Abstract
In human airways, epithelial cells lining the lumen and intraluminal cells (e.g., polymorphonuclear cells) participate in the innate immune response. These cells secrete or express on their surfaces arginine-specific ADP ribosyltransferases. Defensins, antimicrobial proteins secreted by immune cells, are arginine-rich, leading us to hypothesize that ADP ribosylation could modify their biological activities. We found that an arginine-specific ADP ribosyltransferase-1 present on airway epithelial cells modifies Arg-14 of α defensin-1. ADP-ribosylated defensin-1 had decreased antimicrobial and cytotoxic activities but still stimulated T cell chemotaxis and IL-8 release from A549 cells. Further, ADP-ribosylated defensin-1 inhibited cytotoxic and antimicrobial activities of unmodified defensin-1. We identified ADP-ribosylated defensin-1 in bronchoalveolar lavage fluid from smokers but not from nonsmokers, confirming its existence in vivo. Thus, airway mono-ADP-ribosyltransferases could have an important regulatory role in the innate immune response through modification of α defensin-1 and perhaps other basic molecules, with alteration of their biological properties.
In the human airway, epithelial cells, in concert with intraluminal cells (e.g., macrophages), contribute to the innate immune response. Activation of airway epithelial cells releases proinflammatory mediators and chemotaxins, leading to the recruitment of inflammatory cells (e.g., neutrophils) (1, 2). Activated neutrophils, in turn, release azurophilic granules that contain human neutrophil peptides (HNP), small cationic peptides ranging in size from 29–33 aa (3). HNPs are characterized by a high arginine content and contain three pairs of disulfide-linked cysteines (4). They have a broad spectrum of antimicrobial and cytotoxic activities and play an important role in the innate immune response against both Gram-positive and -negative bacteria, fungi, and certain enveloped viruses (5–8). High HNP levels have been found in airway secretions of patients with inflammatory lung diseases (9–11).
A family of eukaryotic and prokaryotic mono-ADP ribosyltransferases (ARTs) catalyze the transfer of ADP ribose from NAD+ to arginine residues in proteins (12). Among the mono-ARTs are bacterial products (e.g., cholera toxin), which are responsible for the clinical syndrome in disease (13). A family of mammalian ARTs that are secreted or localized on the cell surface through glycosylphosphatidylinositol anchors (14–19) are expressed preferentially on epithelial and inflammatory cells such as lymphocytes and neutrophils (17, 20). Substrates of the five known mammalian ARTs (ART-1–5) include proteins that are involved in critical cellular events (e.g., lymphocyte activation, neutrophil chemotaxis) (21–23). Two of these transferases, ART-1 and -5, specifically modify arginine residues in proteins. The finding of ARTs on airway epithelial and inflammatory cells prompted us to evaluate potential substrates for ADP ribosylation in the airway (24). Because of the high arginine content of defensins and the substrate specificity of ARTs, we investigated whether HNP, specifically HNP-1, could be a substrate for ART-1, whether ADP-ribosylation of HNP altered its activities, and whether ADP-ribosylated HNP-1 exists in vivo. Our findings indicate a potentially important function for ADP ribosylation in regulating the activity of basic proteins such as HNP and demonstrate that ADP-ribosylated defensin has unique biological properties.
Methods
Cloning, Expression, and Purification of ART-1.
We cloned wild-type ART-1 from a rabbit muscle cDNA library by PCR by using forward (5′-ACGTACAAGCTTAGCCACCTG GTGACACGTCGAGAC) and reverse (5′-ACGTACGGTACCGTCCAGGTGGC AGGGCCTAGACTT) primers. PCR products were digested with HindIII and KpnI (Roche Molecular Biochemicals), and then subcloned into a pFLAG-MAC (Sigma) expression vector that was used to transform Escherichia coli ampicillin-resistant BL-21 (DE 3) competent cells (Novagen). Single colonies were grown in LB medium with ampicillin (50 μg/ml) to an optical density at 600 nm of 0.4 before adding isopropyl-d thiogalactoside (final concentration, 0.5 mM). After incubation for 2 h at 30°C, bacteria were pelleted by centrifugation (15 min, 3,200 × g, 4°C), resuspended in 50 mM Tris (pH 8.0)/5 mM EDTA/lysozyme (0.25 mg/ml)/1.5 M NaCl/0.1 M MgCl2/DNase I (0.02 μg/ml)/protease inhibitor mixture (Boehringer Mannheim) (50 μg/ml), sonified, and centrifuged (12,500 × g, 45 min, 4°C). Supernatants were applied to anti-FLAG M2 affinity gel (Sigma) and eluted following the manufacturer's instructions. Eluates were concentrated at 4°C, using Centricon 10 columns (Millipore), with centrifugation at 4,300 × g.
ART Assay.
The ART activity of ART-1 was assayed as described (17). For HNP modification by ART-1, we incubated synthetic HNP-1 (58 pmol, 2 μg) (Bachem) in 200 μl of 50 mM potassium phosphate, pH 7.5, with ART-1 (4.5 mmol) and 3 mM NAD +. After overnight incubation at 30°C, the mixture was solubilized in 6 M guanidine and applied to a Vydac (Hesperia, CA) C18 column (The Nest Group, Southborough, MA) equilibrated with solution A (HPLC grade water, 0.1% trifluoroacetic acid). ADP-ribose-HNP-1 was separated from the unmodified form by gradient elution with solution B (100% isopropanol, 0.2% trifluoroacetic acid) at a flow rate of 1 ml/min: 100% solution A, 0–20 min, linear gradient 0–60% of solution B 20–80 min, and 95% solution B 60–85 min. Fractions were analyzed by matrix-assisted laser desorption ionization (MALDI)-MS, as described (25).
Electrospray MS Analysis of ADP-Ribosylated HNP-1.
Samples (58 pmol, 2 μg) of ADP-ribosyl-HNP-1 or HNP-1 were reduced by incubation in 50 μl of 100 mM Tris⋅HCl (pH 8)/1 mM EDTA/20 mM DTT for 1 h at 37°C before incubation for 6 h at 37°C with trypsin (1 μg) (Sigma) plus 100 mM NH4HCO3 (total volume, 100 μl). The modified site was identified by electrospray MS, as described (25).
Antimicrobial and Chromium Release Assays.
Antibacterial activity of HNP-1 and ADP-ribosyl-HNP-1 on E. coli ATCC43827 (American Type Culture Collection, Rockville, MD) was evaluated by a radial diffusion assay (26).
Chromium-labeled A549 cells (American Type Culture Collection) were incubated (18 h, 37°C) in 100 μl of serum-free RPMI medium (GIBCO) containing HNP-1 or ADP-ribosyl-HNP-1 (1.5 to 24 μM) for quantification of defensin cytotoxicity assessed by chromium release from the cells (11).
Effects of ADP-Ribosyl-HNP-1 on HNP-1 Antimicrobial and Cytotoxic Activity.
HNP-1 (100 nM) was incubated (1 h, 37°C) with ADP-ribosyl-HNP-1 (0–800 nM), before assay of antimicrobial activity on E. coli.
HNP-1 (12 μM) was incubated with ADP-ribosyl-HNP-1 (1.5–12 μM) for 1 h at 37°C before assay of chromium release. As control, we used HNP-1 and ADP-ribosyl-HNP-1 (1.5–12 μM).
IL-8 Production by A549 Cells.
A549 cells (3 × 104 cells per well) were incubated in a 96-well plate in 200 μl of serum-free RPMI medium (GIBCO) containing ADP-ribosyl-HNP-1 or HNP-1 (0.25, 0.75, 1.5, or 3 μM). IL-8 in samples of medium taken after 12 and 24 h was measured by indirect ELISA according to the manufacturer's instructions (R & D Systems).
Chemotaxis Assay.
CD3+ T cells were isolated from human peripheral blood prepared by leukopheresis (National Institutes of Health, Department of Transfusion Medicine, Bethesda, MD), as described (27), and suspended in migration medium (RPMI 1640/0.5% BSA/25 mM Hepes). Inserts coated with collagen IV (Becton Dickinson) were placed in 24-well culture plates, forming upper and lower chambers, which were equilibrated with migration medium for 1 h at 37°C in an atmosphere of 5% CO2. After aspiration of wetting medium, migration medium (300 μl) containing 5 × 105 cells and migration medium (500 μl) containing ADP-ribosyl-HNP-1 or HNP-1 (0.025–25 nM) were added to the upper and lower chambers, respectively. Plates were incubated at 37°C in 5% CO2 for 4 h. Lymphocytes that migrated to the lower chamber were suspended in medium, collected by centrifugation and counted in a hemacytometer.
Isolation of ADP-Ribosyl-HNP-1 from Bronchoalveolar Lavage (BAL).
Samples (10 ml) of BAL fluids from 18 cigarette smokers (National Heart, Lung, and Blood Institute Institutional Review Board protocol no. 95-H-167) (total cells: 127 ± 15 million) and 10 healthy nonsmokers (total cells: 33 ± 5.8 million) (Table 1) were applied to SepPack C-18 cartridges (Waters) equilibrated in 10% isopropanol/0.1% trifluoroacetic acid (TFA), washed with the same buffer, and eluted with 50% isopropanol/0.1% TFA (1.5 ml). The eluted proteins were concentrated to 600 μl in a vacuum centrifuge (Savant), adjusted to 6 M guanidine/0.1% acetic acid and separated on a Vydac column C18 HPLC column. Absorbance (210 nm) peaks corresponding to the retention times of HNP-1 and ADP-ribosyl-HNP-1 were analyzed by MALDI-MS. Molecules with masses corresponding to ADP-ribosyl-HNP-1 were further characterized. We incubated ADP-ribosyl-HNP-1 (6 pmol/0.2 μg) in 200 μl of 250 mM NaHCO3/25 mM MgCl2, alkaline phosphatase (5 μg), and pyrophosphatase (5 μg) (Sigma) for 30 min at 37°C before termination of the reaction with 6 M guanidine, separation of reaction products by RP-HPLC, and analysis by MALDI-MS. We also incubated ADP-ribosyl-HNP-1 (11 pmol, 0.42 μg) overnight at 37°C with recombinant human ADP-ribosylarginine hydrolase (1 μg) in 100 μl of 50 mM Tris (pH 7.5)/5 mM DTT/10 mM Mg Cl2 (28), followed by desalting and concentration using Zip-TipC18 (Millipore), and analysis by MALDI-MS.
Table 1.
Characteristics of smoker population and BAL data
| Population
|
BAL
|
|||||
|---|---|---|---|---|---|---|
| No | Smoking history* | FEV1† | Total cells‡ | PMN§ | HNP-1 | ADP-ribose- HNP-1 |
| 1 | 40/d 30 yr | 138 | 245 | 2.1 | + | + |
| 2 | 30/d 11 yr | 73 | 126 | 1.8 | + | − |
| 3 | 40/d 5 yr | 93 | 140 | 1.6 | − | − |
| 4 | 10/d 37 yr | 113 | 55 | 1 | + | − |
| 5 | 40/d 12 yr | 110 | 95 | 1 | − | − |
| 6 | 20/d 29 yr | 109 | 103 | 1.9 | − | − |
| 7 | 20/d 25 yr | 126 | 166 | 3 | + | + |
| 8 | 20/d 15 yr | 95 | 108 | 3.5 | + | − |
| 9 | 20/d 20 yr | 96 | 169 | 2 | + | − |
| 10 | 10/d 9 yr | 143 | 46 | 3 | − | − |
| 11 | 40/d 16 yr | 115 | 150 | 0.5 | + | − |
| 12 | 20/d 36 yr | 130 | 140 | 1.3 | − | − |
| 13 | 30/d 15 yr | 80 | 280 | 8 | + | − |
| 14 | 20/d 30 yr | 96 | 135 | 0.8 | + | − |
| 15 | 20/d 10 yr | 109 | 105 | 1 | + | − |
| 16 | 30/d 26 yr | 97 | 119 | 0.5 | + | + |
| 17 | 30/d 30 yr | 66 | 44 | 0.5 | + | + |
| 18 | 15/d 15 yr | 103 | 52 | 0.5 | − | − |
Cigarettes per day, duration.
Percent of predicted.
Total cells ×106.
Percent of total cells.
Results
Modification of HNP-1 by an Arginine-Specific Mono-ADP-Ribosyltransferase (ART-1).
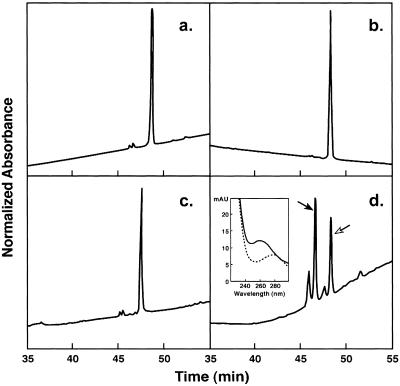
After incubation of HNP-1 with NAD+ and/or ART-1, the reaction products were separated by RP-HPLC, and analyzed by MALDI-MS (Fig. 1). We identified peptides of mass 3,441 Da (retention time 48.5 min), consistent with HNP-1 (calculated 3,442 Da) and of 3,983 Da (retention time 46.5 min), consistent with mono-ADP-ribosyl-HNP (calculated 3,983 Da). As expected, modified HNP exhibited an absorbance peak at 258 nm not seen with HNP (Fig. 1d Inset). The ADP ribose was released by incubation with ADP-ribosylarginine hydrolase, generating the unmodified defensin (data not shown).
Figure 1.
Separation of products of ART-1-catalyzed ADP-ribosylation of HNP-1 by RP-HPLC. HNP were incubated with: (a) reaction buffer alone; (b) NAD+; (c) ART-1; or (d) ART-1 and NAD+, and then analyzed by RP-HPLC. Absorbance at 210 nm (normalized to 100 full scale) is shown as a function of elution time. For a and b, full scale is 750; for c and d, 650 and 250, respectively. ADP-ribosyl-HNP-1 and HNP-1 are indicated by solid and open arrows, respectively. (Inset) Absorbance spectra of ADP-ribosyl-HNP-1 (solid line) and HNP-1 (dotted line), recorded during elution by an inline array detector.
To identify the modified amino acid in HNP-1, reduced ADP-ribosyl-HNP-1 was digested with trypsin, and the fragments were analyzed by MS. A peak of mass of 1,626.3 Da compatible with ADP-ribosylated fragment P6–15 (calculated 1,626.3 Da) was observed. This peak, unique to ADP-ribosylated HNP-1, was generated by inhibition of trypsin cleavage as a result of ADP ribosylation of Arg-14. Spectral analysis was consistent with the presence of adenosine.
Effects of Mono-ADP-Ribosylation of HNP-1 on Antimicrobial and Cytotoxic Activities, Stimulation of T Cell Chemotaxis, and IL-8 Release from A549 Cells.
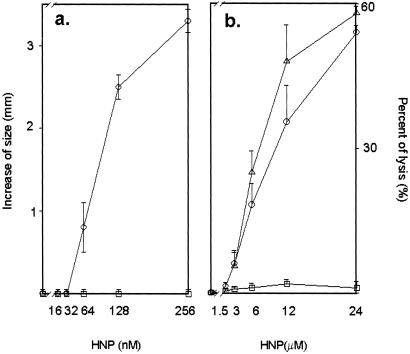
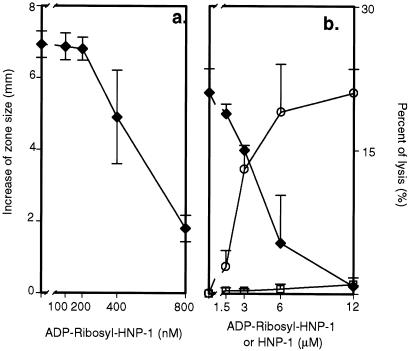
We compared the antimicrobial and cytotoxic activities of HNP-1 and ADP-ribosylated HNP-1, synthesized as defined in Methods, by radial diffusion and chromium-release assays, respectively. ADP-ribosyl-HNP-1 was inactive on E. coli ATCC 43827 cells in the radial diffusion assay (Fig. 2a) and in the cytotoxicity assay using A549 cells (Fig. 2b). ADP-ribosyl-HNP-1 blocked antimicrobial and cytotoxic activities of HNP-1 in a concentration-dependent manner (Fig. 3 a and b, respectively).
Figure 2.
Antimicrobial and cytotoxic activities of unmodified HNP-1 and ADP-ribosyl-HNP-1. (a) Difference in zone size (corresponding to antimicrobial activity) is the diameter of the zone of E. coli growth cleared with the indicated concentration of HNP (○) or ADP-ribosyl-HNP-1 (□) minus the diameter of the central well (3 mm). Data are means ± 1/2 range of values from two experiments. (b) A549 cells were incubated with the indicated concentrations HNP (○), ADP-ribosyl-HNP-1 (□), or synthetic HNP-1 (sHNP, Δ). Percent of lysis (cytotoxic activity) was calculated as (cpmexp − cpmspont)/(cpmmax − cpmspont) × 100; exp, experimental release; max, maximal release; spont, spontaneous release. Data are means ± SEM of values from four experiments.
Figure 3.
Effects of ADP-ribosyl-HNP-1 on HNP-1 antimicrobial and cytotoxic activity. (a) HNP-1 (100 nM) was incubated with the indicated concentration of ADP-ribosyl-HNP-1 before addition to E. coli ATCC 43827 for radial diffusion assay. Difference in zone size was calculated as in Fig. 2 (⧫). Data are means ± SEM of values from three experiments. (b) HNP-1 (12 μM) was incubated with the indicated concentration of ADP-ribosyl-HNP-1 before addition to 51Cr-labeled A549 cells (⧫). Percentage of cell lysis was determined as in Fig. 2 to compare effects of HNP-1(○) and ADP-ribosyl-HNP-1 (□). Data are means ± 1/2 range of values from two experiments. Results were similar in another experiment by using 24 μM HNP-1.
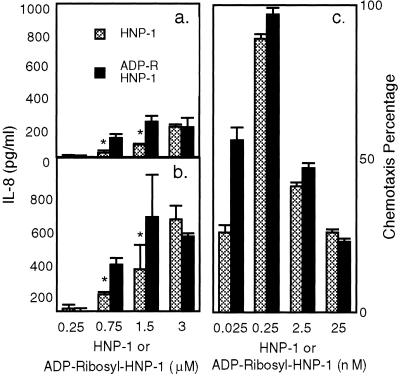
We measured IL-8 released into the medium after incubation of A549 cells with ADP-ribosyl-HNP-1 or HNP-1 for 12 (Fig. 4a) or 24 h (Fig. 4b). At concentrations of 0.75 and 1.5 μM, IL-8 release was significantly higher with ADP-ribosyl-HNP-1 than with the unmodified peptide. Using 1.5 μM of peptide, we confirmed significantly larger amounts of IL-8 in medium after incubation with ADP-ribosyl-HNP-1 (P = 0.01) (data not shown). At a peptide concentration of 3 μM, no differences were observed.
Figure 4.
Effects of ADP-ribosyl-HNP-1 and HNP-1 on IL-8 release by A549 cells and on T cell chemotaxis. Cells were incubated for (a) 12 or (b) 24 h with the indicated concentration of HNP-1 or ADP-ribosyl-HNP-1 before analysis of medium. IL-8 concentration in medium from cells incubated without defensins has been subtracted (at 12 h, 36.4 ± 6 pg/ml; at 24 h, 85 ± 34 pg/ml). Data are means ± 1/2 range of values from two experiments, each performed in triplicate. *, P < 0.05 for difference between IL-8 release stimulated by HNP-1 and ADP-ribosyl-HNP-1. CD3+ cells were incubated with the indicated concentrations of HNP-1 or ADP-ribosyl HNP-1 (c). We used MIP-1β (5 ng/ml) in migration medium, or migration medium alone, as positive and negative controls, respectively. Chemotaxis percentage was calculated as: (number of cells migrated to the lower chamber in the experimental conditions − number of cells migrated in the negative control)/(number of cells migrated in the positive control − number of cells migrated in negative control) × 100. Data are means ± SEM values from four experiments, each performed in duplicate.
We also investigated the chemotactic activity of ADP-ribosyl-HNP-1 toward CD3+ lymphocytes. ADP-ribosyl-HNP-1 retained its ability to recruit T cells, at a level similar to that noted with unmodified HNP-1 (Fig. 4c).
Identification of ADP-Ribosyl-HNP-1 in BAL Fluid.
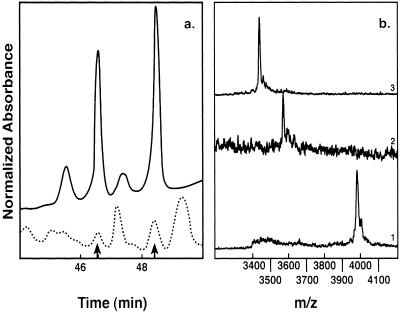
To identify ADP-ribosyl-HNP-1 in humans, we performed BAL on 18 smokers and 10 nonsmokers, none of whom had signs of pulmonary infection. Characteristics of the smokers and lavage findings are summarized in Table 1. The two populations did not differ significantly in age, gender, or pulmonary function (data not shown). The total number of cells recovered from BAL of smokers was 4-fold that of controls (P < 0.001), with a significantly greater absolute number of neutrophils (data not shown). RP-HPLC analysis of BAL fluid from 18 smokers revealed peaks with the same elution time as authentic HNP in 12; 4 of these 12 samples showed peaks with elution times identical to that of ADP-ribosyl-HNP-1 (Fig. 5a). MALDI-MS analysis demonstrated a molecule with a mass of 3,983 Da, confirming the presence of ADP-ribosyl-HNP-1(calculated 3,983 Da) (Fig. 5b1). Defensins were not detected in BAL fluids from the nonsmoker group.
Figure 5.
Characterization of ADP-ribosyl-HNP-1 from BAL fluid by RP-HPLC, MALDI, and enzymatic digestion. (a) Alignment of RP-HPLC chromatograms of products of ART-1-catalyzed ADP-ribosylation of HNP-1 (continuous line) and a chromatogram of a BAL sample from a smoker (dotted line). Arrows indicate elution times of ADP-ribosyl-HNP-1 and HNP-1. (b) (i) The peak with elution time compatible with ADP-ribosyl-HNP-1 (46.5 min), analyzed by MALDI-MS, shows a mass of 3,983 Da, consistent with ADP-ribosyl-HNP-1 (calculated 3,983 Da). (ii) Incubation of ADP-ribosyl-HNP-1 with pyrophosphatase and alkaline phosphatase produced ribosyl-HNP-1 (calculated 3,574 Da). (iii) ADP-ribosylarginine hydrolase cleaved the ribose-arginine linkage to release HNP-1 (calculated 3,443 Da). m/z is on the x axis.
We further characterized the putative ADP-ribosyl-HNP-1 by incubating it with pyrophosphatase and alkaline phosphatase to remove the ADP moiety, producing ribosyl-HNP, or with ADP-ribosylarginine hydrolase, which cleaves the ribose–arginine linkage, releasing HNP. After incubation with pyrophosphatase and alkaline phosphatase, MALDI-MS analysis of the peak separated by RP-HPLC showed a molecule of 3,575 Da, consistent with ribosyl-HNP-1, produced by removal of ADP from the ADP-ribosyl-HNP-1 (Fig. 5b2) (calculated 3,574 Da). The site of linkage of ADP-ribose to HNP was confirmed after incubation with ADP-ribosylarginine hydrolase, which released a molecule of 3,444 Da, consistent with HNP-1 (calculated 3,443 Da) (Fig. 5b3).
Discussion
Glycosylphosphatidylinositol-anchored ARTs (ART-1, 3, 4) are located on the apical surface of airway epithelial cells, as well as on lymphocytes and neutrophils, consistent with the possibility that ARTs could have a regulatory role in airway inflammation. Because these transferases use arginine as an ADP-ribose acceptor, defensins, arginine-rich cationic peptides are potential substrates. In this study, we demonstrated that, in vitro, synthetic HNP-1 is a substrate for ART-1. ADP ribosylation of HNP-1 significantly reduced both its antimicrobial and cytotoxic activities but enhanced its ability to release IL-8 from A549 cells. Further, ADP-ribosyl HNP-1 retained its activity as a chemoattractant for T lymphocytes. Importantly, ADP-ribosyl-HNP-1 inhibited the antimicrobial and cytotoxic activities of HNP-1. Thus, ADP-ribosylation of Arg-14 in HNP-1 yielded a molecule with unique biological properties, which could modulate defensin activity as well as T cell chemotaxis and neutrophil recruitment via IL-8 release from epithelial cells.
Because transferases require NAD+ for ADP ribosylation, a supply of extracellular NAD+ is necessary at the cell surface. NAD+ may be released into the airway lumen during inflammation, cell lysis, and/or apoptosis, and could be transiently and locally available for ADP ribosylation. We questioned whether there would be sufficient NAD+ in the extracellular space for the transferase-catalyzed modification and whether our in vitro studies were relevant to the human airway in vivo. Cigarette smoking results in pulmonary inflammation with increased numbers of neutrophils in the lung. We showed by HPLC, MALDI-MS, and enzymatic modification the presence of ADP-ribosyl-(arginine) HNP-1 in BAL fluids from habitual cigarette smokers. Consistent with the observation that HNP-1 is the most abundant antimicrobial factor in BAL fluids (29), we found that the major modified α defensin was HNP-1.
Defensins exhibit a wide range of antimicrobial activities but are also cytotoxic for several types of mammalian cells, including human epithelial and endothelial cells (30, 31). During lung inflammation, neutrophil defensins reach concentrations that, in vitro, are cytotoxic for airway epithelial cells (9–11, 30). Several mechanisms could modify their cytotoxic and antimicrobial activities. Defensins are synthesized as inactive propeptides (32). In intestinal Paneth cells, the metalloprotease matrilysin cleaves the propeptide, releasing the anionic inhibitory segment and generating active defensin (33). In the airway, binding of defensins to α1 antitrypsin and α1 antichymotrypsin inhibited their cytotoxicity for airway epithelial cells (11). In cystic fibrosis, the high salt concentration of the epithelial lining fluid may block defensin antimicrobial action to the detriment of the host (34, 35). Our study suggests that ADP ribosylation can also modify defensin activities. Because ADP-ribosyl-HNP-1 inhibited antimicrobial and cytotoxic activities of HNP-1, while retaining its T cell chemotactic properties and promoting neutrophil recruitment, it may have an important role as a regulator of the inflammatory response. Such properties could make ADP-ribosylated HNP-1 useful as a therapeutic agent in pulmonary inflammation and lung diseases.
Acknowledgments
We thank Drs. Martha Vaughan, Vincent Manganiello, and Gustavo Pacheco-Rodriguez for helpful discussion and critical review of the manuscript, and Mrs. Eleanor Cavanaugh for technical assistance.
Abbreviations
- HNP
human neutrophil peptides
- ARTs
ADP ribosyltransferases
- MALDI
matrix-assisted laser desorption ionization
- BAL
bronchoalveolar lavage
References
- 1.Nakamura H, Yoshimura K, Jaffe H A, Crystal R G. J Biol Chem. 1991;266:19611–19617. [PubMed] [Google Scholar]
- 2.Rihman-Eisenstat J B, Jorens P G, Herbert C A, Ueki I, Nadel J A. Am J Physiol. 1993;264:L413–L418. doi: 10.1152/ajplung.1993.264.4.L413. [DOI] [PubMed] [Google Scholar]
- 3.Ganz T. Infect Immun. 1987;55:568–571. doi: 10.1128/iai.55.3.568-571.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardi A, Zhang X L, Selsted M E, Skalicky J J, Yip P F. Biochemistry. 1992;31:11357–11364. doi: 10.1021/bi00161a013. [DOI] [PubMed] [Google Scholar]
- 5.Kagan B L, Selsted M E, Ganz T, Leherer R I. Proc Natl Acad Sci USA. 1990;87:210–214. doi: 10.1073/pnas.87.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehrer R I, Ganz T. Ann NY Acad Sci. 1996;797:228–239. doi: 10.1111/j.1749-6632.1996.tb52963.x. [DOI] [PubMed] [Google Scholar]
- 7.Ganz T. Science. 1999;286:420–421. doi: 10.1126/science.286.5439.420. [DOI] [PubMed] [Google Scholar]
- 8.Kagan B L, Ganz T, Leher R I. Toxicology. 1994;87:131–149. doi: 10.1016/0300-483x(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 9.Soong L B, Ganz T, Ellison A, Caughey G H. Inflamm Res. 1997;46:98–102. doi: 10.1007/s000110050114. [DOI] [PubMed] [Google Scholar]
- 10.Ashitani J, Mukae H, Nakazato M, Ihi T, Mashimoto H, Kadota J, Kohno S, Matsukura S. Eur Respir J. 1998;11:104–111. doi: 10.1183/09031936.98.11010104. [DOI] [PubMed] [Google Scholar]
- 11.Panyutich A V, Hiemstra P S, Van Vetering S, Ganz T. Am J Respir Cell Mol Biol. 1995;12:351–357. doi: 10.1165/ajrcmb.12.3.7873202. [DOI] [PubMed] [Google Scholar]
- 12.Okazaki I J, Moss J. Adv Pharmacol. 1996;35:247–280. doi: 10.1016/s1054-3589(08)60277-x. [DOI] [PubMed] [Google Scholar]
- 13.Moss J, Vaughan M. Adv Enzymol. 1988;6:303–379. doi: 10.1002/9780470123072.ch6. [DOI] [PubMed] [Google Scholar]
- 14.Koch F, Haag F, Kashan A, Thiele H G. Proc Natl Acad Sci USA. 1990;87:964–967. doi: 10.1073/pnas.87.3.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy I, Wu Y Q, Roeckel N, Bulle F, Pawlak A, Siegrist S, Mattei M G, Guellaen G. FEBS Lett. 1996;382:276–280. doi: 10.1016/0014-5793(96)00183-4. [DOI] [PubMed] [Google Scholar]
- 16.Koch-Nolte F, Haag F, Braren R, Kuhl M, Hoovers J, Balasubramanian S, Bazan F, Thiele H G. Genomics. 1997;39:370–376. doi: 10.1006/geno.1996.4520. [DOI] [PubMed] [Google Scholar]
- 17.Okazaki I J, Kim H J, McElvaney H G, Lesma E, Moss J. Blood. 1996;88:915–921. [PubMed] [Google Scholar]
- 18.Okazaki I, Kim H J, Moss J. J Biol Chem. 1996;271:22052–22057. doi: 10.1074/jbc.271.36.22052. [DOI] [PubMed] [Google Scholar]
- 19.Zolkiewska A, Nightingale M S, Moss J. Proc Natl Acad Sci USA. 1992;89:11352–11356. doi: 10.1073/pnas.89.23.11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnelly L E, Rendell N B, Murray S, Allport J R, Lo G, Kefalas P, Taylor G W, MacDermot J. Biochem J. 1996;315:635–641. doi: 10.1042/bj3150635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kharadia S V, Huiatt T W, Huang H Y, Peterson J E, Graves D J. Exp Cell Res. 1992;201:33–42. doi: 10.1016/0014-4827(92)90345-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Nemoto E, Kots A Y, Kaslow H R, Dennert G. J Immunol. 1994;153:4048–4058. [PubMed] [Google Scholar]
- 23.Allport JR, Donnelly L E, Kefalas P, Lo G, Nunn A, Yadollahi-Farsani M, Rendell N B, Murray S, Taylor G W, MacDermot J. Br J Clin Pharmacol. 1996;42:99–106. doi: 10.1046/j.1365-2125.1996.37014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balducci E, Horiba K, Usuki J, Park M, Ferrans V J, Moss J. Am J Respir Cell Mol Biol. 1999;21:337–346. doi: 10.1165/ajrcmb.21.3.3638. [DOI] [PubMed] [Google Scholar]
- 25.Rivera-Nieves J, Thompson W C, Levine R L, Moss J. J Biol Chem. 1999;274:19525–19531. doi: 10.1074/jbc.274.28.19525. [DOI] [PubMed] [Google Scholar]
- 26.Takemura H, Kaku M, Kohno S, Hirakata Y, Tanaka H, Yoshida R, Tomono K, Koga H, Wada A, Hirayama T, Kamihira S. Antimicrob Agents Chemother. 1996;40:2280–2284. doi: 10.1128/aac.40.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chertov O, Michiel D F, Xu L, Wang J M, Tani K, Murphy W J, Longo D L, Taub D D, Oppenheim J J. J Biol Chem. 1996;271:2935–2940. doi: 10.1074/jbc.271.6.2935. [DOI] [PubMed] [Google Scholar]
- 28.Moss J, Jacobson M K, Stanley S J. Proc Natl Acad Sci USA. 1985;82:5603–5607. doi: 10.1073/pnas.82.17.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnapp D, Harris D. Am J Respir Cell Mol Biol. 1998;19:352–356. doi: 10.1165/ajrcmb.19.3.3384. [DOI] [PubMed] [Google Scholar]
- 30.Okrent D G, Lichtenstein AK, Ganz T. Am Rev Respir Dis. 1990;141:179–185. doi: 10.1164/ajrccm/141.1.179. [DOI] [PubMed] [Google Scholar]
- 31.Lichtenstein A, Ganz T, Selsted M, Lehrer R I. Blood. 1986;68:1407–1410. [PubMed] [Google Scholar]
- 32.Valore E, Martin E, Harwig S, Ganz T. J Clin Invest. 1996;97:1624–1629. doi: 10.1172/JCI118588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson CL, Ouellette A J, Satchell D P, Ayabe T, Lopez-Boado Y S, Stratman J L, Hultgren S J, Matrisian L M, Parks W C. Science. 1999;286:113–116. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 34.Goldman MJ, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 35.Yu Q, Lehrer R I, Tam J P. J Biol Chem. 2000;275:3943–3949. doi: 10.1074/jbc.275.6.3943. [DOI] [PubMed] [Google Scholar]