Abstract
Mutations in the DAX-1 [dosage-sensitive sex reversal–adrenal hypoplasia congenita (AHC) critical region on the X chromosome; NR0B1] gene cause X-linked AHC associated with hypogonadotropic hypogonadism. DAX-1 encodes an unusual orphan member of the nuclear hormone receptor superfamily, acting as a transcriptional repressor of genes involved in the steroidogenic pathway. All DAX-1 mutations found in AHC patients alter the protein C terminus, which shares similarity to the ligand binding domain of nuclear hormone receptors and bears transcriptional repressor activity. This property is invariably impaired in DAX-1 AHC mutants. Here we show that the localization of DAX-1 AHC mutant proteins is drastically shifted toward the cytoplasm, even if their nuclear localization signal, which resides in the N terminal of the protein, is intact. Cytoplasmic localization of DAX-1 AHC mutants correlates with an impairment in their transcriptional repression activity. These results reveal a critical role of an intact C terminus in determining DAX-1 subcellular localization and constitute an important example of a defect in human organogenesis caused by impaired nuclear localization of a transcription factor.
Adrenal hypoplasia congenita (AHC) is an inherited disorder of the adrenal cortex commonly manifested as an early-onset adrenal insufficiency syndrome. Two different forms of this disease have been recognized: a X-linked form and a rare autosomal recessive or sporadic form (1). AHC is a lethal disease if untreated, mainly because of mineralocorticoid deficit. Although it can present a wide clinical spectrum (1–3), a constant feature of the X-linked syndrome is the association with hypogonadotropic hypogonadism (HHG). It has been shown that HHG is caused by combined hypothalamic failure in gonadotropin-releasing hormone release and pituitary defect in gonadotropin production (2). Mutations in the DAX-1 (dosage-sensitive sex reversal-AHC critical region on the X chromosome gene 1; NR0B1) gene, situated in Xp21, are responsible for the X-linked form of AHC/HHG (4, 5).
DAX-1 encodes an unusual orphan member of the nuclear hormone receptor superfamily (4–7) whose expression is restricted to the adrenal cortex, testis, ovary, pituitary gonadotropes, and hypothalamic ventromedial nucleus (8, 9). This pattern of expression closely matches the characteristics of the disease and suggests that DAX-1 also plays a role in reproductive function. Strikingly, inactivation of the mouse Ahch homologue does not produce adrenal hypoplasia, but male sterility caused by progressive testicular seminiferous tubules degeneration (10). Other links with reproductive function and phenotypical sex determination are provided by the findings that the DAX-1 gene lies inside the critical region on the X chromosome whose duplication causes dosage-sensitive sex reversal in XY individuals (11, 12), and that overexpression of DAX-1 in particular mouse strains causes phenotypical sex reversal (13). Furthermore, mouse Dax-1 has been shown to antagonize the synergistic action of steroidogenic factor-1 (SF-1) and WT-1 -KTS isoforms in activating expression of the Müllerian inhibiting substance gene promoter (14).
DAX-1 is a transcriptional repressor of several genes involved in steroid hormone metabolism (15–19). A bipartite transcriptional repression domain lies in the C terminus of the DAX-1 protein (20, 21). Transcriptional repression by DAX-1 is thought to be mediated by its interaction with the N-CoR (22) and Alien (23) corepressors. Importantly, all DAX-1 mutations identified to date in AHC patients have in common the ability to alter the sequence of the protein C-terminal domain and impair its transcriptional repression activity (3–5, 20–24). Importantly, DAX-1 is a nucleo-cytoplasmic shuttling protein associated with ribonucleoprotein structures in the nucleus and polyribosomes in the cytoplasm (25). These data suggest that DAX-1 plays additional regulatory functions in posttranscriptional processes.
Here we show that the subcellular localization of DAX-1 AHC mutant proteins is invariably shifted toward the cytoplasm, even if their nuclear localization signal (NLS), which resides in the protein N terminus, is intact. The effect of AHC mutations is also evident when the DAX-1 C terminus is fused to a heterologous NLS-containing DNA binding domain (DBD). Cytoplasmic localization of DAX-1 AHC mutants directly correlates with the magnitude of the reduction of their transcriptional repression activity. These results indicate that AHC is a defect in adrenal gland organogenesis caused by an impairment of DAX-1 localization in the nucleus.
Materials and Methods
DNA Clones.
DAX-1 deletion constructs were PCR-generated from the pSV.DAX-1 expression vector (25), to encode amino acids 1–69 (R1), 1–135 (R1-R2), 1–205 (R1-R3), and 205–470 (C) in the pSG5 vector. Mutations were introduced into pSV.DAX-1 by the QuickChange mutagenesis system (Stratagene). All clones were verified by sequencing. GAL4 DBD/DAX-1 fusions have been described (21).
Cell Culture and Transfections.
HeLa cells were cultured in DMEM containing 2.5% calf serum, 2.5% FCS, and antibiotics. COS1 and Y-1 cells were cultured as described (15, 16). Transfections using the human StAR luciferase reporter were performed as described (15, 16), using the calcium phosphate method. Data represent the average of three independent experiments performed in duplicate.
Immunofluorescence.
Cells growing in chamber slides (Nunc) were processed as described (25). The mAb 3E5, which recognizes a unique epitope in the DAX-1 C-terminal domain, was used to assess the distribution of DAX-1 deletion constructs lacking the N-terminal domain. In all other cases, the anti-DAX-1 2F4 antibody (26) was used. GAL4 DBD fusion proteins were detected by mAb 2GV3/3GV2 (27). SF-1 was detected by a polyclonal rabbit antiserum (Upstate Biochemicals, Waltham, MA). No staining was detected in nontransfected cells or when using nonimmune IgG as primary antibody. Subcellular localization was scored as nuclear when the protein was exclusively present in the nucleus in more than 95% of transfected cells; it was scored as cytoplasmic when exclusively nuclear staining was present in less than 5% of transfected cells and nucleo-cytoplasmic in all other cases (28). Data were computed from 2–11 experiments, with each experiment scoring at least 200 transfected cells.
Immunoblot.
It was performed as described (15, 16), using 10 μg per sample of total cell extract from transfected HeLa cells. Expression of wild-type and mutant DAX-1 proteins was quantitated by densitometry and corrected by the amount of the endogenous lactate dehydrogenase protein revealed with a specific antibody (Sigma).
Pull-Down Assay.
Glutathione S-transferase (GST) and GST/N-CoR (1629–2453) were expressed in BL21 Escherichia coli and purified on glutathione-Sepharose beads following the manufacturer's protocol (Amersham Pharmacia). 35S-labeled chicken TRα and DAX-1 proteins were expressed in vitro by using the TNT system (Promega). Pull-down assays were performed by mixing immobilized GST or GST N-CoR (1629–2453) with 35S-labeled proteins in a buffer containing 20 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EGTA, 1% Triton X-100. cTRα interaction with GST N-CoR (1629–2453) was assayed in the presence and the absence of 10−6 M 3,3′,5-triiodothyroacetic acid. After 1 h incubation at 4°C with rocking, resin was washed three times with the same buffer. Retained proteins were analyzed by SDS/PAGE and fluorography. Binding quantitation was performed by densitometry.
Results
The N-Terminal Region of DAX-1 Contains the NLS.
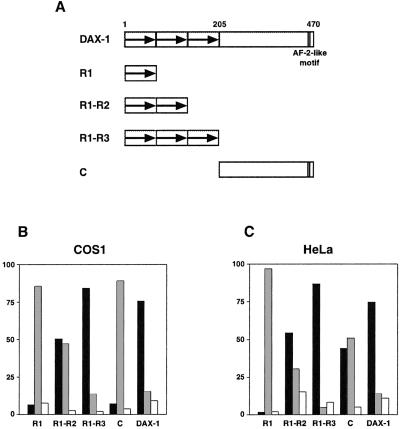
Three conserved repeats are present in the DAX-1 N terminus, preceding a region that shares high similarity to the nuclear hormone receptor LBD (4, 21) (Fig. 1). DAX-1 lacks the zinc finger DBD (domain C) and the hinge region (domain D), present in most members of the nuclear hormone receptor superfamily (6). Thereby, the sequences known to function as proto-NLSs in steroid receptors and cooperate with the ligand-inducible NLS present in the ligand binding domain (28–30), are absent in DAX-1. To identify the role of the domains directing DAX-1 subcellular localization, we studied the distribution of the full-length protein and a number of deletion constructs in HeLa and COS-1 cells. These constructs correspond to the N-terminal R1, R1-R2, and R1-R3 repeats and the C-terminal domain (Fig. 1). The DAX-1 N-terminal repeats have an additive effect in triggering nuclear accumulation (Fig. 1). The isolated C-terminal domain is prevalently nucleo-cytoplasmic in both cell lines, with transfected HeLa cells showing a higher incidence of nuclear localization of this domain (Fig. 1). Full-length DAX-1 is prevalently localized in the nucleus, with a distribution very similar to the R1-R3 construct, but in a minority of cases it has a nucleo-cytoplasmic or cytoplasmic distribution (Fig. 1). DAX-1 localization was not influenced by the presence or absence of serum and phenol red, which might contain putative DAX-1 ligands, in the culture medium (ref. 30; data not shown). These results show that the DAX-1 NLS resides in its N-terminal region and are consistent with previous data showing nuclear accumulation of a construct encoding Dax-1 amino acids 1–99 (14).
Figure 1.
Protein domains directing subcellular localization in DAX-1. (A) Structure of the DAX-1 protein and scheme depicting the constructs used in transfection studies. The three N-terminal repeats are indicated by an arrow. The position of the AF-2-like motif at the C terminal is indicated. Distribution of DAX-1 deletion constructs in transfected COS-1 (B) and HeLa (C) cells. Percentage of nuclear localization is indicated with black histograms, nucleo-cytoplasmic localization in gray, and cytoplasmic localization in white.
Critical Role of DAX-1 C-Terminal Residues in Determining Its Subcellular Localization.
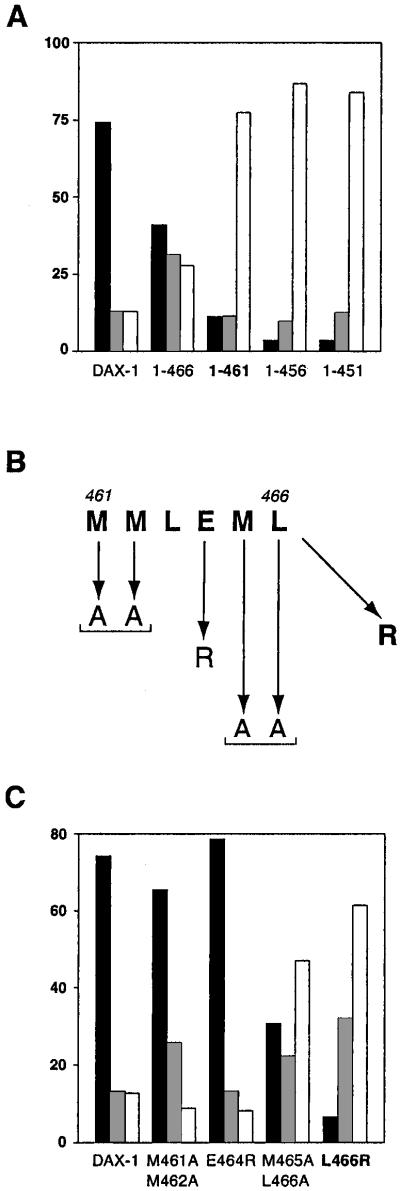
Homology models of the DAX-1 C-terminal domain reveal the presence of all features constituting the nuclear receptor LBD fold (21, 31). In particular, DAX-1 contains an amphipathic α-helical motif in the C-terminal putative H12 that in other nuclear receptors is essential for ligand-dependent transcriptional activation (AF-2) (32, 33). It is remarkable that two DAX-1 mutations that have been described in AHC kindreds affect the putative AF-2 domain (M462stop; L466R) (34, 35).
To investigate whether AHC mutations affect DAX-1 protein localization, we first analyzed truncations at the C terminus. This type of mutation is the most frequent cause of AHC (4, 5, 31). Surprisingly, deletion of the last nine amino acids (1–461), a mutation also found in one clinical mutant (34), is sufficient to shift the distribution of the protein almost entirely to the cytoplasm (Fig. 2). Further truncations to amino acids 456 and 451 have no additional effect. Conversely, DAX-1 1–466, which retains its putative AF-2 domain, is still prevalently nuclear, even if the percentage of transfected cells showing a nucleo-cytoplasmic or a cytoplasmic distribution is considerably increased (Fig. 2). To assess the role of the putative AF-2 domain in determining DAX-1 localization, we introduced amino acid substitutions known to impair ligand-dependent transcriptional activation in nuclear hormone receptors (32, 33) (Fig. 2). Double substitution of M461 and M462 with alanine, as well as the E464R mutation, had only very mild effects on DAX-1 localization (Fig. 2). Conversely, the M465A/L466A mutant was considerably more nucleo-cytoplasmic and cytoplasmic than wild-type DAX-1. Remarkably, the AHC L466R mutation considerably shifted the protein toward a cytoplasmic localization (Fig. 2).
Figure 2.
Distribution of DAX-1 C-terminal mutants. (A) Subcellular localization of full-length DAX-1 and the 1–466, 1–461, 1–456, and 1–451 deletions in HeLa cells. (B) Sequence of the AF-2-like motif at the DAX-1 C terminus and mutations introduced. (C) Localization of the DAX-1 M461A/M462A, E464R, M465A/L466A, and L466R mutants. Percentage of nuclear localization is indicated with black histograms, nucleo-cytoplasmic localization in gray, and cytoplasmic localization in white. DAX-1 AHC mutants are indicated in bold.
These results indicate that the sequence corresponding to the C-terminal putative H12 plays a critical role in determining DAX-1 nuclear localization, in a manner independent of the N-terminal NLS. In particular, the nature of the amino acid residue at position 466 appears critical for determining the intracellular localization of DAX-1. Interestingly, equivalent mutations in AF-2 are known to impair ligand-dependent transactivation in nuclear hormone receptors (32, 33).
Localization of DAX-1 AHC Mutants Is Systematically Shifted Toward the Cytoplasm.
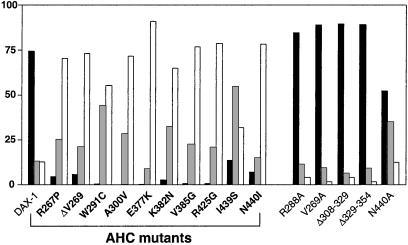
We have analyzed the distribution of 10 additional AHC mutants carrying single amino acid substitutions at multiple sites of the DAX-1 putative ligand binding domain. In all cases the distribution of the mutant proteins was drastically shifted toward the cytoplasm (Fig. 3). Remarkably, the mutant whose localization was least affected was I439S, originally identified in a patient with adult-onset adrenal insufficiency and incomplete hypogonadotropic hypogonadism (3). Alanine substitutions of two residues affected by AHC mutations (V269 and N440) have only a very mild effect, showing that the nature of the mutant amino acid is critical for the altered distribution of the protein. No effect on localization has been found for the R288A, Δ308–329, and Δ329–354 artificial mutants (Fig. 3).
Figure 3.
Localization of DAX-1 mutants in HeLa cells. Percentage of nuclear localization is indicated with black histograms, nucleo-cytoplasmic localization in gray, and cytoplasmic localization in white. DAX-1 AHC mutants are indicated in bold.
Effect of AHC Mutations on DAX-1 Fusion Proteins Containing a Heterologous NLS.
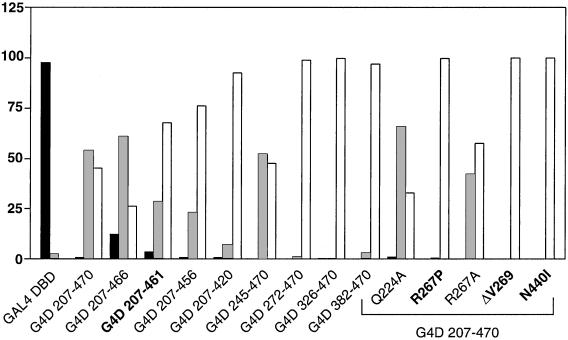
The property of DAX-1 AHC mutations to impair nuclear localization is striking, especially because an intact NLS is still present in the protein N terminus. To assess whether these mutations would also affect the intracellular distribution of proteins containing a heterologous NLS, we analyzed a battery of GAL4 DBD DAX-1 C-terminal fusion proteins in HeLa cells (Fig. 4). The GAL4 DBD protein is endowed with its own NLS (36) and was localized in the nucleus, as expected. Surprisingly, the GAL4 DBD/DAX-1 207–470 fusion protein, which includes the entire DAX-1 C-terminal domain, has a prevalent nucleo-cytoplasmic and cytoplasmic distribution (Fig. 4). Although GAL4 DBD/DAX-1 207–466 still retains a prevalent nucleo-cytoplasmic distribution, truncation to amino acid 461 nearly completely shifted the distribution to the cytoplasm. Proteins with deletions to amino acids 456 and 420 were also cytoplasmic. Conversely, a protein with a 245–470 N-terminal deletion had the same distribution as the 207–470 fusion protein, but when the deletion was extended to amino acid 272 and further the fusion proteins always localized in the cytoplasm (Fig. 4). Furthermore, AHC mutations R267P, ΔV269, and N440I induced total redistribution of the fusion GAL4 DBD/DAX-1 207–470 protein to the cytoplasm, whereas no effect was produced by the R267A and the Q224A mutations (Fig. 4).
Figure 4.
Localization of GAL4 DBD/DAX-1 C-terminal fusion proteins. Percentage of nuclear localization is indicated with black histograms, nucleo-cytoplasmic localization in gray, and cytoplasmic localization in white. DAX-1 AHC mutants are indicated in bold.
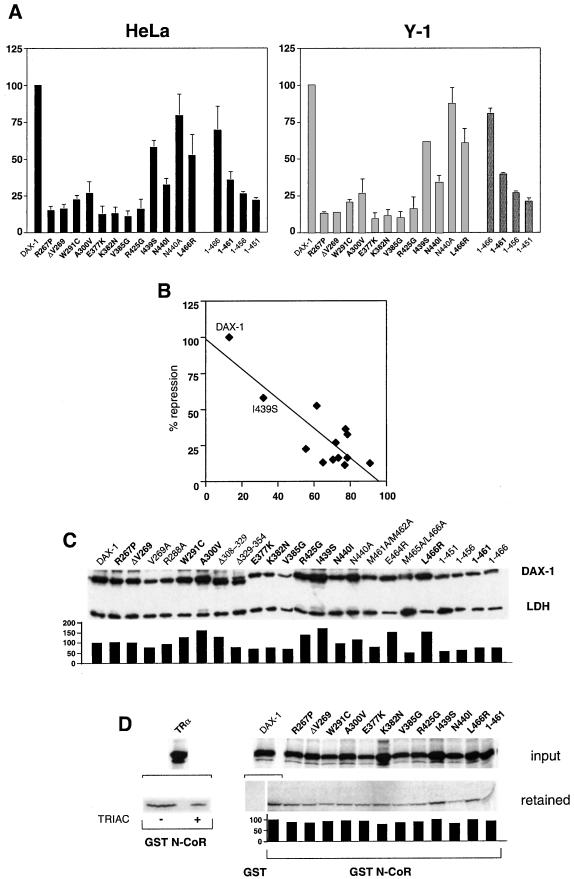
Cytoplasmic Localization of DAX-1 AHC Mutants Correlates with Their Transcriptional Repression Impairment.
All DAX-1 AHC mutants investigated here have lost their ability to repress StAR promoter activity (Fig. 5A). Remarkably, I439S DAX-1 displays higher repression activity compared with the other AHC mutants, as also shown previously (3, 24). An inverse correlation exists between repression activity of DAX-1 AHC mutants and the extent of their exclusively cytoplasmic localization (Fig. 5B). No relationship is present between the amount of protein expressed and transcriptional repression potency (Fig. 5C). Conversely, DAX-1 displays weak interaction with the N-CoR corepressor in vitro, which is only very marginally affected by AHC mutations (Fig. 5D). Furthermore, we have already shown that DAX-1 AHC mutants bind their DNA hairpin binding site in the StAR promoter with the same affinity as the wild-type protein (21). Taken together, these data indicate that the major determinant for the impaired biological activity of DAX-1 AHC mutants is the shift in the intracellular localization of the protein, notwithstanding the presence of an intact NLS.
Figure 5.
Loss of transcriptional repression by DAX-1 AHC mutants correlates with their cytoplasmic localization. (A) Histogram showing repression of StAR promoter activity by wild-type and mutant DAX-1 proteins in HeLa (Left) and Y-1 (Right) cells. StAR expression was activated by cotransfection of a SF-1 expression vector (HeLa) or forskolin (10 μg/ml) treatment (Y-1). Repression by DAX-1 mutants is expressed as percentage of the repression by wild-type DAX-1, which is set equal to 100. SEM is indicated. DAX-1 AHC mutants are indicated in bold. (B) Graph showing the relationship between repression activity (y axis) and percentage of cytoplasmic localization (x axis) of wild-type and AHC DAX-1 proteins. The position of the I439S mutant, whose repression activity is the least impaired, is indicated. r = 0.85. (C) Immunoblot showing expression of the various DAX-1 proteins examined. DAX-1 AHC mutants are indicated in bold. Expression levels were quantified by densitometry and normalized by the amount of endogenous lactate dehydrogenase (LDH) protein, with the expression of wild-type DAX-1 set equal to 100. (D) Pull-down assay showing interaction of DAX-1 proteins with GST (wild-type DAX-1) and GST N-CoR (1629–2453) (wild-type and mutant DAX-1 proteins). As a control, cTRα interaction with GST N-CoR (1629–2453) is shown in the absence (−) and the presence (+) of 10−6 M 3,3′,5-triiodothyroacetic acid (TRIAC). Fluorography exposure times were overnight for cTRα and 10 days for wild-type and mutant DAX-1 proteins. The ratio of bound to input protein is shown in the histogram, with wild-type DAX-1 set equal to 100.
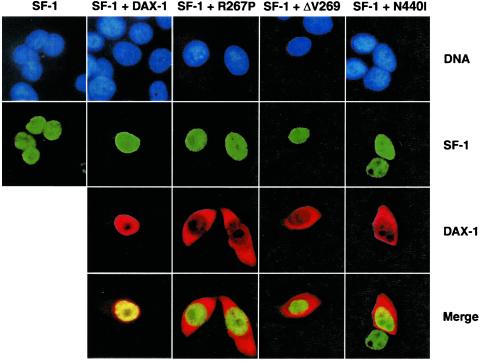
SF-1 Nuclear Localization Is Not Affected by Cotransfection of DAX-1 AHC Mutants.
DAX-1 has been shown to interact with and modulate the function of SF-1, an orphan nuclear receptor that is an essential regulator of adrenal gland and gonad organogenesis (14, 20, 22, 37). One possibility for AHC pathogenesis is that DAX-1 mutants, which all retain the sequences required for interaction with SF-1 (20), trigger its relocalization to the cytoplasm. However, SF-1 remains a nuclear protein whether cotransfected with either wild-type DAX-1 or with the R267P, ΔV269, and N440I mutants (Fig. 6).
Figure 6.
SF-1 is a nuclear protein independently of the presence of wild-type or AHC mutants DAX-1. Double-color immunofluorescence showing localization of transfected SF-1 (green) in HeLa cells, in the absence and the presence of transfected wild-type, R267P, ΔV269, and N440I DAX-1 (red). DNA was stained by the Hoechst 33342 dye (blue). (Magnification: ×600.)
Discussion
Regulation of nuclear import constitutes an important control level for a number of proteins involved in gene expression (38). More specifically, subcellular distribution of nuclear hormone receptors is a tightly regulated step that is critical for appropriate modulation of their target gene expression (reviewed in ref. 39).
Here we have shown that DAX-1 AHC mutant proteins have an impaired nuclear localization. The abnormal intracellular localization of DAX-1 AHC mutants has a direct effect on the transcriptional regulatory function of the protein, so that the mutant proteins are not able to repress target gene expression, being retained in the cytoplasm. AHC is an example of a congenital human disease caused by impaired nuclear localization of a transcription factor. The effect of AHC mutations is remarkable as the NLS of DAX-1 is always intact in all AHC mutants. Thus, cytoplasmic retention of DAX-1 AHC mutants overrides the activity of the NLS, which lies in the N-terminal part of the molecule and is not directly affected by AHC mutations (Fig. 3). Strikingly, the effect of the AHC mutation also is exerted on a heterologous NLS (Fig. 4).
Our results indicate that the integrity of a bipartite domain within the DAX-1 C terminus, which we have previously identified as essential for transcriptional repression activity (21), is required for nuclear localization. We hypothesize that AHC mutations induce a misfolded conformation of the DAX-1 C terminus, which exposes new molecular surfaces able to interact with cytoplasmic anchoring sites. This notion is reminiscent of the case of a simian virus 40 large T antigen frameshift mutant that is cytoplasmic even in the presence of an intact NLS, because of the presence of multiple hydrophobic residues in its novel C terminus (40). The nature of the cytoplasmic sites of interaction for DAX-1 AHC mutants is at the moment unknown. We have not found interaction of either wild-type DAX-1 or AHC mutants with components of the heat shock protein complex (data not shown).
Our findings unravel an important aspect of the molecular pathways by which AHC develops. Although DAX-1 AHC mutants have an abnormal intracellular distribution, AHC pathogenesis must occur with a mechanism independent from SF-1 mislocalization. These data are consistent with recent findings showing nonoverlapping functions in development between the DAX-1 and SF-1 orphan receptors (41). The AHC-induced changes in DAX-1 intracellular distribution represent an exquisite example of how the tight control of protein nuclear localization is essential in the regulation of developmental processes (42).
Acknowledgments
We thank B. Bardoni, K. Ohe, J.-P. Renaud, and J.-M. Wurtz for discussions, E. Heitz for technical assistance, and K. Essabri and T. Lerouge for the gift of reagents and cell culture, oligonucleotide-peptide synthesis, and sequencing facilities. This work was supported by the Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Universitaire Régional, Fondation de la Recherche Médicale and Association pour la Recherche sur le Cancer, Centre Hospitalier Universitaire Régional, Human Frontiers Science Program, and Organon (Akzo/Nobel).
Abbreviations
- AHC
adrenal hypoplasia congenita
- DAX-1
dosage-sensitive sex reversal-AHC critical region on the X chromosome gene 1
- SF-1
steroidogenic factor-1
- NLS
nuclear localization signal
- GST
glutathione S-transferase
- DBD
DNA-binding domain
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kletter G B, Gorski J L, Kelch R P. Trends Endocrinol Metab. 1991;2:123–128. [Google Scholar]
- 2.Habiby R L, Boepple P, Nachtigall L, Sluss P M, Crowley W F, Jr, Jameson J L. J Clin Invest. 1996;98:1055–1062. doi: 10.1172/JCI118866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabarin A, Achermann J C, Recan D, Bex V, Bertagna X, Christin-Maitre S, Ito M, Jameson J L, Bouchard P. J Clin Invest. 2000;105:321–328. doi: 10.1172/JCI7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanaria E, Muscatelli F, Bardoni B, Strom T M, Guioli S, Guo W, Lalli E, Moser C, Walker A P, McCabe E R B, et al. Nature (London) 1994;372:635–641. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]
- 5.Muscatelli F, Strom T M, Walker A P, Zanaria E, Récan D, Meindl A, Bardoni B, Guioli S, Zehetner G, Rabi W, et al. Nature (London) 1994;372:672–676. doi: 10.1038/372672a0. [DOI] [PubMed] [Google Scholar]
- 6.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lalli E, Sassone-Corsi P. Curr Opin Endocrinol Diabetes. 1999;6:185–190. [Google Scholar]
- 8.Ikeda Y, Swain A, Weber T J, Hentges K E, Zanaria E, Lalli E, Tamai K T, Sassone-Corsi P, Lovell-Badge R, Camerino G, et al. Mol Endocrinol. 1996;10:1261–1272. doi: 10.1210/mend.10.10.9121493. [DOI] [PubMed] [Google Scholar]
- 9.Swain A, Zanaria E, Hacker A, Lovell-Badge R, Camerino G. Nat Genet. 1996;12:404–409. doi: 10.1038/ng0496-404. [DOI] [PubMed] [Google Scholar]
- 10.Yu R, Ito M, Saunders T L, Camper S A, Jameson J L. Nat Genet. 1998;20:353–357. doi: 10.1038/3822. [DOI] [PubMed] [Google Scholar]
- 11.Bardoni B, Zanaria E, Guioli S, Floridia G, Worley K C, Tonini G, Ferrante E, Chiumello G, McCabe E R B, Fraccaro M, et al. Nat Genet. 1994;7:497–501. doi: 10.1038/ng0894-497. [DOI] [PubMed] [Google Scholar]
- 12.Roberts L M, Shen J, Ingraham H A. Am J Hum Genet. 1999;65:933–942. doi: 10.1086/302601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swain A, Narvaez V, Burgoyne P, Camerino G, Lovell-Badge R. Nature (London) 1998;391:761–767. doi: 10.1038/35799. [DOI] [PubMed] [Google Scholar]
- 14.Nachtigal M W, Hirokawa Y, Enyeart-VanHouten D L, Flanagan J N, Hammer G D, Ingraham H A. Cell. 1998;93:445–454. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 15.Zazopoulos E, Lalli E, Stocco D M, Sassone-Corsi P. Nature (London) 1997;390:311–315. doi: 10.1038/36899. [DOI] [PubMed] [Google Scholar]
- 16.Lalli E, Melner M H, Stocco D M, Sassone-Corsi P. Endocrinology. 1998;139:4237–4243. doi: 10.1210/endo.139.10.6217. [DOI] [PubMed] [Google Scholar]
- 17.Hanley N A, Rainey W E, Wilson D I, Ball S G, Parker K L. Mol Endocrinol. 2001;15:57–68. doi: 10.1210/mend.15.1.0585. [DOI] [PubMed] [Google Scholar]
- 18.Aigueperse C, Val P, Pacot C, Darne C, Lalli E, Sassone-Corsi P, Veyssiere G, Jean C, Martinez A. Mol Endocrinol. 2001;15:93–111. doi: 10.1210/mend.15.1.0577. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z J, Jeffs B, Ito M, Achermann J C, Yu R N, Hales D B, Jameson J L. Proc Natl Acad Sci USA. 2001;98:7988–7993. doi: 10.1073/pnas.141543298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito M, Yu R, Jameson J L. Mol Cell Biol. 1997;17:1476–1483. doi: 10.1128/mcb.17.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lalli E, Bardoni B, Zazopoulos E, Wurtz J-M, Strom T M, Moras D, Sassone-Corsi P. Mol Endocrinol. 1997;11:1950–1960. doi: 10.1210/mend.11.13.0038. [DOI] [PubMed] [Google Scholar]
- 22.Crawford P A, Dorn C, Sadovsky Y, Milbrandt J. Mol Cell Biol. 1998;18:2949–2956. doi: 10.1128/mcb.18.5.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altincicek B, Tenbaum S P, Dressel U, Thormeyer D, Renkawitz R, Baniahmad A. J Biol Chem. 2000;275:7662–7667. doi: 10.1074/jbc.275.11.7662. [DOI] [PubMed] [Google Scholar]
- 24.Achermann J C, Ito M, Silverman B L, Habiby R L, Pang S, Rosler A, Jameson J L. J Clin Endocrinol Metab. 2001;86:3171–3175. doi: 10.1210/jcem.86.7.7660. [DOI] [PubMed] [Google Scholar]
- 25.Lalli E, Ohe K, Hindelang C, Sassone-Corsi P. Mol Cell Biol. 2000;20:4910–4921. doi: 10.1128/mcb.20.13.4910-4921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamai K T, Monaco L, Alastalo T-P, Lalli E, Parvinen M, Sassone-Corsi P. Mol Endocrinol. 1996;10:1561–1569. doi: 10.1210/mend.10.12.8961266. [DOI] [PubMed] [Google Scholar]
- 27.White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ylikomi T, Bocquel M T, Berry M, Gronemeyer H, Chambon P. EMBO J. 1992;11:3681–3694. doi: 10.1002/j.1460-2075.1992.tb05453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guiochon-Mantel A, Loosfelt H, Lescop P, Sar S, Atger M, Perrot-Applanat M, Milgrom E. Cell. 1989;57:1147–1154. doi: 10.1016/0092-8674(89)90052-4. [DOI] [PubMed] [Google Scholar]
- 30.Picard D, Yamamoto K R. EMBO J. 1990;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y-H, Guo W, Wagner R L, Huang B-L, McCabe L, Vilain E, Burris T P, Anyane-Yeboa K, Burghes A H M, Chitayat D, et al. Am J Hum Genet. 1998;62:855–864. doi: 10.1086/301782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danielan P S, White R, Lees J A, Parker M G. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakae J, Tajima T, Kusuda S, Kohda N, Okabe T, Shinohara N, Kato M, Murashita M, Mukai T, Imanaka K, et al. J Clin Endocrinol Metab. 1996;81:3680–3685. doi: 10.1210/jcem.81.10.8855822. [DOI] [PubMed] [Google Scholar]
- 35.Abe S, Nakae J, Yasoshima K, Tajima T, Shinohara N, Murashita M, Satoh K, Koike A, Takahashi Y, Fujieda K. Am J Med Genet. 1999;84:87–89. [PubMed] [Google Scholar]
- 36.Silver P A, Keegan L P, Ptashne M. Proc Natl Acad Sci USA. 1984;81:5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen W H, Moore C C, Ikeda Y, Parker K L, Ingraham H A. Cell. 1994;77:651–661. doi: 10.1016/0092-8674(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 38.Kaffman A, O'Shea E K. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 39.Yamashita S. Histol Histopathol. 1998;13:255–270. doi: 10.14670/HH-13.255. [DOI] [PubMed] [Google Scholar]
- 40.Schneider J, Schindewolf C, van Zee K, Fanning E. Cell. 1988;54:117–125. doi: 10.1016/0092-8674(88)90185-7. [DOI] [PubMed] [Google Scholar]
- 41.Ikeda Y, Takeda Y, Shikayama T, Mukai T, Hisano S, Morohashi K-I. Dev Dyn. 2001;220:363–376. doi: 10.1002/dvdy.1116. [DOI] [PubMed] [Google Scholar]
- 42.Affolter M, Marty T, Vigano M A. Genes Dev. 1999;13:913–915. doi: 10.1101/gad.13.8.913. [DOI] [PubMed] [Google Scholar]