Abstract
Inactivating mutations of the adenomatous polyposis coli gene (APC) or activating mutations of the β-catenin gene (CTNNB1) initiate colorectal neoplasia. To address the biochemical and physiologic effects of mutant β-catenin, we disrupted either the mutant or wild-type CTNNB1 allele in a human colorectal cancer cell line. Cells with only wild-type β-catenin had decreased colony-forming ability when plated at low density, although their growth was similar to that of parental cells when passaged under routine conditions. Immunohistochemistry and cell-fractionation studies suggested that mutant β-catenin activity was distinguished primarily by cellular localization and not by protein degradation. Surprisingly, we found mutant β-catenin bound less well to E-cadherin than did wild-type β-catenin, and the membranous localization of wild-type and mutant β-catenin was accordingly distinct. These findings pose several challenges to current models of APC/β-catenin function.
With an estimated 130,000 new cases and 58,000 deaths per year, colorectal cancer is the second leading cause of cancer death in the United States (1). Most of these tumors are initiated by truncating mutations of the adenomatous polyposis coli (APC) tumor suppressor (reviewed in ref. 2). Among the dozen or more proteins that associate with full length APC, β-catenin seems to play an especially important role (3, 4), as indicated by the identification of oncogenic mutations of the β-catenin gene (CTNNB1) in colorectal cancers that lack APC mutations (5–8). Understanding the interaction of APC and β-catenin has therefore been a major focus of study.
A variety of studies suggest that APC acts to inhibit the function of β-catenin (reviewed in refs. 2 and 9). Accordingly, APC has been demonstrated to inhibit tumor cell growth (10–13), whereas mutant β-catenin has been shown to promote neoplastic transformation (14, 15). Beyond simple binding and possible sequestration (3, 4), APC could modulate β-catenin activity in several ways. In one scenario, APC binding facilitates the phosphorylation of β-catenin by the serine/threonine kinase GSK3β (16), leading to the degradation of β-catenin (17) by ubiquitin-dependent proteolysis (18). A distinct but not mutually exclusive scenario is suggested by the ability of APC to act as a nuclear exporter and thereby regulate β-catenin by promoting its translocation out of the nucleus (19–21). However, it remains unclear which of these mechanisms plays a predominant role in the regulation of β-catenin by APC.
The β-catenin protein has itself been implicated in diverse cellular processes ranging from cell adhesion to transcription. At the transcriptional level, β-catenin can form heterodimers with members of the Tcf family of transcription factors and activate genes containing Tcf-binding sites (22, 23). This β-catenin/Tcf-4-regulated transcription (CRT) is inhibited by intact APC, but this inhibition is lost in mutant APC (5, 24). Moreover, oncogenic mutations of β-catenin render it resistant to APC inhibition of CRT (5, 24, 25). Thus, one common feature of APC or β-catenin mutation is constitutive activation of CRT. Accordingly, several targets of CRT have been identified and found to be expressed at elevated levels in colorectal cancer cells (26–33), but their role and the role of CRT in β-catenin-mediated transformation have yet to be fully defined.
Although much attention has recently focused on β-catenin's role as a transcriptional regulator, β-catenin was originally identified through its association with E-cadherin and its role in cell adhesion (reviewed in ref. 34). E-cadherin is an adhesion molecule that acts as a tumor suppressor in several neoplasms (35–37), and β-catenin is required for the proper function of E-cadherin (reviewed in ref. 34). Mutations in APC and the E-cadherin gene are synergistic in intestinal tumor initiation in mice (38). However, it is unknown whether mutations in β-catenin result in altered E-cadherin/β-catenin interaction, and if so, whether this interaction has phenotypic consequences.
Although a great deal has been learned about β-catenin, the precise effects of mutant β-catenin in human colorectal cancer cells are not known, and likewise, the relationship of these effects to the various biochemical activities described above is unclear. To address these questions, we generated human colorectal cancer cells with only wild-type (WT) or mutant β-catenin by using gene-targeting technology and assessed the effect of gene disruption on the physiologic and biochemical properties of the cells.
Materials and Methods
Disruption of CTNNB1.
The targeting construct for CTNNB1 was constructed by using methods described (39, 40). A bacterial artificial chromosome containing CTNNB1 was obtained from Research Genetics (Huntsville, AL). Two fragments of the bacterial artificial chromosome clone, one 2.4 kb and the second 6 kb, were used to construct the 5′ and 3′ arms of the targeting vector, respectively. The 2.4-kb fragment was derived from a sequence within intron 1, and the 6-kb fragment was derived from sequences in intron 7 of CTNNB1 (Fig. 1) and cloned into pBluescript SK(−) (Stratagene). A sequence (5′-CCAGTACTTGAAAACTAACGAT-3′), derived from a region of CTNNB1 deleted by the targeting construct, was ligated to the 5′ end of a hygromycin-resistance/thymidine kinase (hyg/tk) gene. This sequence served as an internal primer site for PCR-based screening. Other details of the constructs are available from the authors upon request.
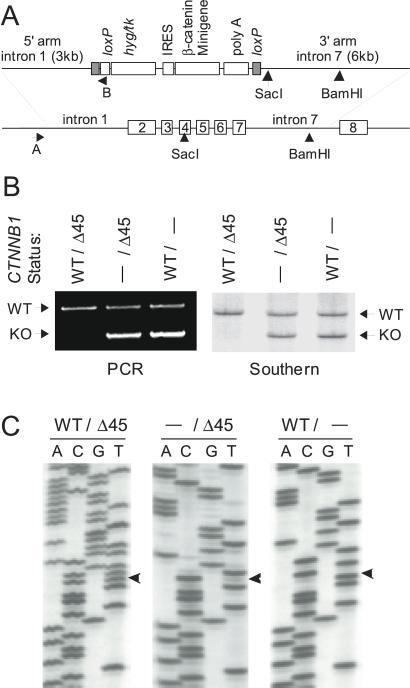
Figure 1.
CTNNB1 targeting. (A) The upper map shows the targeting construct used to disrupt CTNNB1. The 5′ and 3′ arms were obtained from a human bacterial artificial chromosome library and ligated to a selection cassette flanked by two loxP sites. These loxP sites enabled the efficient removal of the cassette after successful targeting events. The β-catenin minigene encodes a S33Y mutant CTNNB1, and the primer site B was incorporated to facilitate rapid PCR identification of knockout (KO) cells. The lower map shows the regions of CTNNB1 that were targeted by the KO construct. (B) Rapid PCR screening was used to identify clones with successful targeting events at the CTNNB1 (PCR), and targeting events were confirmed by Southern analysis (Southern). The WT and KO alleles are labeled accordingly. (C) CTNNB1 was sequenced in KO clones. Parental HCT116 cells (WT/Δ45) possess both mutant (Δ45) and WT CTNNB1. WT KO clones (−/Δ45) possess only mutant CTNNB1, whereas mutant KO clones (WT/−) have only WT CTNNB1. The first base of codon 45 is labeled with an arrowhead.
HCT116 cells were obtained from the American Type Culture Collection and cultured in McCoy's medium supplemented with 10% FBS (GIBCO). Tranfections were performed with PacI-linearized targeting vectors and Lipofectamine as directed by the manufacturer (GIBCO). To generate cells with disrupted CTNNB1, HCT116 human colon cancer cells were transfected with the targeting construct and selected in 0.1 mg/ml hygromycin (GIBCO). After transfection, cells were diluted in selection media and distributed in 96-well plates. After selection, genomic DNA was prepared from the drug-resistant clones by using the QiaAmp column system (Qiagen, Chatsworth, CA). Clones with a successfully targeted allele were identified by PCR with the primers 5′-GACCTTTGATCTCCTGAATTGATCG-3′ and 5′-ATCGTTAGTTTTCAAGTACTGG-3′ and Taq Platinum (Invitrogen). Adenovirus-mediated expression of Cre was used to remove the selection cassette as described (39). Southern blot and sequencing analysis were used to confirm the CTNNB1 genotype as described (5, 40).
Reporter Assays.
β-Catenin/Tcf reporter assays were performed as described by using the vectors pOT and pOF (41), which are low-background reporters containing Tcf-4-binding sites (24). Cells of various CTNNB1 genotypes were transfected with reporter plasmid (1 μg) and a vector containing β-galactosidase under control of the cytomegalovirus (CMV) promoter (pCMV-β, Promega) (0.25 μg). Cells were then collected, and luciferase activity was measured by using the Luciferase Activity Assay (Promega) and normalized for transfection efficiency by using β-galactosidase activity.
Colony Formation Assays and Cell-Mixing Experiments.
Subconfluent cells were trypsinized, counted, and plated in flasks with fresh media or in 96-well plates. Equal numbers of cells were plated in all cases. Cells were grown for 10 days and stained with crystal violet as described (42). For mixing experiments, cells were mixed in at 1:5 ratio (CTNNB1−/Δ45:CTNNB1WT/−) and grown for 14 days before harvest. Genomic DNA was prepared as described above and PCR was performed with a fluorescence-labeled primer, 5′-TTTGATGGAGTTGGACATGG-3′, and an unlabeled primer, 5′-CAGGACTTGGGAGGTATCCA-3′. The PCR product from the mutant allele is 3 bp shorter than the product from the WT allele. The mutant and WT alleles were separated and quantified by capillary gel electrophoresis (SpectruMedix 9600, State College, PA).
Cell Fractionation.
Cell fractionation was performed essentially as described (43). A protease inhibitor mixture (Complete, Boehringer Mannheim) was added to all solutions used for fractionation. Cells were harvested by scraping into cold PBS, rinsed two times with cold PBS, and then resuspended in L-buffer (1× PBS/0.1% Triton X-100/0.1% Nonidet P-40). Cells were then incubated on ice for 10 min until >99% lysed as determined by trypan blue exclusion. Nuclei were pelleted by centrifugation at 1,000 × g for 10 min at 4°C and the supernatant was used as the cytoplasmic fraction. The nuclear pellet was purified further from membrane contaminants by rinsing two times in L-buffer, passaging through a 22-gauge needle, and centrifuging through a 0.85 M sucrose cushion (20,000 × g for 15 min). Nuclei in the pellet were sonicated to make a lysate. Protein concentrations of the cytoplasmic and nuclear lysates were determined by using the Bradford Protein Assay (Bio-Rad), and equal amounts of protein were used for Western blot analyses.
Immunohistochemistry.
Cells were rinsed twice with PBS and then fixed with Leucoperm (Serotec), permeabilized with 1% Nonidet P-40 in PBS, and blocked in goat serum for 1 h. Anti-β-catenin antibody (Transduction Laboratories, Lexington, KY) was applied in GT (goat serum containing 0.05% Tween 20). After washing in PBST (PBS with 0.05% Tween 20), a fluorochrome-labeled secondary antibody (Molecular Probes) was applied in GT for 1 h. Cells were washed three times (5 min each) in PBST. Slides were mounted in DAPCO/glycerol and analyzed with a Nikon Eclipse E800 microscope equipped with a charge-coupled device camera (Photometrics, Tucson, AZ) and IPLAB software (Signal Analytics, Fairfax, VA). High-resolution images were obtained by using a Nikon confocal microscope.
Immunoprecipitation and Western Blot Analysis.
Attached cells from a confluent T25 flask were rinsed with PBS and incubated in 1.5 ml of IP buffer (150 mM NaCl/50 mM Tris⋅HCl, pH 7.5/0.5% Nonidet P-40/complete protease inhibitor mixture) for 10 min on ice. Cells were scraped and precleared with protein A-agarose beads (Boehringer Mannheim) for 30 min at 4°C. Proteins were then incubated with antibody for 15 h at 4°C and precipitated by using protein A agarose beads that had been blocked with 3% powdered milk. Beads were washed four times with 1 ml of IP buffer and then mixed with 2× Laemmli sample buffer. Western blotting was performed essentially as described (39). Antibodies used included anti-E-cadherin antibody (Santa Cruz Biotechnology), anti-β-catenin antibody (Transduction Laboratories), and anti-lamin B (Santa Cruz Biotechnology).
Results
Targeted Deletion of CTNNB1.
The human colorectal cancer cell line HCT116 was chosen for these studies because it is heterozygous (CTNNB1WT/Δ45), containing one WT allele and one mutant allele with a 3-bp deletion that eliminates the serine residue at codon 45 (5, 8). The targeting vector for β-catenin was constructed as described in Materials and Methods and used to generate cells lacking either the WT or mutant alleles (Fig. 1). This targeting vector included a mutant CTNNB1 in case loss of the mutant β-catenin resulted in negative selection (Fig. 1A). However, this exogenous mutant CTNNB1 did not seem to be expressed in the knockout (KO) clones. To ensure that the exogenous CTNNB1 gene did not play a role, we excised it from all KO clones by cre-mediated recombination with the flanking loxP sites. PCR and Southern blot analyses were used to identify clones with successful targeting events (Fig. 1B). To identify whether the WT or the mutant CTNNB1 was knocked out, genomic DNA was prepared from each clone and the CTNNB1 was sequenced (Fig. 1C). Two independently derived WT βCTNNB1 KO clones (CTNNB1−/Δ45, clones W1, W2) and four mutant CTNNB1 KO clones (CTNNB1WT/−, clones M1–M4) were obtained and used for the studies described below. All clones of the same genotype behaved similarly in the assays described below.
Elimination of Mutant CTNNB1 Results in Decreased CRT Activity.
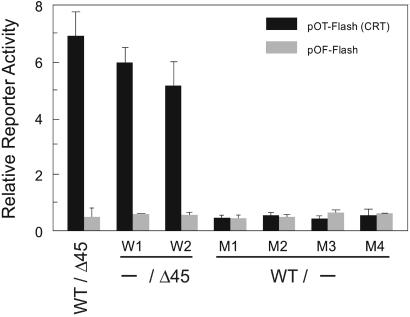
Human colon cancer cells with either mutant APC or mutant β-catenin have constitutively active CRT activity (5, 24). To determine whether disruption of CTNNB1 altered this activity, we measured CRT by using an appropriate reporter containing Tcf-4-binding sites (41). Parental HCT116 cells (CTNNB1WT/Δ45) and both clones with their WT CTNNB1 disrupted (CTNNB1−/Δ45) had equally high CRT reporter activity (pOT, Fig. 2). In contrast, all four clones with mutant CTNNB1 disrupted (CTNNB1WT/−) had low CRT reporter activity that was indistinguishable from the activity measured with an inactive reporter containing mutated Tcf-4-binding sites (pOF, Fig. 2). These results show that removal of the mutant β-catenin was sufficient to abrogate constitutive CRT in HCT116 colon cancer cells.
Figure 2.
CRT in CTNNB1 KO cells. CRT was measured in cells with the indicated genotypes. Parental cells and WT KO clones (W1, W2) have mutant CTNNB1 and elevated CRT activity, whereas mutant KO clones (M1–M4) lack mutant CTNNB1 and have no measurable CRT. The graph shows the average of duplicate experiments with error bars corresponding to one standard deviation.
Elimination of Mutant CTNNB1 Results in Decreased Clonogenicity Under Defined Conditions.
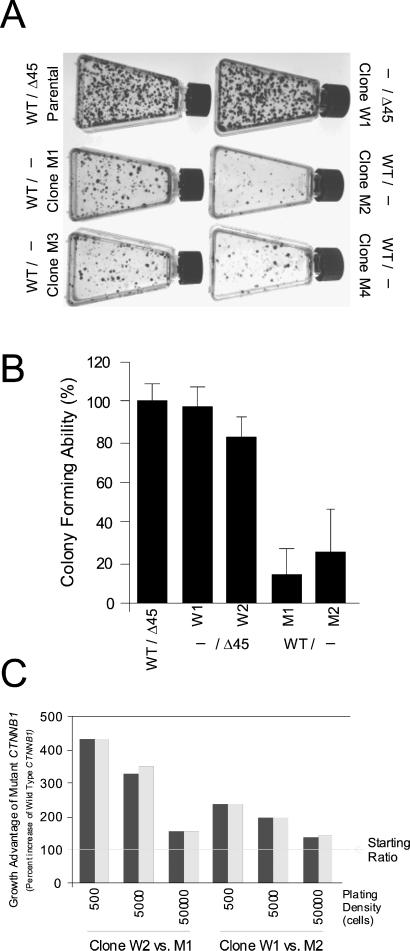
Surprisingly, cells with their mutant CTNNB1 disrupted survived and continued to grow in culture. When passaged routinely, their growth rates were similar to those of parental cells or cells with their WT CTNNB1 disrupted. However, when cells were plated at low density, differences between these cell types were apparent. As shown in Fig. 3A, all four cell lines with disrupted mutant CTNNB1 (CTNNB1WT/−) had significantly decreased colony-forming ability compared with parental cells (CTNNB1WT/Δ45) or cells with a disrupted WT CTNNB1 (CTNNB1−/Δ45).
Figure 3.
Deletion of mutant CTNNB1 results in decreased growth and survival. (A) Clones with the indicated CTNNB1 genotypes were diluted and plated in flasks. Clones without mutant CTNNB1 (M1–M4) had markedly decreased clonogenic survival compared with cells with mutant CTNNB1 (Parental, W1, W2). (B) Clones with the indicated CTNNB1 genotypes were diluted and plated in 96-well plates so that on average less than one cell grew per well. The numbers of colonies that grew for each cell type were then counted. Cells lacking mutant CTNNB1 had significantly decreased clonogenic survival. Data points are expressed as a percentage of the number of colonies obtained with the parental cell line (WT/Δ45). Data are expressed as the average of at least three experiments with error bars corresponding to one standard deviation. (C) Disrupted WT (W1, W2) or mutant (M1, M2) CTNNB1 cells were cocultured for 14 days. The growth advantage of the mutant CTNNB1-containing cells (W1, W2) was expressed as percentage increase in the fraction of mutant CTNBB1-containing cells as determined by sequencing. The increase is expressed relative to the starting ratio of mutant- to WT-containing cells (1:5, ≈17% mutant cells) and is indicated as 100%. Mixtures of cells were plated at 500, 5,000, or 50,000 cells per T25 flask as indicated. For example, the 430% growth increase indicates that the mutant CTNNB1-containing cells increased from ≈20% of the plated population to more than 85%. Paired bars represent the results of duplicate experiments. Similar behavior was observed in all four possible combinations of the two WT and mutant CTNNB1 KO clones tested.
Two independent experimental approaches were used to validate this specific growth defect. First, cells were inoculated in 96-well plates at a density of one cell per well, and the number of wells in which clones grew was quantified. As shown in Fig. 3B, cells with a disrupted mutant CTNB1 (CTNNB1WT/−) were able to form colonies at frequencies only 20–30% that of parental cells (CTNNB1WT/Δ45) or cells with a disrupted WT CTNNB1 (CTNNB1−/Δ45).
Second, cells with disruption of either the WT or mutant CTNNB1 were mixed at a 1:5 ratio, plated, and allowed to grow for 14 days. DNA was harvested from the cells and PCR was used to amplify the CTNNB1. Because the mutant allele harbored a 3-bp deletion of CTNNB1, the PCR products from the two alleles could be easily distinguished and used to determine the ratio of the two cell types in culture. When the mixed cells were plated at low density, the relative abundance of mutant CTNNB1 cells (CTNNB1−/Δ45) increased by 235–430% (Fig. 3C). This increase was attenuated when cells were plated at higher densities (Fig. 3C). The ratio of the two cell types after mixed growth was consistent with that predicted from the data in Fig. 3 A and B, wherein the cell types were analyzed individually.
We also tested the growth of these lines under anchorage-independent conditions in vitro and as s.c. xenografts in nude mice. All cell lines, whatever the CTNNB1 allele disrupted, grew in soft agar and formed tumors in nude mice. Although some quantitative differences were apparent, these differences were in general accord with the less robust growth of cells with a disrupted mutant CTNNB1 observed in culture.
Regulation of WT and Mutant β-Catenin.
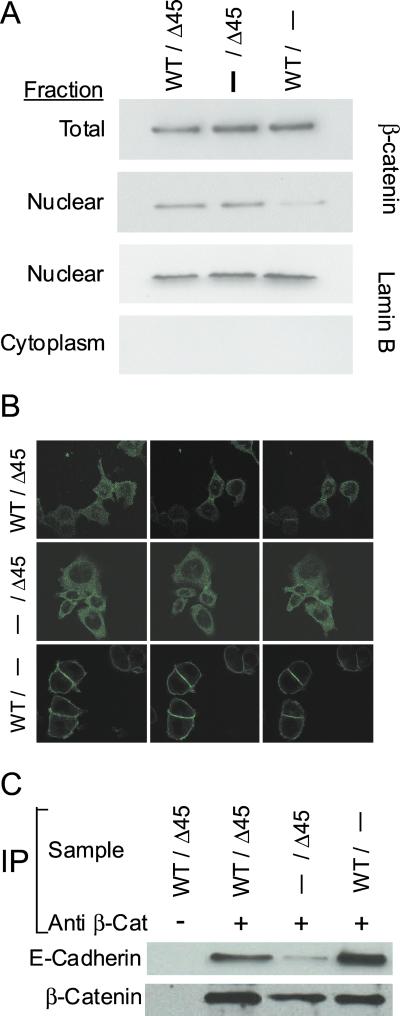
β-Catenin is regulated by both degradation and subcellular localization (17, 19–21). To determine the relative levels of β-catenin in the WT and mutant KO, we performed Western blot analyses with anti-β-catenin antibodies. As shown in Fig. 4A, cells with disrupted mutant CTNNB1 (CTNNB1WT/−) contained as much β-catenin as cells with disrupted WT CTNNB1 (CTNNB1−/Δ45) or parental cells (CTNNB1WT/Δ45).
Figure 4.
Mutant β-catenin displays abnormal subcellular localization and decreased binding to E-cadherin. (A) Cells with the indicated genotypes were fractionated and analyzed by Western blotting as indicated. The total amount of β-catenin is similar in parental, WT (−/Δ45), and mutant (WT/−) KO clones. WT β-catenin is largely excluded from the nuclear fraction, whereas mutant β-catenin is not. Western blot analysis of the nuclear protein lamin B controlled for the quality of the fractionation. (B) Parental (WT/Δ45), WT (−/Δ45), and mutant (WT/−) CTNNB1 KO cells were stained with anti-β-catenin antibody and imaged by using a confocal microscope. Confocal images at three different levels are shown for each cell type. (C) Lysates from KO cells were immunoprecipitated with anti-β-catenin antibody as indicated. The immunoprecipitated complexes were then analyzed by Western blotting using anti-E-cadherin antibody or anti-β-catenin. Mutant β-catenin has decreased binding to E-cadherin (compare −/Δ45 with WT/− and WT/Δ45).
Given the same total levels of β-catenin protein in the cell, why did the cells with disrupted mutant CTNNB1 not have constitutively high CRT? Cell-fractionation studies were performed to address this issue. Nuclear fractions from each of the cell types were generated and analyzed by Western blotting. Cells whose mutant CTNNB1 alleles were disrupted were found to have significantly less β-catenin in the nuclear fraction than parental cells or cells with disrupted WT CTNNB1 alleles (Fig. 4A). Western blots with anti-lamin B antibody were performed to confirm the quality of the cellular fractionation and to control for loading (Fig. 4A).
To investigate the subcellular distribution of β-catenin further, we stained the cells with an anti-β-catenin antibody. Disruption of mutant CTNNB1 resulted in a striking redistribution of β-catenin protein, so that virtually all of it was localized to cell membranes, particularly at cell junctions (Fig. 4B). In contrast, the β-catenin in parental cells and in cells with their WT CTNNB1 disrupted was distributed diffusely throughout the cytoplasm, with faint nuclear staining visible. These differences were more pronounced in cells plated sparsely than in confluent cells; in confluent cells, membranous localization of β-catenin was detectable in all cell types. The distribution of β-catenin and APC has been shown to be dependent on cell density (44–46).
Finally, we determined whether mutation of β-catenin was associated with a decreased ability to bind to E-cadherin. Immunoprecipitation experiments with cells containing only CTNNB1 (CTNNB1−/Δ45) showed that less mutant β-catenin was bound to E-cadherin than WT β-catenin (Fig. 4C). This observation is consistent with the localization of mutant and WT β-catenin observed on immunostaining (Fig. 4B).
Discussion
Our results revealed several important aspects of β-catenin regulation and function, some of which were unexpected on the basis of previous experiments. These can be summarized as follows.
Catenin-Regulated Transcription.
On the basis of previous experiments, it had been assumed that the increased CRT activity observed in colorectal cancer cells was due to either mutant β-catenin or mutant APC. However, the possibility that this increased activity was due to other cellular aberrations could not be excluded. Our results definitively establish that the CRT activity observed with standard reporters is completely dependent on the endogenous mutant β-catenin in HCT116 cells.
On the other hand, the expression of CRT-regulated endogenous genes was not affected by disruption of the mutant CTNNB1. Thus, the expression of c-MYC and cyclin D1 gene was unchanged in the various KO cell lines generated in this study (data not shown), suggesting that the regulation of these genes in these cells is not β-catenin-dependent under the conditions tested or they are not physiological targets. However, it is also possible that these genes are critical mediators of β-catenin function under in vivo conditions only. Likewise, it is possible that these genes are under complex controls and their expression levels are somehow compensated for in vitro when mutant β-catenin is deleted.
Posttranscriptional Regulation of β-Catenin.
Several studies have suggested that mutant β-catenin accumulates in cells that contain either a CTNNB1 or APC mutation. Such mutations are predicted to interfere with phosphorylation and subsequent ubiquitinization and proteasome-mediated degradation. Our results revealed that the total amount of β-catenin in cells was independent of the nature of β-catenin (WT or mutant). The mutation present in HCT116 cells (Δ45) is predicted to have the most dramatic consequences on phosphorylation and degradation of β-catenin of any yet studied (47). Our current data and previous studies (12) with inducible APC suggest that increases in total cellular β-catenin concentration are not a major consequence of APC or β-catenin mutations, at least in the cell lines studied. In contrast, our results support the idea that cellular localization is a critical determinant of β-catenin function. The results of Fig. 4A show that mutation of β-catenin is associated with nuclear localization, consistent with the idea that nuclear transport is a critical determinant of β-catenin function. Moreover, this altered localization not only involves partitioning between nucleus and cytoplasm, but also affects the membranous localization of the protein, particularly in cells that are sparsely plated (Fig. 4B).
Physiologic Effects of β-Catenin Mutation.
Two separate functions have been proposed for β-catenin. As noted in the Introduction, β-catenin was originally identified as a protein that bound to E-cadherin and connected it to the cytoskeleton. Later, β-catenin was identified as part of a heterodimeric transcription complex that functions in the nucleus and is manifest as CRT activity. It is currently believed that β-catenin's role in neoplasia is largely dependent on this latter function. Part of the reason for this belief is that mutations of CTNNB1, like mutations of APC, result in increased CRT activity, whereas no prior evidence has shown that such subtle mutations alter the interaction between β-catenin and E-cadherin. Indeed, the mutations in β-catenin that have been observed in cancer cells are not in the domain of β-catenin that has been reported to interact with E-cadherin (48), although deletion of residues 28–134 of β-catenin abrogated E-cadherin-dependent cell–cell adhesion in a gastric cancer line (49). One interesting observation in our study was that the β-catenin missense mutation we studied clearly affected its interaction with E-cadherin. This finding was documented by coimmunoprecipitation experiments and was supported by striking differences in membrane localization between WT and mutant β-catenin (Fig. 4C). Our results suggest that a critical consequence of CTNNB1 mutations in neoplasia might involve E-cadherin. Consistent with this, recent studies have indicated that E-APC in Drosophila can alter adhesion by acting through the Drosophila homolog of CTNNB1 (50). Further studies will be required to determine whether CTNNB1 functions through E-cadherin, through Tcf-4, or through both these pathways.
“Hit and Run” Pathways.
Previous experiments with Myc and Ras have demonstrated that tumor maintenance requires the continuing presence of the mutated oncogenes that contributed to tumorigenesis (51–53). It has been widely assumed that the continued inactivation of tumor suppressor genes would also be required for continued tumor growth. This assumption has been supported by studies showing that the overexpression of Rb, p53, and APC (for example) all inhibit growth. However, these studies have all used overexpression rather than re-creation of WT tumor suppressor gene expressed at normal, physiologic levels.
The β-catenin gene is a surrogate for the tumor-suppressor gene APC, because mutations in these two genes are mutually exclusive and are thought to operate in a single pathway. Our results demonstrate that elimination of β-catenin does not prevent growth of the tumor cells, either in vitro or as tumors in nude mice, although it does have an impact on growth at clonal density. Several possible interpretations of these results are as follows: First, our assays for growth may not mimic those encountered in vivo, and β-catenin (or APC) mutations might be absolutely required for continued growth of tumors in their natural microenvironment. Second, it is possible that such mutations might be required for initiation of tumors (“gatekeeper” function) but not for their continued growth once they acquire mutations in other growth-regulating genes (like those in c-Ki-RAS and TGFβ-RII). Third, it is possible that mutation of other genes in addition to CTNNB1 are required in lieu of a APC mutations and that β-catenin plays a role in only part of APC's tumor-suppressive activities. Additional studies to evaluate these possibilities, both in human and mouse systems, should be informative.
Acknowledgments
We thank members of the Molecular Genetics Laboratory of The Sidney Kimmel Comprehensive Cancer Center for their helpful comments and suggestions during the course of this work. This work was supported by grants from the National Institutes of Health (CA57345 and CA43460). K.W.K. received research funding from Genzyme Molecular Oncology (Genzyme) and K.W.K. is a consultant to Genzyme. The university and researchers (B.V., K.W.K.) own Genzyme stock, which is subject to certain restrictions under university policy. The terms of these arrangements are being managed by the university in accordance with its conflict-of-interest policies.
Abbreviations
- APC
adenomatous polyposis coli
- CRT
β-catenin/Tcf-4–regulated transcription
- KO
knockout
- WT
wild type
References
- 1.American Cancer Society. Cancer Facts & Figures. Atlanta, GA: Am. Cancer Soc.; 2001. [Google Scholar]
- 2.Kinzler K W, Vogelstein B. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 2002. pp. 565–587. [Google Scholar]
- 3.Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain S H, Masiarz F R, Munemitsu S, Polakis P. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 4.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 5.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 6.Ilyas M, Tomlinson I P, Rowan A, Pignatelli M, Bodmer W F. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwao K, Nakamori S, Kameyama M, Imaoka S, Kinoshita M, Fukui T, Ishiguro S, Nakamura Y, Miyoshi Y. Cancer Res. 1998;58:1021–1026. [PubMed] [Google Scholar]
- 8.Sparks A B, Morin P J, Vogelstein B, Kinzler K W. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 9.Polakis P. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 10.Baeg G-H, Matsumine A, Kuroda T, Bhattacharjee R N, Miyashiro I, Toyoshima K, Akiyama T. EMBO J. 1995;14:5618–5625. doi: 10.1002/j.1460-2075.1995.tb00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groden J, Joslyn G, Samowitz W, Jones D, Bhattacharyya N, Spirio L, Thliveris A, Robertson M, Egan S, Meuth M, White R. Cancer Res. 1995;55:1531–1539. [PubMed] [Google Scholar]
- 12.Morin P J, Vogelstein B, Kinzler K W. Proc Natl Acad Sci USA. 1996;93:7950–7954. doi: 10.1073/pnas.93.15.7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shih I M, Yu J, He T C, Vogelstein B, Kinzler K W. Cancer Res. 2000;60:1671–1676. [PubMed] [Google Scholar]
- 14.Whitehead I, Kirk H, Kay R. Mol Cell Biol. 1995;15:704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolligs F T, Hu G, Dang C V, Fearon E R. Mol Cell Biol. 1999;19:5696–5706. doi: 10.1128/mcb.19.8.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272:1023–1025. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 17.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosin-Arbesfeld R, Townsley F, Bienz M. Nature (London) 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- 20.Henderson B R. Nat Cell Biol. 2000;2:653–660. doi: 10.1038/35023605. [DOI] [PubMed] [Google Scholar]
- 21.Neufeld K L, Nix D A, Bogerd H, Kang Y, Beckerle M C, Cullen B R, White R L. Proc Natl Acad Sci USA. 2000;97:12085–12090. doi: 10.1073/pnas.220401797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 23.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 24.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 25.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 26.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 27.Tetsu O, McCormick F. Nature (London) 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 28.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He T C, Chan T A, Vogelstein B, Kinzler K W. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann B, Gelos M, Siedow A, Hanski M L, Gratchev A, Ilyas M, Bodmer W F, Moyer M P, Riecken E O, Buhr H J, Hanski C. Proc Natl Acad Sci USA. 1999;96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford H C, Fingleton B M, Rudolph-Owen L A, Goss K J, Rubinfeld B, Polakis P, Matrisian L M. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 32.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 33.Xu L, Corcoran R B, Welsh J W, Pennica D, Levine A J. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]
- 34.Aberle H, Schwartz H, Kemler R. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 35.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve A E. Nature (London) 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 36.Richards F M, McKee S A, Rajpar M H, Cole T R, Evans D G, Jankowski J A, McKeown C, Sanders D S, Maher E R. Hum Mol Genet. 1999;8:607–610. doi: 10.1093/hmg/8.4.607. [DOI] [PubMed] [Google Scholar]
- 37.Okegawa T, Li Y, Pong R C, Hsieh J T. J Urol. 2002;167:1836–1843. [PubMed] [Google Scholar]
- 38.Smits R, Ruiz P, Diaz-Cano S, Luz A, Jagmohan-Changur S, Breukel C, Birchmeier C, Birchmeier W, Fodde R. Gastroenterology. 2000;119:1045–1053. doi: 10.1053/gast.2000.18162. [DOI] [PubMed] [Google Scholar]
- 39.Chan T A, Hermeking H, Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 40.Waldman T, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 41.da Costa L T, He T C, Yu J, Sparks A B, Morin P J, Polyak K, Laken S, Vogelstein B, Kinzler K W. Oncogene. 1999;18:5010–5014. doi: 10.1038/sj.onc.1202872. [DOI] [PubMed] [Google Scholar]
- 42.Chan T A, Hwang P M, Hermeking H, Kinzler K W, Vogelstein B. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- 43.Neufeld K L, White R L. Proc Natl Acad Sci USA. 1997;94:3034–3039. doi: 10.1073/pnas.94.7.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brocardo M G, Bianchini M, Radrizzani M, Reyes G B, Dugour A V, Taminelli G L, Gonzalez Solveyra C, Santa-Coloma T A. Biochem Biophys Res Commun. 2001;284:982–986. doi: 10.1006/bbrc.2001.5066. [DOI] [PubMed] [Google Scholar]
- 45.Zhang F, White R L, Neufeld K L. Mol Cell Biol. 2001;21:8143–8156. doi: 10.1128/MCB.21.23.8143-8156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dietrich C, Scherwat J, Faust D, Oesch F. Biochem Biophys Res Commun. 2002;292:195–199. doi: 10.1006/bbrc.2002.6625. [DOI] [PubMed] [Google Scholar]
- 47.Liu C, Li Y, Semenov M, Han C, Baeg G, Tan Y, Zhang Z, Lin X, He X. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 48.Hulsken J, Birchmeier W, Behrens J. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawanishi J, Kato J, Sasaki K, Fujii S, Watanabe N, Niitsu Y. Mol Cell Biol. 1995;15:1175–1181. doi: 10.1128/mcb.15.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamada F, Bienz M. Nat Cell Biol. 2002;4:208–213. doi: 10.1038/ncb755. [DOI] [PubMed] [Google Scholar]
- 51.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 52.Felsher D W, Bishop J M. Proc Natl Acad Sci USA. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O'Hagan R, Pantginis J, Zhou H, et al. Nature (London) 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]