Summary
Mothers against decapentaplegic homolog 5 (SMAD5) is a transcriptional regulator that functions within the TGF-β signaling cascade. Evidence from animal studies show that it is crucial for dorsoventral patterning, left-right asymmetry, cardiac looping, and other embryonic processes. However, its role in human development has not been explored, and the contribution of SMAD5 variants to congenital disease is unknown. Here, we report SMAD5 variants identified in six unrelated families with seven individuals presenting with congenital heart disease (CHD). Isolated congenital heart defects are observed in six individuals who carry de novo or inherited missense, nonsense, frameshift, or copy-number variants in SMAD5. A multi-organ phenotype is observed in one individual with a de novo SMAD5 variant that alters an amino acid crucial for SMAD5 multimerization. Septal defects, identified in four individuals, are the most common cardiac lesion in our cohort, with hypoplastic left heart also observed in two individuals. In silico assessment of SMAD5 missense variants predicts disrupted binding to co-factors, and in vitro functional assessment shows changes in SMAD5 gene and protein expression, as well as impaired activation of a BMP4-responsive promoter by the variants. Our findings suggest haploinsufficiency as the underlying molecular mechanism in five of the six families, resulting in isolated CHD, with a SMAD5 dominant-negative variant identified in one family leading to multiple congenital defects. Here, we provide evidence that SMAD5 variants lead to CHD and offer a basis for future exploration of SMAD5 variants in both CHD and post-natal disease.
Keywords: congenital heart disease, cardiac septal defects, hypoplastic left heart, SMAD5 signaling, TGF-β signaling, BMP signaling
Graphical abstract

We report germline SMAD5 variants associated with congenital heart disease in seven individuals from six families. Through in silico and functional assessments, our findings imply that these heterozygous missense, nonsense, frameshift, and copy-number deletions in SMAD5 lead to haploinsufficiency and cardiac abnormalities.
Main text
Congenital heart disease (CHD), affecting up to 1% of newborns, represents a global health burden for pediatric, adolescent, and adult populations.1 The likelihood of receiving a genetic diagnosis for an individual with CHD can vary between 10% to 41%, and improving the rates of genetic diagnosis by discovering new gene-disease associations continues to be an international effort.2 At present, pathogenic variants in 192 genes are associated with CHD, with hundreds more predicted to be involved.3 As many of these genes have known associations with postnatal diseases such as neurodevelopmental disorders, cancer, or cardiovascular disease,4 identification of a clinically actionable variant also provides essential guidance regarding patient prognostication. This is especially relevant as 97% of patients with CHD reach adulthood, and adult cases now outnumber pediatric CHD cases.5
The transforming growth factor β (TGF-β) superfamily encompasses intracellular signaling cascades downstream of receptor-ligand interactions and regulates development, differentiation, and survival (Figure S1).6 TGF-β and bone morphogenic protein (BMP) signaling pathways form the two main branches of the superfamily, and pathogenic variants in genes encoding TGF-β/BMP ligands and intracellular effectors are implicated in both germline and somatic disease.6 The cardiovascular system is frequently affected, with CHD caused by variants in genes: ACVR2B (MIM: 613751), BMP2 (MIM: 617877), CFC1 (MIM: 605376), SMAD1 (MIM: 601595), SMAD2 (MIM: 619657), SMAD4 (MIM: 139210), SMAD6 (MIM: 614823), and others,7,8,9,10,11,12,13 suggesting that disruption of additional factors within this pathway may also disrupt development of the heart. SMAD proteins are the intracellular components of TGF-β/BMP pathways, and include receptor regulated R-SMADs (SMAD1, SMAD2, SMAD3, SMAD5, and SMAD8), which are phosphorylated by membrane-bound TGF-β or BMP receptors,14 common-SMAD, SMAD4, which trimerizes with R-SMADs to produce transcriptional regulatory complexes, and SMAD6 and SMAD7, which are inhibitory SMADs (i-SMADs), that inhibit BMP and TGF-β cascades, respectively.14 Of the SMAD group of transcription factors, variants in SMAD5 (MIM: 603110), SMAD7 (MIM: 602932), and SMAD8 (MIM: 603295) are currently not associated with human congenital defects (Figure S1).
Through sequencing of Australian cohorts and GeneMatcher,15 we identified SMAD5 variants in six unrelated families affected by CHD (Figure 1A; Table 1). To our knowledge, pathogenic SMAD5 variants have not been reported in individuals with congenital defects. All families gave informed consent, and the study was approved by ethics committees of corresponding institutions.
Figure 1.
Pedigrees of families with SMAD5 variants
(A) Genotypes of sequenced individuals are displayed below each family member. +/+ indicates absence of SMAD5 variant. Affected individuals are indicated by filled shapes. Black arrows indicate the proband of each family (F). Individuals who carry a variant without a CHD diagnosis are indicated by a black circle within a white shape. Gray-filled shapes indicate individuals with non-descriptive CHD phenotypes. The Sanger sequencing chromograph for F2 can be found in Figure S2.
(B) Schematic of the chromosomal deletion observed in affected individual in F3 (hg37). Numbers are assigned to protein-coding genes deleted within the region (1: SLC25A48; 2: IL9; 3: FBXL21; 4: LECT2; 5: TGFβ1; 6: SMAD5; 7: TRPC7; 8: SPOCK1). Arrows indicate directionality of the gene.
(C) Missense and protein-truncating variants observed in affected individuals in this cohort are positioned on a schematic of SMAD5 protein sequence (ENST00000545279.6, NM_005903.7, NP_005894.3). L3 loop (F416–T432) is shown within the MH2 domain.
(D) Missense variant constraint across the SMAD5 sequence is derived from gnomAD (version 4.1.0). 0.0 is intolerant to missense variation, and 1.0+ is tolerant to missense variation.
(E) Sequence conservation between selected vertebrates of residues affected by the missense variants in our cohort.
(F) Sequence conservation between all human SMAD proteins of residues affected by the missense variants in our cohort. Residue impacted by the variant in MH1 domain (p.V78F) is highlighted in pink, and the residues affected by the variants in MH2 domain (p.N361D and p.T430I) are highlighted in blue. Sequence alignments were created using the Uniprot Align tool.
ASD, atrial septal defect; AVSD, atrioventricular septal defect; HLH, hypoplastic left heart; MH1, Mad homolog 1; MH2, Mad homolog 2; ToF, tetralogy of Fallot; VSD, ventricular septal defect.
Table 1.
Clinical characteristics of individuals with SMAD5 variants
| Case (sex, vital status) | F1.3 (male, deceased) | F2.3 (male, alive) | F2.5 (male, alive) | F3.3 (male, alive) | F4.3 (male, deceased) | F5.3 (female, alive) | F6.2 (male, alive) |
|---|---|---|---|---|---|---|---|
| SMAD5 variant (NM_005903.7) | c.1289C>T | c.1202del | c.1202del | 5q31.1-q31.2 (135172706–136488443)x1 | c.232G>T | c.1081A>G | c.781C>T |
| Protein | p.Thr430Ile | p.Ala402Glufsa13 | p.Ala402Glufsa13 | NA | p.Val78Phe | p.Asn361Asp | p.Glu261a |
| GnomAD allelesa | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Inheritance | de novo | inherited | inherited | de novo | de novo | de novo | inherited (suspected) |
| Cardiac | ToF | HLH, VSD | ASD, VSD | AVSD | HLH, ASD | AV canal defect | ASD |
| Neurodevelopment | NA | + | NA | NA | NA | – | NA |
| Skeletal | + | – | NA | NA | – | – | NA |
| Gastrointestinal | + | + | NA | NA | – | – | NA |
| Facial dysmorphology | + | – | NA | – | – | – | – |
| Urogenital | + | – | NA | NA | – | – | – |
| Other phenotypic findings | + | – | NA | NA | – | – | + |
ASD, atrial septal defect, AV canal defect, atrioventricular canal defect; AVSD, atrioventricular septal defect; HLH, hypoplastic left heart; NA, data not assessed; ToF, tetralogy of Fallot; VSD, ventricular septal defect.
GnomAD version 4.1.0.
Family 1 (F1), family 2 (F2), and family 6 (F6) were recruited to Australian CHD sequencing cohorts (n = 363 trios and singletons). F1 underwent genome sequencing. F2.3 was recruited as a singleton and underwent exome sequencing (ES), and other members of F2 were Sanger sequenced (Figure S2). F6.2 underwent ES, and other members of F6 were unavailable for sequencing. F3, F4, and F5 were recruited to the study via GeneMatcher.15 F3.3 was recruited to the 'Untangling the genetics of congenital heart defects' study at Ghent University Hospital (n = 535 trios and singletons) and initially assessed via comparative genomic hybridization and single-nucleotide polymorphism microarray. Later, ES was performed for all members of the family. F4 and F5 were part of the Pediatric Cardiac Genomics Consortium (PCGC) study (n = 763 trios),16 and underwent ES.
Isolated heart defects were identified in 5/6 families, and septal defects were the most prevalent structural lesion (Figure S3). Atrioventricular canal defects were identified in two individuals (F3.3 and F5.3), an atrial septal defect was identified in one individual (F6.2), and individual F2.5 was diagnosed with both a ventricular and an atrial septal defect. Individual F6.5 presented with an atrioventricular canal defect; however, whether she carries the SMAD5 variant that was identified in her father (F6.2) could not be confirmed. Hypoplastic left heart (HLH) was observed in two individuals (F2.3 and F4.3), both of whom were also diagnosed with septal defects. Individuals F6.1, F6.4, and F6.6 were reported as CHD affected, but further phenotypic details were unavailable. F1.3 was a terminated pregnancy with tetralogy of Fallot (TOF) diagnosed in utero and extracardiac anomalies in craniofacial, urogenital, renal, limb, and vertebral systems observed during autopsy examination. F2.3 presented with neurodevelopmental and gastrointestinal phenotypes during post-natal development. F6.2, who underwent genetic assessment as an adult, was diagnosed with adult-onset epilepsy and skin cancer at 40 years of age and sebaceous carcinoma at 68 years. Clinical features of the cohort are summarized in Table 1 and Figure S3 and described in detail in the supplemental information.
Four families in the cohort carried novel, de novo variants in SMAD5, three of which were missense variants that were predicted to be damaging by in silico metrics (Figure 1A; Table S1). Variant c.1289C>T (p.T430I) (GenBank: NM_005903.7) was observed in F1.3, c.232G>T (p.V78F) was identified in F4.3, and c.1081A>G (p.N361D) was found in F5.3. F3.3 had a novel, de novo copy-number deletion encompassing SMAD5 on chromosome 5 (Chr5(GRCh37):135172706-136488443del) (Figure 1B; Table S2). Of the seven protein-coding genes that were deleted in the 1.3-Mb region, which included TGFB1, there were none associated with CHD or congenital defects, and only SMAD5 was constrained against loss of function (LOF) in the Genome Aggregation Database (gnomAD version 4.1.0) (Table S2). A novel, inherited SMAD5 variant was identified in F2. F2.3 and F2.5, who were diagnosed with CHD, inherited c.1202del (p.A402Qfs∗13) from the mother (F2.2), who was presumed to be asymptomatic but was not formally assessed (Figure 1A). Only F2.3 underwent ES, and the variant was identified in other family members by Sanger sequencing (Figure S2). Phenotyping information on members of the extended family of F2 could not be obtained. The novel SMAD5 variant in F6, c.781C>T (p.Q261∗), has a 50% chance of being inherited. This and the presence of four affected family members (F6.1, F6.4, F6.5, and F6.6) suggest that they share the heterozygous variant. However, since only F6.2 underwent ES, this is unconfirmed as the other affected individuals were unavailable for testing, and they were not counted as SMAD5 variant carriers. For all cases, clinically relevant variants in known CHD genes were not identified.3
All SMAD proteins share an N-terminal Mad-homolog 1 (MH1) domain, responsible for DNA binding, a linker region that may regulate subcellular localization, and a C-terminal MH2 domain, which regulates homo- and heterotrimerization, receptor interaction, and transactivation.14 Variant p.V78F (F4) occurs within the N-terminal MH1 domain, p.Q261∗ (F6) occurs within the linker region, and p.N361D (F5), p.A402Qfs∗13 (F2), and p.T430I (F1) are within the C-terminal MH2 domain (Figure 1C). Protein truncating variants in F2 and F6 are expected to be subject to nonsense-mediated decay and result in LOF; and along with copy-number loss in F3, these would result in SMAD5 haploinsufficiency. Missense variants from F1 (p.T430I), F4 (p.V78F), and F5 (p.N361D) occur in regions that are intolerant toward missense variation, highly conserved across multiple species, and predicted to impact protein function by in silico metrics (Figures 1D–1F; Table S1). To determine the molecular impact of these missense variants, we further assessed them by in silico modeling and in vitro assays.
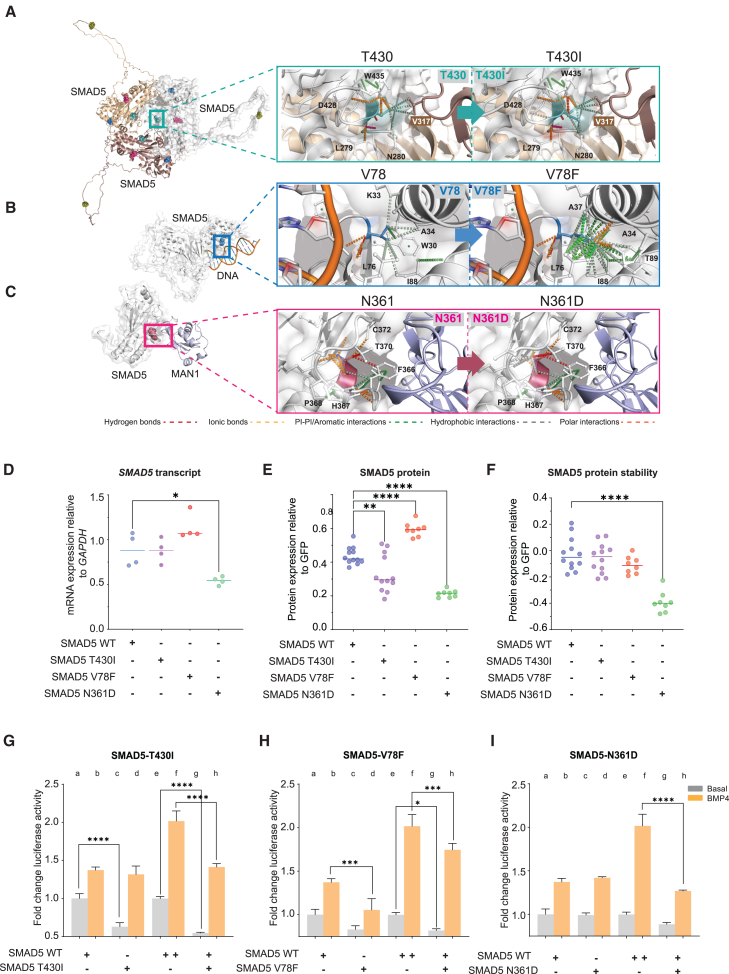
A highly conserved residue, T430 resides within the L3 loop of the SMAD5 MH2 domain (residues F416–T432; Figures 1C–1F) and participates in R-SMAD homo- and heterotrimerization.17,18 Given the lack of experimental crystal structures available describing SMAD5 trimerization as a homotrimer and heterotrimer (with one SMAD4 monomer), these complexes were generated using default parameters in AlphaFold2.19 We compared the interaction profiles between reference p.T430 and variant p.I430 (F1.3) at position 430 within the predicted SMAD5 homotrimer (Figure 2A). This highlighted an extensive hydrophobic interaction spanning D428, I430, and N280, which is predicted to shift the orientation of the variant side chain toward N280, leading to destabilization and reduced affinity to other trimer subunits (Table S3). To further understand how the missense variants affect gene expression, protein expression and stability, and transcription factor activity of SMAD5, the variants were transiently transfected into HEK293T cells, with vectors carrying human wild-type (WT) or variant SMAD5 cDNA. Transfection with SMAD5-T430I led to a decrease in SMAD5 protein levels, although its transcript levels and protein stability were unchanged (Figures 2D–2F). To assess transcription activation ability of the variants, we utilized the Xvent2-luc promoter.20 Xvent2 is a Xenopus homeobox gene that is a direct target of BMP4 and regulates dorsoventral patterning downstream of BMP4 and, subsequently, SMAD5.20,21,22 The promoter has been utilized previously to assess promoter activation by BMP pathway components such as SMAD1 and SMAD4.20,23 Activation of the Xvent2-luc promoter was decreased in response to SMAD5-T430I in basal conditions relative to WT-SMAD5 in HEK293T cells (Figure 2G, columns a–d). Addition of BMP4 restored promoter activity to WT levels. We also assessed whether SMAD5-T430I can act on WT-SMAD5 within our transactivation system by co-transfecting the variant with WT-SMAD5. We observed that SMAD5-T430I was able to significantly reduce promoter activation by WT-SMAD5 in both basal and BMP4+-treated cells (Figure 2G, columns e–h). These data suggest that SMAD5-T430I acts dominantly on WT-SMAD5, which, as indicated by our in silico predictions, could be due to interference with SMAD5 trimerization.
Figure 2.
Structural and functional analysis of the SMAD5 missense variants
(A–C) Impact of missense variants p.T430I (A), p.V78F (B), and p.N361D (C) on SMAD5 homotrimer (predicted), SMAD5-DNA (predicted), and SMAD5-cofactor (predicted) complexes.
(A) p.T430 (teal) predominantly interacts with neighboring monomers within the homotrimer, and p.T430I slightly shifts the residue side chain away from the binding partner to reduce affinity.
(B) p.V78F (blue) stabilizes the conformation of SMAD5 away from DNA binding, reducing the possibility of steric hindrance and enabling higher affinity to DNA.
(C) p.N361D (raspberry) reduces the polar bonds directing its orientation away from co-factor, MAN1, binding, leading to a higher affinity to MAN1. The models were generated using PDB structures 6TBZ, 5ZOK, and AlphaFold2, and analyses were performed using PyMOL (version 2.5.4).
(D) Expression of SMAD5 mRNA in cultured HEK293T cells transiently transfected with wild-type (WT) SMAD5 or variant plasmids. n = 4; ∗p < 0.05; one-way ANOVA with Tukey’s post hoc test.
(E) Expression of SMAD5 protein in cultured HEK293T cells transiently co-transfected with FLAG-SMAD5 variant plasmids and GFP. Cells were lysed and assessed for levels of FLAG-SMAD5 variants by ELISA. FLAG-SMAD5 levels were normalized to GFP. n = 8–12; ∗∗p < 0.01; ∗∗∗∗p < 0.0001; one-way ANOVA with Tukey’s post hoc test.
(F) Protein stability of SMAD5 variants in cultured HEK293T cells co-transfected with FLAG-SMAD5 variants and GFP. Cells were treated with cycloheximide or vehicle for 8 h, lysed, and assessed for levels of FLAG-SMAD5 variants by ELISA. FLAG-SMAD5 levels were normalized to GFP. Protein levels of FLAG-SMAD5 variants are presented relative to their levels in vehicle-treated cell lysates. n = 8–12; ∗∗∗∗p < 0.0001; one-way ANOVA with Tukey’s post hoc test.
(G–I) Transcriptional activation ability of the SMAD5 variants p.T430I, p.V78F, and p.N361D was tested on a BMP-activated (Xvent2-luc) promoter (columns a–d) in HEK293T cells. Impact of the variants on WT-SMAD5 activation of the Xvent2-luc promoter was also assessed (columns e–h), where cells were transfected with 2× WT-SMAD5 or co-transfected with WT-SMAD5 and variant. Fold change was calculated by normalizing variant activity over vector-only activity. n = 4–5; ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; one-way ANOVA with Tukey’s post hoc test.
All data are presented as mean ± SD.
The de novo variant identified in F4.3, p.V78F, occurs at the DNA-binding interface of the SMAD5 MH1 domain (Figure 1C). This region is highly conserved (Figures 1D–1F) and is responsible for recognizing target DNA sequences, a process that is restricted by the spatial requirements of the MH1 domain when SMAD5 binds to DNA as a trimer.24 To model the SMAD5/DNA complex for analyses of p.V78F, an interaction model of SMAD5 with DNA was created using the SMAD5 AlphaFold2 structure and PDB structure 6TBZ. V78 did not directly contact DNA, and the change from V78 to F78 led to penetration of the residue further into the pocket lined by L76, I88, T89, A34, and A37 (Figure 2B). Formation of more hydrophobic, polar, and aromatic interactions through its larger, hydrophobic, side chain stabilized the side chain in this orientation (Figure 2B; Table S4). This reduced the possibility of steric hindrance and enabled higher-affinity binding of SMAD5 with DNA, suggesting that the p.V78F substitution may improve the spatial organization of the MH1 domain. We observed a significant increase in SMAD5 protein levels in SMAD5-V78F-transfected HEK293T cells, with no observed impact on protein stability (Figures 2D–2F). Activation of the Xvent2-luc promoter was reduced in cells transfected with SMAD5-V78F following BMP4+ treatment relative to WT-SMAD5 (Figure 2H, columns a–d). A similar reduction in promoter activation was observed in WT and variant co-transfected cells (Figure 2H, columns e–h). This reduction in activity suggests that although SMAD5-V78F may have higher affinity to DNA as a monomer, as predicted in silico, its ability to initiate transcription in vitro is compromised. These findings point to reduced SMAD5 transcriptional activity as a mechanism of p.V78F action.
The residue mutated in F5.3, p.N361, occurs within a conserved region of the SMAD5 MH2 domain (Figures 1C–1F), which in other R-SMADs has been observed to interact with co-factors.25 To assess the impact of p.N361D on SMAD5 co-factor interaction, we modeled the protein-protein interaction between SMAD5 and MAN1, which has been observed in vitro,26 using the available SMAD1:MAN1 structure (PDB: 5ZOK). The change in residue from p.N361 to p.D361 led to a loss of polar interactions that direct the protein backbone away from MAN1 within the WT structure, resulting in a higher affinity to MAN1 (Figure 2C; Table S4). There were significant reductions in SMAD5 transcript and protein levels and protein stability in SMAD5-N361D transfected cells relative to WT transfected cells (Figures 2D–2F). Activation of the Xvent2-luc promoter was not affected in cells transfected with SMAD5-N361D in basal or BMP4+ conditions relative to WT-SMAD5 (Figure 2I, columns a–d). However, transfection with SMAD5-N361D significantly reduced the promoter activation response of WT-SMAD5 in BMP4+treated cells when co-transfected (Figure 2I, columns e–h). The reduction in expression and stability of SMAD5-N361D suggests that the variant may be hypomorphic, and that the decrease in SMAD5 molecules available to trimerize may underlie the inability of WT-SMAD5 to activate the Xvent2-luc promoter in the presence of SMAD5-N361D.
Our findings suggest that functional haploinsufficiency is the disease mechanism for five out of six SMAD5 variants identified in our study. Whether by predicted loss of transcript, protein product, or protein function, SMAD5 haploinsufficiency leads to isolated congenital heart defects in the individuals who carry these variants. Population frequency data from the gnomAD reference population reveal that SMAD5 is under strict selection against LOF variation, exemplified by an LOEUF (observed/expected LOF variants) score of 0.174 and an LOF intolerance (pLI) score of 1.27 These values indicate that the loss of SMAD5 function is likely to be detrimental towards human survival. Similar observations have been made for individuals carrying LOF variants in other SMAD genes such as SMAD1 and SMAD4, both with pLI scores of 1, where haploinsufficiency has been linked to isolated CHD.10,28 In mice, Smad5 null embryos do not survive past mid-gestation.29,30,31,32 Homozygous null embryos fail to turn and die by embryonic day 11.0 due to amnion, gut, heart, craniofacial, and neural tube defects. Observed heart defects include ectopic or underdeveloped heart, abnormal heart looping, and misshapen dorsal aorta. Heterozygous embryos develop normally, suggesting that the level of SMAD5 required for normal development differs between human and mouse. It is plausible that in the majority of affected individuals in our cohort, the threshold requirement for SMAD5 functionality during heart development was not met by the action of the single functional SMAD5 allele, whereas it was sufficient for the normal development of other organs. This is not unique for SMAD5, however, and individuals with heterozygous, pathogenic variants in SMAD1, SMAD4, and SMAD6 develop congenital defects, while mice heterozygous for null alleles develop without defects.33,34,35 Given that Smad5 null embryos die at mid-gestation, additional heart phenotypes such as septal defects, the most prevalent type of cardiac lesion in our cohort, have not been observed in mice. This reduces our ability to understand the biological processes that are affected by SMAD5 haploinsufficiency during heart development. However, observations from studying BMP signaling during cardiac septation in mice show that the pathway is required for orchestrating the migration and differentiation of second heart field and cardiac neural crest cells that contribute to septum formation.36 Additionally, individuals with pathogenic variants in SMAD1, paralog of SMAD5 downstream of BMP ligands, frequently present with septal defects,10 signifying the necessity for SMAD signaling during human cardiac septation. Thus, the developmental consequences of disrupted BMP signaling appear consistent with the main phenotypic outcomes of our cohort.
F1.3 is the only individual with multiple congenital defects in our cohort. There were no variants identified in 770 genes (in house list) associated with CHD or CHD with extracardiac features. Functional assessment of the de novo SMAD5 variant identified in F1.3, p.T430I, showed dominant-negative activity. Attempts to produce a CRISPR knockin mouse model of this variant were unsuccessful (4 attempts, 109 embryos screened). However, a zebrafish model of this variant was created and studied previously. The orthologous missense variant in zebrafish (T429I, termed somitabun) disrupts dorsoventral patterning and results in severe embryo dorsalization via dominant-negative activity.37,38 The variant leads to severe developmental defects and lethality. Heart development is strongly impaired, with the heart tubes failing to join at the midline and resulting in two separate heart chambers that lack beating cells.38 This phenotype could be rescued by the addition of BMP4, similar to our luciferase assay observations, with BMP4 restoring the reduced promoter activation by SMAD5-T430I to wild-type levels in HEK293T cells (Figure 2G). The F1.3 variant, p.T430I, occurs in the L3 loop of the MH2 domain, which is highly intolerant to missense variation and is depleted of missense variants in the gnomAD reference population (Figures 1C and 1D). This region controls trimerization of SMADs and may also determine the interaction with membrane-bound receptors.17 Mutation of the paralogous residue in SMAD2, T432, to lysine or alanine, inhibits SMAD2 interaction with the TGF-β receptor.39 Thus, it is possible that p.T430I may also affect interaction with its receptors, which may contribute to its functional impact. This, however, does not account for the observed dominant-negative effects of the variant. If the main mechanism of p.T430I is loss of receptor interaction, then addition of WT-SMAD5 would be expected to rescue the impact on promoter transactivation. As this is not the case (Figure 2G), we expect that the principal effect of this variant may occur through disrupted trimerization. By disrupting trimerization, it is anticipated that p.T430I impacts the function of SMAD5 protein encoded by the remaining SMAD5 allele, resulting in a level of SMAD5 activity that is not sufficient to maintain SMAD5-mediated biological processes within the embryo. This then leads to a phenotypic outcome that approaches complete LOF of SMAD5 during human embryonic development, which, similar to observations in animal models, affects multiple organ systems and is incompatible with survival.
The identification of two individuals with HLH in our cohort suggests a potential role for SMAD5 in this severe form of CHD, which remains largely genetically unresolved. Studies in mice have shown that BMP signaling, through SMAD1/5, acts upstream of cardiac myosins MYH6 and MYH7, which are implicated in HLH, left ventricular non-compaction, and cardiomyopathies (MIM: 160710, 160760).40 Examination of heart tissue from patients with HLH have revealed changes in BMP signaling components, linking disruption of the pathway to formation of a hypoplastic left heart.41,42 Thus, acting downstream of BMP ligands, SMAD5 may regulate components whose functions are necessary for proper left ventricle formation.
In one of the HLH cases, F2, we observe incomplete penetrance and variable expressivity of a SMAD5 LOF variant. In this family, the proband (F2.3) is diagnosed with HLH, the brother (F2.5) with septal defects, and the mother (F2.2) appears to be an unaffected carrier of the SMAD5 frameshift variant p.A402Qfs∗13. Variable expressivity has been reported for familial genetic cases of CHD, and co-occurrence of malformations within families has been observed to be highly variable.43 In previous studies of familial cases of HLH, echocardiography of 'unaffected' parents, who had not reported or suspected a heart defect, revealed minor cardiac defects in 12% of examined individuals.44,45,46 Since the mother (F2.2) could not be assessed for cardiac malformation, it is not possible to assess penetrance of this SMAD5 LOF variant.
Of the seven CHD-affected individuals in our cohort carrying SMAD5 variants, five are surviving. This enables consideration of post-natal onset phenotypes that may be associated with carrying a pathogenic SMAD5 variant. However, we are limited by a small cohort as well as our current understanding of the roles SMAD5 plays during human development. Within our cohort, F6.2 was recruited as an adult survivor of CHD, who was diagnosed with adult-onset epilepsy, skin cancer, and sebaceous adenoma. Although we currently do not have evidence to link the SMAD5 variant to the proband’s adult-onset phenotypes, there are tentative links in the literature that connect non-canonical SMAD5 signaling to several types of cancers (lung and breast).47,48 Additionally, disruption of SMAD5 function in human bone marrow cells has been linked to leukemogenesis, where SMAD5 is involved in TGF-β-mediated inhibition of hematopoietic progenitor proliferation.49 A recent publication has identified SMAD5 variants in two patients with pulmonary arterial hypertension (PAH).50 The reported individuals were diagnosed with PAH at 29 and 42 years of age, with one individual also presenting with a ventricular septal defect. Furthermore, BMP-SMAD1/5/8 signaling has been linked to several other respiratory diseases, including chronic obstructive pulmonary disease and pulmonary fibrosis.51 Therefore, longitudinal monitoring of the surviving individuals as well as identification of SMAD5 variants in other disease cohorts may clarify a role for SMAD5 variants in post-natal disease and may provide a rationale for monitoring individuals with SMAD5 variants for development of further disease phenotypes.
We present compelling clinical, genetic, and molecular evidence that congenital defects can be caused by heterozygous SMAD5 variants through haploinsufficient or dominant-negative mechanisms. These variants were identified in a cohort primarily diagnosed with isolated CHD, with septal defects or HLH observed in multiple individuals. This work further expands on the role played by the TGF-β superfamily in human development and reinforces its significance in human congenital disease. Larger numbers of affected individuals will be required to reveal the full spectrum of developmental and post-natal impacts of SMAD5 variants and to define further mechanisms of variant pathogenicity.
Data and code availability
All SMAD5 variants reported in the manuscript have been deposited to ClinVar with reserved accessions (ClinVar: SCV005849048–SCV005849053). The patient genotype data for F4 and F5 are available via the PCGC study (dbGaP: phs001194.v4.p3). Genotype data for the other cases have not been deposited to a public repository due to consent restrictions, but can be made available from the lead author upon reasonable request.
Consortia
Congenital Heart Disease Synergy Study Group members: Sally L. Dunwoodie, David S. Winlaw, Eleni Giannoulatou, Natasha Nassar, Edwin P. Kirk, Gavin Chapman, Gillian M. Blue, Samantha Lain, and Gary Sholler.
Australian Genomics Cardiovascular Genomics Disorders Flagship members: Lesley Ades, Mohammad Al-Shinnag, John J. Atherton, Rachel Austin, Richard D. Bagnall, Chris Barnett, Gillian M. Blue, Simon Bodek, Kirsten Boggs, Michael Bogwitz, Tiffany Boughtwood, Alessandra Bray, Marie-Jo Brion, Jaye Brown, Rob Bryson Richardson, Charlotte Burns, Michelle Cao, Sarah Casauria, Heather Chalinor, Yuchen Chang, Gavin Chapman, Belinda Chong, Felicity Collins, Gemma Correnti, Kathy Cox, Fiona Cunningham, Debjani Das, Andrew Davis, Jason Davis, Paul De Fazio, Sophie Devery, Sally L. Dunwoodie, Nathan Dwyer, Stefanie Elbracht-Leong, David Elliott, Annabelle Enriquez, Diane Fatkin, Miriam Fine, Keri Finlay, Denisse Garza, Eleni Giannoulatou, Laura Gongolidis, Belinda Gray, Cassie Greer, Eric Haan, Mathilda Haas, Bernadette Hanna, Richard Harvey, Janette Hayward, Carmen Herrera, Georgie Hollingsworth, Ari E. Horton, Jodie Ingles, Joanne Isbister, Matilda Jackson, Paul James, Sarah Jane-Pantaleo, Renee Johnson, Andrew Kelly, Edwin Kirk, Jonathon Lipton, Sebastian Lunke, Ivan Macciocca, Paul MacIntyre, Evanthia O. Madelli, Amali Mallawaarachchi, Julia Mansour, Ellenore Martin, Jacob Mathew, Tessa Mattiske, Julie McGaughran, Alison McLean, Caroline Medi, Alejandro Metke, Di Milnes, Michael Milward, Ansley Morrish, Jim Morwood, Helen Mountain, David Mowat, Natalie Nowak, Noelia Nunez Martinez, Sinead O’Sullivan, Angela Overkov, Nicholas Pachter, Chirag Patel, Mark Perrin, Andreas Pflaumer, Rachel Pope-Couston, Nicola K. Poplawski, Preeti Punni, Michael C.J. Quinn, Sulekha Rajagopalan, Hariharan Raju, Emma M. Rath, Matthew Regan, Jonathan Rogers, Mark Ryan, Sarah Sandaradura, Paul Scuffham, Chris Semsarian, Isabella Sherburn, Mary-Clare Sherlock, Michelle de Silva, Emma Singer, Magdalena Soka, Carla Smerdon, Janine Smith, Renee Smyth, Zornitza Stark, Raymond Sy, Jessica Taylor, Shelby Taylor, Michel Tchan, Tina Thompson, Alison Trainer, Giulia Valente, Karin van Spaendonck-Zwarts, Kunal Verma, Miranda Vidgen, Jitendra Vohra, Kathryn Waddel-Smith, Mathew Wallis, Robert G. Weintraub, Meredith Wilson, David Winlaw, Lisa Worgan, Linda Wornham, Kathy Wu, Laura Yeates, and Dominica Zentner.
Acknowledgments
This research was funded by the Australian National Health and Medical Research Council (NHMRC) Synergy Grant (ID1181325, to S.L.D., D.S.W., E.G., and E.P.K.), Australian Genomics NHMRC Grants (GNT1113531 and GNT2000001), Australian Genomics Cardiovascular Genetic Disorders Flagship supported by the Medical Research Future Fund (EPCD000028), the NSW Health Cardiovascular Research Capacity Program Senior Researcher Grant (to S.L.D. and R.P.H.), and the Australian Medical Research Future Fund Phenomics Translation Initiative (to G.C. and S.L.D.), and with philanthropic support from the Kelley Foundation (to S.L.D.) and the Cornish Foundation (to D.S.W. and S.L.D.). S.L.D. was supported by NHMRC Fellowship and Investigator grants (ID1135886 and ID2007896). R.P.H. was supported by NHMRC Investigator grant no. ID2008743 and Australian Research Council Discovery Project grant no. DP210102134. E.G. was supported by NHMRC Investigator grant no. ID2018360. W.K.C. was supported by NIH U01 HL153009. G.M.B. was supported by an NSW Health Cardiovascular Research Capacity Program Early-Mid Career Researcher grant and an NSW CVRN Career Advancement grant. The authors thank the patients and their families and Desiree Hilton, Bridget O’Malley, Krzysztof Bernatowicz, Kathryn E. Waddell-Smith, Ilse Meerschaut, and Preeti Punni for their efforts in patient recruitment and management. We thank Dr. Justin Szot for critical assessment of the manuscript. We acknowledge support from the Victor Chang Cardiac Research Institute Innovation Center, funded by the NSW government. Figures S1 and S3 and the graphical abstract were created in BioRender.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2025.100478.
Contributor Information
Dimuthu Alankarage, Email: d.alankarage@victorchang.edu.au.
Sally L. Dunwoodie, Email: s.dunwoodie@victorchang.edu.au.
Web resources
CHDgene website: Chdgene.victorchang.edu.au
Supplemental information
References
- 1.van der Linde D., Konings E.E.M., Slager M.A., Witsenburg M., Helbing W.A., Takkenberg J.J.M., Roos-Hesselink J.W. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J. Am. Coll. Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Morrish A.M., Smith J., Enriquez A., Sholler G.F., Mervis J., Dunwoodie S.L., Kirk E.P., Winlaw D.S., Blue G.M. A new era of genetic testing in congenital heart disease: A review. Trends Cardiovasc. Med. 2022;32:311–319. doi: 10.1016/j.tcm.2021.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Yang A., Alankarage D., Cuny H., Ip E.K.K., Almog M., Lu J., Das D., Enriquez A., Szot J.O., Humphreys D.T., et al. CHDgene: A Curated Database for Congenital Heart Disease Genes. Circ. Genom. Precis. Med. 2022;15 doi: 10.1161/circgen.121.003539. [DOI] [PubMed] [Google Scholar]
- 4.Morton S.U., Quiat D., Seidman J.G., Seidman C.E. Genomic frontiers in congenital heart disease. Nat. Rev. Cardiol. 2022;19:26–42. doi: 10.1038/s41569-021-00587-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dellborg M., Giang K.W., Eriksson P., Liden H., Fedchenko M., Ahnfelt A., Rosengren A., Mandalenakis Z. Adults With Congenital Heart Disease: Trends in Event-Free Survival Past Middle Age. Circulation. 2023;147:930–938. doi: 10.1161/circulationaha.122.060834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Massagué J., Sheppard D. TGF-β signaling in health and disease. Cell. 2023;186:4007–4037. doi: 10.1016/j.cell.2023.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kosaki R., Gebbia M., Kosaki K., Lewin M., Bowers P., Towbin J.A., Casey B. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am. J. Med. Genet. 1999;82:70–76. doi: 10.1002/(Sici)1096-8628(19990101)82:1<70::Aid-Ajmg14>3.0.Co;2-Y. [DOI] [PubMed] [Google Scholar]
- 8.Tan T.Y., Gonzaga-Jauregui C., Bhoj E.J., Strauss K.A., Brigatti K., Puffenberger E., Li D., Xie L., Das N., Skubas I., et al. Monoallelic BMP2 Variants Predicted to Result in Haploinsufficiency Cause Craniofacial, Skeletal, and Cardiac Features Overlapping Those of 20p12 Deletions. Am. J. Hum. Genet. 2017;101:985–994. doi: 10.1016/j.ajhg.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamford R.N., Roessler E., Burdine R.D., Saplakoğlu U., dela Cruz J., Splitt M., Goodship J.A., Towbin J., Bowers P., Ferrero G.B., et al. Loss-of-function mutations in the EGF-CFC gene CFC1 are associated with human left-right laterality defects. Nat. Genet. 2000;26:365–369. doi: 10.1038/81695. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z., Qiao X.H., Xu Y.J., Liu X.Y., Huang R.T., Xue S., Qiu H.Y., Yang Y.Q. SMAD1 Loss-of-Function Variant Responsible for Congenital Heart Disease. BioMed Res. Int. 2022;2022 doi: 10.1155/2022/9916325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granadillo J.L., Chung W.K., Hecht L., Corsten-Janssen N., Wegner D., Nij Bijvank S.W.A., Toler T.L., Pineda-Alvarez D.E., Douglas G., Murphy J.J., et al. Variable cardiovascular phenotypes associated with SMAD2 pathogenic variants. Hum. Mutat. 2018;39:1875–1884. doi: 10.1002/humu.23627. [DOI] [PubMed] [Google Scholar]
- 12.Le Goff C., Mahaut C., Abhyankar A., Le Goff W., Serre V., Afenjar A., Destrée A., di Rocco M., Héron D., Jacquemont S., et al. Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat. Genet. 2011;44:85–88. doi: 10.1038/ng.1016. [DOI] [PubMed] [Google Scholar]
- 13.Musfee F.I., Guo D., Pinard A.C., Hostetler E.M., Blue E.E., Nickerson D.A., University of Washington Center for Mendelian Genomics UW-CMG. Bamshad M.J., Milewicz D.M., Prakash S.K. Rare deleterious variants of NOTCH1, GATA4, SMAD6, and ROBO4 are enriched in BAV with early onset complications but not in BAV with heritable thoracic aortic disease. Mol. Genet. Genomic Med. 2020;8 doi: 10.1002/mgg3.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross S., Hill C.S. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pediatric Cardiac Genomics Consortium, Gelb B., Brueckner M., Chung W., Goldmuntz E., Kaltman J., Kaski J.P., Kim R., Kline J., Mercer-Rosa L., et al. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circ. Res. 2013;112:698–706. doi: 10.1161/circresaha.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chacko B.M., Qin B.Y., Tiwari A., Shi G., Lam S., Hayward L.J., De Caestecker M., Lin K. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol. Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Kawabata M., Inoue H., Hanyu A., Imamura T., Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H.S., Park M.J., Lee S.Y., Hwang Y.S., Lee H., Roh D.H., Kim J.I., Park J.B., Lee J.Y., Kung H.F., Kim J. Transcriptional regulation of Xbr-1a/Xvent-2 homeobox gene: analysis of its promoter region. Biochem. Biophys. Res. Commun. 2002;298:815–823. doi: 10.1016/s0006-291x(02)02570-6. [DOI] [PubMed] [Google Scholar]
- 21.Onichtchouk D., Gawantka V., Dosch R., Delius H., Hirschfeld K., Blumenstock C., Niehrs C. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling [correction of controling] dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki A., Chang C., Yingling J.M., Wang X.F., Hemmati-Brivanlou A. Smad5 induces ventral fates in Xenopus embryo. Dev. Biol. 1997;184:402–405. doi: 10.1006/dbio.1997.8548. [DOI] [PubMed] [Google Scholar]
- 23.Henningfeld K.A., Rastegar S., Adler G., Knöchel W. Smad1 and Smad4 are components of the bone morphogenetic protein-4 (BMP-4)-induced transcription complex of the Xvent-2B promoter. J. Biol. Chem. 2000;275:21827–21835. doi: 10.1074/jbc.M000978200. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz L., Kaczmarska Z., Gomes T., Aragon E., Torner C., Freier R., Baginski B., Martin-Malpartida P., de Martin Garrido N., Marquez J.A., et al. Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes. Comput. Struct. Biotechnol. J. 2021;19:632–646. doi: 10.1016/j.csbj.2020.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazono K.I., Ohno Y., Wada H., Ito T., Fukatsu Y., Kurisaki A., Asashima M., Tanokura M. Structural basis for receptor-regulated SMAD recognition by MAN1. Nucleic Acids Res. 2018;46:12139–12153. doi: 10.1093/nar/gky925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan D., Estévez-Salmerón L.D., Stroschein S.L., Zhu X., He J., Zhou S., Luo K. The integral inner nuclear membrane protein MAN1 physically interacts with the R-Smad proteins to repress signaling by the transforming growth factor-{beta} superfamily of cytokines. J. Biol. Chem. 2005;280:15992–16001. doi: 10.1074/jbc.M411234200. [DOI] [PubMed] [Google Scholar]
- 27.Chen S., Francioli L.C., Goodrich J.K., Collins R.L., Kanai M., Wang Q., Alföldi J., Watts N.A., Vittal C., Gauthier L.D., et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2024;625:92–100. doi: 10.1038/s41586-023-06045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., Xu Y.J., Yang C.X., Huang R.T., Xue S., Yuan F., Yang Y.Q. SMAD4 loss-of-function mutation predisposes to congenital heart disease. Eur. J. Med. Genet. 2023;66 doi: 10.1016/j.ejmg.2022.104677. [DOI] [PubMed] [Google Scholar]
- 29.Chang H., Huylebroeck D., Verschueren K., Guo Q., Matzuk M.M., Zwijsen A. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development. 1999;126:1631–1642. doi: 10.1242/dev.126.8.1631. [DOI] [PubMed] [Google Scholar]
- 30.Umans L., Vermeire L., Francis A., Chang H., Huylebroeck D., Zwijsen A. Generation of a floxed allele of Smad5 for cre-mediated conditional knockout in the mouse. Genesis. 2003;37:5–11. doi: 10.1002/gene.10219. [DOI] [PubMed] [Google Scholar]
- 31.Yang X., Castilla L.H., Xu X., Li C., Gotay J., Weinstein M., Liu P.P., Deng C.X. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development. 1999;126:1571–1580. doi: 10.1242/dev.126.8.1571. [DOI] [PubMed] [Google Scholar]
- 32.Chang H., Zwijsen A., Vogel H., Huylebroeck D., Matzuk M.M. Smad5 is essential for left-right asymmetry in mice. Dev. Biol. 2000;219:71–78. doi: 10.1006/dbio.1999.9594. [DOI] [PubMed] [Google Scholar]
- 33.Galvin K.M., Donovan M.J., Lynch C.A., Meyer R.I., Paul R.J., Lorenz J.N., Fairchild-Huntress V., Dixon K.L., Dunmore J.H., Gimbrone M.A., Jr., et al. A role for smad6 in development and homeostasis of the cardiovascular system. Nat. Genet. 2000;24:171–174. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- 34.Tremblay K.D., Dunn N.R., Robertson E.J. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- 35.Sirard C., de la Pompa J.L., Elia A., Itie A., Mirtsos C., Cheung A., Hahn S., Wakeham A., Schwartz L., Kern S.E., et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCulley D.J., Kang J.O., Martin J.F., Black B.L. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev. Dyn. 2008;237:3200–3209. doi: 10.1002/dvdy.21743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hild M., Dick A., Rauch G.J., Meier A., Bouwmeester T., Haffter P., Hammerschmidt M. The smad5 mutation somitabun blocks Bmp2b signaling during early dorsoventral patterning of the zebrafish embryo. Development. 1999;126:2149–2159. doi: 10.1242/dev.126.10.2149. [DOI] [PubMed] [Google Scholar]
- 38.Mullins M.C., Hammerschmidt M., Kane D.A., Odenthal J., Brand M., van Eeden F.J., Furutani-Seiki M., Granato M., Haffter P., Heisenberg C.P., et al. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development. 1996;123:81–93. doi: 10.1242/dev.123.1.81. [DOI] [PubMed] [Google Scholar]
- 39.Lo R.S., Chen Y.G., Shi Y., Pavletich N.P., Massagué J. The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF-beta receptors. EMBO J. 1998;17:996–1005. doi: 10.1093/emboj/17.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J., Greene S.B., Bonilla-Claudio M., Tao Y., Zhang J., Bai Y., Huang Z., Black B.L., Wang F., Martin J.F. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev. Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miao Y., Tian L., Martin M., Paige S.L., Galdos F.X., Li J., Klein A., Zhang H., Ma N., Wei Y., et al. Intrinsic Endocardial Defects Contribute to Hypoplastic Left Heart Syndrome. Cell Stem Cell. 2020;27:574–589.e8. doi: 10.1016/j.stem.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricci M., Mohapatra B., Urbiztondo A., Birusingh R.J., Morgado M., Rodriguez M.M., Lincoln J., Vatta M. Differential changes in TGF-β/BMP signaling pathway in the right ventricular myocardium of newborns with hypoplastic left heart syndrome. J. Card. Fail. 2010;16:628–634. doi: 10.1016/j.cardfail.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Ellesøe S.G., Workman C.T., Bouvagnet P., Loffredo C.A., McBride K.L., Hinton R.B., van Engelen K., Gertsen E.C., Mulder B.J.M., Postma A.V., et al. Familial co-occurrence of congenital heart defects follows distinct patterns. Eur. Heart J. 2018;39:1015–1022. doi: 10.1093/eurheartj/ehx314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brenner J.I., Berg K.A., Schneider D.S., Clark E.B., Boughman J.A. Cardiac malformations in relatives of infants with hypoplastic left-heart syndrome. Am. J. Dis. Child. 1989;143:1492–1494. doi: 10.1001/archpedi.1989.02150240114030. [DOI] [PubMed] [Google Scholar]
- 45.Theis J.L., Hu J.J., Sundsbak R.S., Evans J.M., Bamlet W.R., Qureshi M.Y., O'Leary P.W., Olson T.M. Genetic Association Between Hypoplastic Left Heart Syndrome and Cardiomyopathies. Circ. Genom. Precis. Med. 2021;14 doi: 10.1161/circgen.120.003126. [DOI] [PubMed] [Google Scholar]
- 46.Tomita-Mitchell A., Stamm K.D., Mahnke D.K., Kim M.S., Hidestrand P.M., Liang H.L., Goetsch M.A., Hidestrand M., Simpson P., Pelech A.N., et al. Impact of MYH6 variants in hypoplastic left heart syndrome. Physiol. Genomics. 2016;48:912–921. doi: 10.1152/physiolgenomics.00091.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z., Wang J., Zeng X., Li D., Ding M., Guan R., Yuan L., Zhou Q., Guo M., Xiong M., et al. Two-stage study of lung cancer risk modification by a functional variant in the 3'-untranslated region of SMAD5 based on the bone morphogenetic protein pathway. Mol. Clin. Oncol. 2018;8:38–46. doi: 10.3892/mco.2017.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opyrchal M., Salisbury J.L., Zhang S., McCubrey J., Hawse J., Goetz M.P., Lomberk G.A., Haddad T., Degnim A., Lange C., et al. Aurora-A mitotic kinase induces endocrine resistance through down-regulation of ERα expression in initially ERα+ breast cancer cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruno E., Horrigan S.K., Van Den Berg D., Rozler E., Fitting P.R., Moss S.T., Westbrook C., Hoffman R. The Smad5 gene is involved in the intracellular signaling pathways that mediate the inhibitory effects of transforming growth factor-beta on human hematopoiesis. Blood. 1998;91:1917–1923. [PubMed] [Google Scholar]
- 50.Cao D., Grünig E., Sirenko Y., Radchenko G., Gall H., Ahmed A., Theiß S., Lankeit M., Meder B., Laugsch M., Eichstaedt C.A. SMAD5 as a novel gene for familial pulmonary arterial hypertension. Clin. Sci. 2025;139:15–27. doi: 10.1042/cs20241340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Z., Zhuang J., He L., Zhu S., Kong W., Lu W., Zhang Z. Exploring Smad5: a review to pave the way for a deeper understanding of the pathobiology of common respiratory diseases. Mol. Med. 2024;30:225. doi: 10.1186/s10020-024-00961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All SMAD5 variants reported in the manuscript have been deposited to ClinVar with reserved accessions (ClinVar: SCV005849048–SCV005849053). The patient genotype data for F4 and F5 are available via the PCGC study (dbGaP: phs001194.v4.p3). Genotype data for the other cases have not been deposited to a public repository due to consent restrictions, but can be made available from the lead author upon reasonable request.