence of competition over that in the absence of competition X 100 (%).
Immunohistochemistry.
Paraformaldehyde-fixed paraffin-embedded sections of C. jacchus brain (7 μm) were deparaffinized, hydrated, and treated with a citrate-based antigen-unmasking solution (Vector Laboratories) at high temperature for 20 min. Sections were blocked with 3% normal goat serum (Sigma) in PBS for 1 h at 37°C, washed with PBS-T, and incubated with biotinylated MOG-specific Fab (2.8 μg/ml) for 2 h at 37°C. Additional experiments were performed with the same dilutions of Fab fragments in the presence of rMOG to demonstrate specificity of binding. After incubation with the alkaline phosphatase (AP)-conjugated avidin complex [Vectastain ABC-AP (Vector Laboratories), 30 min, room temperature], fluorescence was revealed by the VectorRed AP substrate (Vector Laboratories), and slides were counterstained with hematoxylin.
Results
Ig Gene Usage of Recombinant MOG-Specific Fab Fragments.
Sixty randomly chosen, MOG-specific Fab-encoding clones were sequenced. The IGHV subgroup usage in this library was limited to IGHV1 and IGHV3, and IGKV usage to IGKV1 and IGKV3. Ninety-four percent (57 clones) of all clones were composed of IGHV1/IGKV3 (representative clones are designated M26, M38, and M45), and 6% were IGHV3/IGKV1 (M3-8, M3-31; 2 clones) or IGHV3/IGKV3 (M3-24; 1 clone). Sequences corresponding to contact residues (CDRs) showed considerable diversity, with variability in the H-CDR3 motifs (Table 1).
Recombinant Fab Fragments Exclusively Recognize Structural Epitopes of MOG.
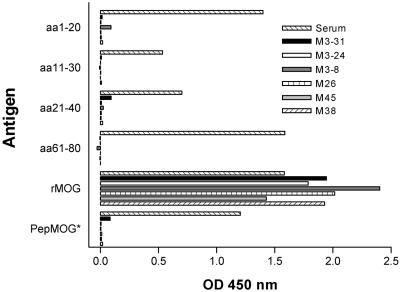
Polyclonal Ab populations present in serum of rMOG-immunized marmosets have been shown to recognize a broad repertoire of specificities, including linear epitopes corresponding to short peptide sequences contained within MOGaa1–125 (12, 22). Surprisingly, however, none of the recombinant Fab fragments studied showed binding to any of these linear, extended epitopes or to PepMOG (Fig. 1). Additional testing with an array of 60 overlapping 12-mer peptides confirmed these results (not shown). Thus, the MOG-specific Fab fragments selected from the combinatorial library exclusively recognized conformation-dependent epitopes.
Figure 1.
Conformational requirements of MOG-specific Fab clones. Means of triplicate or quadruplicate values. (*) PepMOG designates a mixture of overlapping 20-mer peptides spanning the entire sequence of rMOG. For comparison, representative reactivity of rMOG-immune serum Abs is also shown (Serum).
Diversity of Structural Ab Epitopes of MOG.
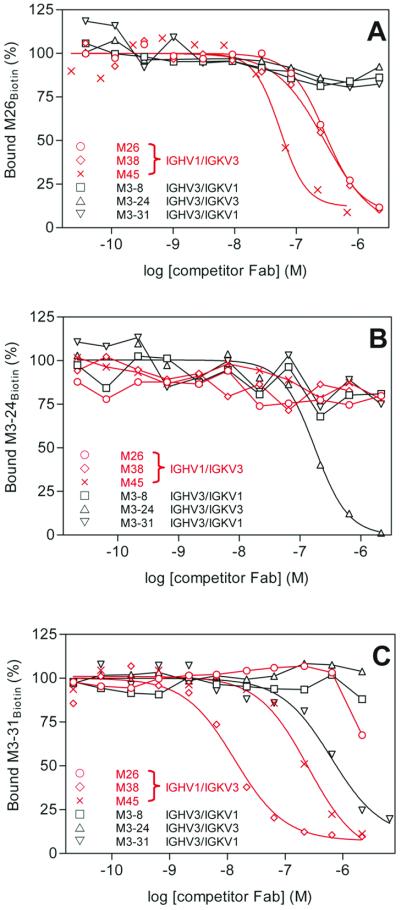
We performed competition experiments between Fab fragments representing all H/L chain combinations to understand the diversity of structural epitopes of rMOG targeted by the recombinant Fab fragments. Increasing concentrations of nonbiotinylated Fab fragments were allowed to compete in rMOG-coated ELISA wells with individual Fab fragments labeled with biotin. Fig. 2A illustrates the binding of a fixed amount of biotinylated M26 Fab (M26Biotin, IGHV1/IGKV3) in the presence of increasing concentrations of all other representative Fab fragments. Despite the variability in the CDR motifs (Table 1), all Fabs encoded by IGHV1/IGKV3 (M26, M38, and M45) recognize a similar epitope of rMOG. In contrast, no competition was observed between M26Biotin and M3-8, M3-31(IGHV3/IGKV1), or M3-24 (IGHV3/IGKV3). Fig. 2B illustrates a similar experiment with M3-24Biotin as the displaced Fab, which shows no competition with any of the other Fab fragments. These results indicate that the M3-24 Fab defines an epitope of rMOG that is distinct from that recognized by M26, M38, and M45. A similar lack of competition was observed for the M3-8Biotin Fab (not shown), suggesting that this IGHV3/IGKV1 combination defines another, unique conformational epitope.
Figure 2.
Competition ELISAs with representative Fab fragments. (A) M26Biotin is displaced from rMOG by itself (○), M38 (◊), and M45 (×) but not by M3-8 (□), M3-24 (▵), or M3-31 (▿). (B) M3-24Biotin is displaced only by itself (▵). (C) M3-31Biotin is displaced by itself (▿), M38 (◊), and M45 (×), and also by high concentrations of M26 (○). Highlighted in red are the Fab fragments that tightly cluster within the major immunogenic region of MOG. IGHV and IGKV gene usage is indicated in the legend.
Subtle conformational features on exposed surfaces of MOG may be responsible for a microheterogeneity within the Ab-binding sites. We found that M38 and M45 could displace the Fab M3-31Biotin, whereas only weak displacement by M26 occurred at significantly higher concentrations (Fig. 2C). Noncompetitive inhibition (e.g., steric interference) may play a role in this case and may explain the lack of displacement observed for the reverse experiment (e.g., M26Biotin vs. unlabeled M3-31, shown in Fig. 2A). Whether the M26 and M3-31 Fab fragments define similar or separate epitopes cannot be currently resolved. Nonetheless, these experiments identify at least three distinct conformational epitopes accessible on rMOG. All Fab fragments encoded by IGHV1/IGKV3 seem to recognize similar or closely associated epitopes on a single, major immunogenic region of MOG, which may be partially overlapped by M3-31.
Relevance of Antigenic Specificities Defined by Combinatorial Fab Fragments.
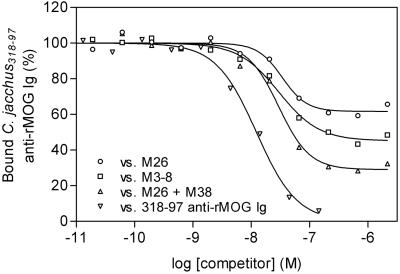
We next examined the ability of the recombinant Fab fragments to displace native anti-MOG Abs from C. jacchus serum, which represent a polyclonal mixture of Ab-specificities against linear determinants, structural determinants, or both. Biotinylated, affinity-purified anti-MOG Abs were incubated in the presence of increasing concentrations of Fabs. Fig. 3 shows representative experiments in which the M26 and M3-8 Fabs were allowed to compete against native, polyclonal auto-Abs from rMOG-immunized marmosets. Combinations of both M26 and M3-8 Fab fragments showed an additive effect for displacement, a finding that supports our hypothesis that the epitopes recognized by the M26 and M3-8 Fab fragments are topographically distinct (Fig. 3). Importantly, the representative Fabs derived from a single animal in this study efficiently displaced serum Abs from four genetically distinct marmosets (not shown).
Figure 3.
Monoclonal MOG-specific Fab fragments are representative of major Ab specificities in outbred marmosets. Displacement of native, polyclonal serum anti-MOG Abs by M26 (○), M3-8 (□), or a combination of M26 and M3-8 (▵) MOG-specific Fab fragments, and unlabeled serum anti-MOG Abs (▿). Shown here are results obtained with marmoset no. 318-97; identical results were obtained with affinity-purified Abs from 3 additional animals.
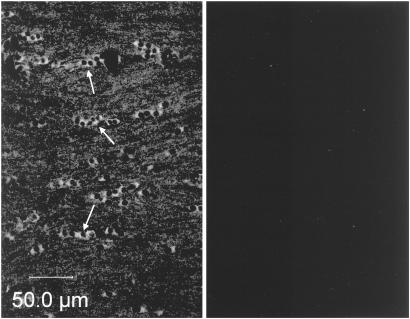
To verify that the Fab fragments were capable of binding to exposed epitopes of MOG on myelin sheaths, we confirmed by immunofluorescence that the recombinant Fab fragments were capable of binding to the MOG protein in situ in CNS white matter. Fig. 4 Left shows strong staining of oligodendrocytes and staining of myelinated fibers in C. jacchus corpus callosum with the biotinylated M26 Fab fragment. Specificity was confirmed by the ability to completely quench the fluorescent signal by addition of rMOG (Fig. 4 Right). Identical results were obtained with the M3-8 Fab fragment.
Figure 4.
Binding of recombinant Fab fragments to MOG in situ on C. jacchus CNS myelin. Fluorescent light micrographs of C. jacchus corpus callosum showing oligodendrocytes and myelinated fibers stained with the biotinylated Fab fragment M26 (Left). Specificity of the staining was confirmed by signal quenching after coincubation with rMOG (Right). Arrows indicate groups of aligned oligodendrocyte cell bodies.
In Vivo Pathogenicity of Conformational Versus Linear Epitope-Specific Anti-MOG Abs.
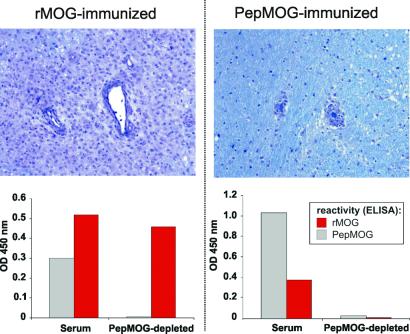
To understand further how epitope recognition influences Ab pathogenicity, we examined the binding characteristics of serum anti-MOG Abs in marmosets immunized with either rMOG (n = 4) or PepMOG (n = 2) before and after removal of the PepMOG-reactive fractions. Consistent with previous experience (19), rMOG-immunized animals developed severe neurological signs corresponding to multifocal, widespread inflammatory infiltrates accompanied by prominent demyelination (Fig. 5). In contrast, animals immunized with PepMOG exhibited reduced disease burden with little or no demyelination. Importantly, we found that the repertoire of MOG-reactive Abs in this group was strictly restricted to linear epitopes, as removal of PepMOG-reactive Abs completely abolished reactivity to MOG. However, the sera from each of the rMOG-immune animals contained residual reactivity against whole rMOG after the complete removal of peptide-specific Abs (Fig. 5), indicative of the presence of immunogenic structural epitopes. Thus, the conformation-dependent Abs are only present in rMOG-immunized animals and seem to be responsible for the extensive demyelination observed in lesions of rMOG-induced EAE.
Figure 5.
Correlations between anti-MOG Ab epitope recognition and neuropathological phenotypes. (Upper) Perivascular mononuclear cell infiltrates in brain white matter of representative rMOG- (Left) and PepMOG-immunized marmosets (Right). Note the large size of the infiltrate and the broad area of demyelination in the rMOG-immunized animal, and the lack of demyelination after PepMOG-immunization. Luxol Fast Blue/periodic acid Schiff, ×200. (Lower) The specificities of serum anti-MOG Abs in these animals were analyzed before (Serum) and after (PepMOG-depleted) removal of Abs binding to PepMOG (see Materials and Methods). rMOG reactivity is clearly retained after removal of the peptide-reactive Abs from rMOG-immune serum (Left), indicating the presence of separate subsets of Abs that react either with linear peptides or conformational rMOG. In marked contrast, no reactivity to MOG remains after removal of PepMOG-specific Abs from the animal immunized with PepMOG (Right), demonstrating that conformation-dependent Abs were not produced. Identical results were obtained in the other rMOG- and PepMOG-immunized monkeys.
Marmoset Fab Fragments Delineate Structural Determinants of the MOG Ab Response in Humans.
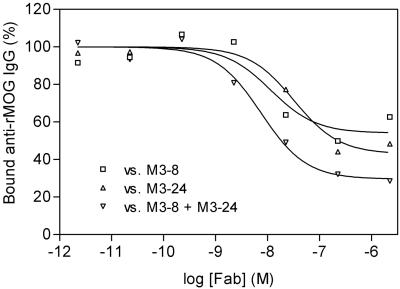
We examined the ability of recombinant marmoset Fab fragments to displace affinity-purified anti-MOG Abs from the sera of three patients with MS (AA, DM, and WS), who were previously shown to be MOG-reactive by ELISA. We found that the M3-8 fragment was able to compete with anti-MOG Abs from all three patients and also found competition with M3-24 for patient AA. Furthermore, similar to marmosets, the combination of M3-8 and M3-24 showed an additive effect (Fig. 6, representative experiment; patient AA). These results indicate that the targets for MOG-specific Abs in humans include conformation-dependent epitopes that are identical to those in marmosets.
Figure 6.
Competition between marmoset Fab fragments and human anti-MOG Abs. Affinity-purified serum anti-MOG Abs from patient AA with MS are displaced by M3-8 (□) and M3-24 (▵) or a combination of and M3-8 and M3-24 (▿).
Discussion
In this report we provide information regarding the molecular complexity of pathogenic auto-Ab responses against exposed domains of MOG in an outbred species.
Previous studies reporting the effects of passive transfer of certain Abs in rodent and marmoset systems (5, 7, 23), and of MOG-DNA vaccination in SJL mice (24), have shown that conformation-dependent anti-MOG Abs are capable of inducing demyelination. In contrast, whether Abs directed at linear determinants of MOG have demyelinating properties has not been unequivocally demonstrated (14, 25). Data obtained from two PepMOG-immunized animals in this study suggest that the presence of MOG-peptide-specific Abs is not associated with widespread demyelination. Indeed, EAE in these animals was reminiscent of the disease phenotype produced by adoptive transfer of MOG-reactive T cells (26). Similar EAE phenotypes could also be reproduced in animals immunized with groups of individual peptides that contain the marmoset immunodominant MOG-T cell epitopes (n = 7) (11). Taken together, these results suggest that Abs against linear peptides are not pathogenic in marmosets and that recognition of conformational features of MOG is a prerequisite for Ab pathogenicity.
Peptide-specific anti-MOG Abs are part of the MOG-immune repertoire in EAE and can be detected in the serum of healthy controls and patients with MS (9, 10). However, because of the stringent conditions applied during the panning process, it is likely that the conformational epitopes of rMOG define binding sites for Abs of higher affinity than MOG-peptide Abs, which were not found in the Fab library. Similar differences in affinity have been described in the case of a different antigen (27, 28). Nonetheless, we show that the Fab fragments specifically bound to native MOG in situ in brain tissue, indicating that our combinatorial approach had yielded Ab fragments that correctly define structural features of MOG that are exposed in vivo. The relevance of these Ab fragments is underlined further by our finding that the monospecific Fab fragments are capable of displacing a significant portion of the polyclonal, native anti-MOG Abs in several marmosets, despite the genetic heterogeneity present between outbred individuals. Thus, despite the fact that our library may not exhaustively include all Ab specificities present in the polyclonal, MOG-specific humoral repertoire, we propose that the MOG-specific Fab fragments represent epitope specificities with demyelinating potential.
Accurate definition of the determinants of MOG that are targets of demyelinating Abs in humans will be of critical importance. Qualitative differences in epitope recognition may be present among anti-MOG Ab populations that are frequently detected in patients with MS and healthy controls (13, 29, 30). For example, T cell mimicry between viral antigens and MOG peptides has been reported (31), but in the absence of exposure of B cells to the whole MOG polypeptide, may only induce production of MOG peptide-specific Abs. These auto-Abs would be detected in standard Ab assays, although they may not be pathogenic. We show here that Ab fragments that define structural determinants of MOG in C. jacchus can be used to specifically detect the presence of MOG-specific idiotypes directed against identical determinants in humans. Although in marmosets the M26 and M3-8 Fab fragments seem to represent important specificities, in serum from MS patients the M3-8 and M3-24 Fabs have thus far been shown to compete against native anti-MOG Abs. Additional experiments are proceeding to identify human Abs capable of competing with the remaining marmoset Fabs.
Clonal B cell expansion with restricted usage of IGHV germ-line genes in CNS lesions and cerebrospinal fluid of patients with MS has been reported (15–18). In this context, it was of interest to find that a limited number of H- (IGHV1 and IGHV3) and κL-chain (IGKV1 and IGKV3) subgroup genes was used in the marmoset MOG-specific Ab repertoire. However, we also found that diverse CDR-encoding gene rearrangements were used to target only three epitopes of MOG. Therefore, the current study extends beyond prior molecular analyses of Ig gene usage, which have not identified target antigens for the clonally expanded immune responses. Competition experiments also demonstrated that the C. jacchus Fab fragments define antigenic determinants of MOG that are commonly targeted in all marmosets, regardless of H- and L-chain usage.
The critical importance of MOG to autoimmune demyelination is a consequence of its restricted expression in the CNS (32), its exposed extracellular domain at the outermost lamellae (33), its high level of encephalitogenicity in multiple species, and tendency to induce pathogenic auto-Ab responses directed against the myelin sheath (reviewed in ref. 11). The finding reported here that related conformational features of MOG are targets of auto-Abs in marmosets and humans highlights the value of nonhuman primate models for dissection of auto-Ab responses relevant to the pathophysiology of CNS tissue damage in MS.
Supplementary Material
Acknowledgments
We thank Sarah M. McLachlan, Basil Rapoport, Carlos F. Barbas III, Don L. Siegel, and Gregg J. Silverman for expert advice, and Scott S. Zamvil for helpful discussion. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants AI43073 (to S.L.H.) and NS1-996-02 (to H.-C.v.B.), the Nancy Davis Foundation, the New York Community Trust, the German Hertie Foundation, the Deutsche Multiple Sklerose Gesellschaft, and National Multiple Sclerosis Society Grant JF2087-A-2 (to C.P.G.).
Abbreviations
- H chain
heavy chain
- L chain
light chain
- CDR
complementarity determining region
- MOG
myelin/oligodendrocyte glycoprotein
- rMOG
rat MOGaa1–125
- PepMOG
mixture of overlapping 20-mer peptides spanning the entire sequence of rMOG
- EAE
experimental allergic encephalomyelitis
- MS
multiple sclerosis
- CNS
central nervous system
- PBS-T
PBS containing 0.05% Tween 20
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF393229–AF393240).
The nomenclature of Ig genes is derived from the HUGO Gene Nomenclature Committee (http://www.gene.ucl.ac.uk/nomenclature): IGHV, Ig H chain variable region; IGKV, Ig κL chain variable region; IGKC, Ig κL chain constant region.
References
- 1.Hohlfeld R. Brain. 1997;120:865–916. doi: 10.1093/brain/120.5.865. [DOI] [PubMed] [Google Scholar]
- 2.Bauer J, Rauschka H, Lassmann H. Glia. 2001;36:235–243. doi: 10.1002/glia.1112. [DOI] [PubMed] [Google Scholar]
- 3.Brosnan C F, Raine C S. Brain Pathol. 1996;6:243–257. doi: 10.1111/j.1750-3639.1996.tb00853.x. [DOI] [PubMed] [Google Scholar]
- 4.Cross A H, Trotter J L, Lyons J. J Neuroimmunol. 2001;112:1–14. doi: 10.1016/s0165-5728(00)00409-4. [DOI] [PubMed] [Google Scholar]
- 5.Genain C P, Nguyen M H, Letvin N L, Pearl R, Davis R L, Adelman M, Lees M B, Linington C, Hauser S L. J Clin Invest. 1995;96:2966–2974. doi: 10.1172/JCI118368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linington C, Berger T, Perry L, Weerth S, Hinze-Selch D, Zhang Y, Lu H C, Lassmann H, Wekerle H. Eur J Immunol. 1993;23:1364–1372. doi: 10.1002/eji.1830230627. [DOI] [PubMed] [Google Scholar]
- 7.Schluesener H J, Sobel R A, Linington C, Weiner H L. J Immunol. 1987;139:4016–4021. [PubMed] [Google Scholar]
- 8.Genain C P, Cannella B, Hauser S L, Raine C S. Nat Med. 1999;5:170–175. doi: 10.1038/5532. [DOI] [PubMed] [Google Scholar]
- 9.Karni A, Bakimer-Kleiner R, Abramsky O, Ben-Nun A. Arch Neurol. 1999;56:311–315. doi: 10.1001/archneur.56.3.311. [DOI] [PubMed] [Google Scholar]
- 10.Xiao B G, Linington C, Link H. J Neuroimmunol. 1991;31:91–96. doi: 10.1016/0165-5728(91)90014-x. [DOI] [PubMed] [Google Scholar]
- 11.von Büdingen H-C, Tanuma N, Villoslada P, Ouallet J-C, Hauser S L, Genain C P. J Clin Immunol. 2001;21:155–170. doi: 10.1023/a:1011031014433. [DOI] [PubMed] [Google Scholar]
- 12.Mesleh M F, Belmac N, Lu C W, Krishnan V V, Maxwell R S, Genain C P, Cosman M. Neurobiol Dis. 2002;9:160–172. doi: 10.1006/nbdi.2001.0474. [DOI] [PubMed] [Google Scholar]
- 13.Haase C G, Guggenmos J, Brehm U, Andersson M, Olsson T, Reindl M, Schneidewind J M, Zettl U K, Heidenreich F, Berger T, et al. J Neuroimmunol. 2001;114:220–225. doi: 10.1016/s0165-5728(00)00462-8. [DOI] [PubMed] [Google Scholar]
- 14.Ichikawa M, Johns T G, Adelmann M, Bernard C C. Int Immunol. 1996;8:1667–1674. doi: 10.1093/intimm/8.11.1667. [DOI] [PubMed] [Google Scholar]
- 15.Qin Y, Duquette P, Zhang Y, Talbot P, Poole R, Antel J. J Clin Invest. 1998;102:1045–1050. doi: 10.1172/JCI3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owens G P, Kraus H, Burgoon M P, Smith-Jensen T, Devlin M E, Gilden D H. Ann Neurol. 1998;43:236–243. doi: 10.1002/ana.410430214. [DOI] [PubMed] [Google Scholar]
- 17.Colombo M, Dono M, Gazzola P, Roncella S, Valetto A, Chiorazzi N, Mancardi G L, Ferrarini M. J Immunol. 2000;164:2782–2789. doi: 10.4049/jimmunol.164.5.2782. [DOI] [PubMed] [Google Scholar]
- 18.Baranzini S E, Jeong M C, Butunoi C, Murray R S, Bernard C C, Oksenberg J R. J Immunol. 1999;163:5133–5144. [PubMed] [Google Scholar]
- 19.Genain C P, Hauser S L. Methods. 1996;10:420–434. doi: 10.1006/meth.1996.0120. [DOI] [PubMed] [Google Scholar]
- 20.Amor S, Groome N, Linington C, Morris M M, Dornmair K, Gardinier M V, Matthieu J M, Baker D. J Immunol. 1994;153:4349–4356. [PubMed] [Google Scholar]
- 21.Barbas C F, III, Burton D R, Scott J K, Silverman G J. Phage Display: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 22.Genain C P, Abel K, Belmar N, Villinger F, Rosenberg D P, Linington C, Raine C S, Hauser S L. Science. 1996;274:2054–2057. doi: 10.1126/science.274.5295.2054. [DOI] [PubMed] [Google Scholar]
- 23.Brehm U, Piddlesden S J, Gardinier M V, Linington C. J Neuroimmunol. 1999;97:9–15. doi: 10.1016/s0165-5728(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 24.Bourquin C, Iglesias A, Berger T, Wekerle H, Linington C. Eur J Immunol. 2000;30:3663–3671. doi: 10.1002/1521-4141(200012)30:12<3663::AID-IMMU3663>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 25.Adelmann M, Wood J, Benzel I, Fiori P, Lassmann H, Matthieu J M, Gardinier M V, Dornmair K, Linington C. J Neuroimmunol. 1995;63:17–27. doi: 10.1016/0165-5728(95)00124-7. [DOI] [PubMed] [Google Scholar]
- 26.Villoslada P, Abel K, Heald N, Goersches R, Hauser S L, Genain C P. Eur J Immunol. 2001;31:2942–2950. doi: 10.1002/1521-4141(2001010)31:10<2942::aid-immu2942>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachs D H, Schechter A N, Eastlake A, Anfinsen C B. Proc Natl Acad Sci USA. 1972;69:3790–3794. doi: 10.1073/pnas.69.12.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jemmerson R, Blankenfeld R. Mol Immunol. 1989;26:301–307. doi: 10.1016/0161-5890(89)90084-9. [DOI] [PubMed] [Google Scholar]
- 29.Reindl M, Linington C, Brehm U, Egg R, Dilitz E, Deisenhammer F, Poewe W, Berger T. Brain. 1999;122:2047–2056. doi: 10.1093/brain/122.11.2047. [DOI] [PubMed] [Google Scholar]
- 30.Sun J, Link H, Olsson T, Xiao B G, Andersson G, Ekre H P, Linington C, Diener P. J Immunol. 1991;146:1490–1495. [PubMed] [Google Scholar]
- 31.Mokhtarian F, Zhang Z, Shi Y, Gonzales E, Sobel R A. J Neuroimmunol. 1999;95:43–54. doi: 10.1016/s0165-5728(98)00254-9. [DOI] [PubMed] [Google Scholar]
- 32.Gardinier M V, Amiguet P, Linington C, Matthieu J M. J Neurosci Res. 1992;33:177–187. doi: 10.1002/jnr.490330123. [DOI] [PubMed] [Google Scholar]
- 33.Brunner C, Lassmann H, Waehneldt T V, Matthieu J M, Linington C. J Neurochem. 1989;52:296–304. doi: 10.1111/j.1471-4159.1989.tb10930.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.