Abstract
We have examined the effect of neutrophil concentration on killing of a clinical isolate of Staphylococcus epidermidis. Human neutrophils at concentrations varying from 105 to 107 per ml were mixed in suspension with S. epidermidis at concentrations varying from 103 to 108 colony-forming units/ml, and the concentration of viable bacteria was assayed after various times at 37°C. The rate of bacterial killing depended on the concentration of neutrophils and not on the ratio of neutrophils to bacteria. Below a critical concentration of neutrophils, bacteria growth was greater than neutrophil killing of bacteria even when the ratio of neutrophils to bacteria was 100:1. We fitted the time course of bacterial concentration and its dependence on neutrophil concentration with an exponential function, the exponent of which is (−kp + g)t, where k is the second-order rate constant for bacterial killing, p is the neutrophil concentration, g is the first-order rate constant for bacterial growth, and t is time. We found that k ≈ 2 × 10−8 ml per neutrophil per min, and g ≈ 8 × 10−3/min. Only when p is greater than g/k, which we call the critical neutrophil concentration, does the bacterial concentration fall. Under optimal assay conditions, the critical neutrophil concentration was 3–4 × 105 per ml, a value very close to that (≤5 × 105 per ml) known to predispose humans to bacterial and fungal infections.
Neutrophils are the first line of defense against bacteria that invade tissues and blood. These cells differentiate in the bone marrow, circulate in the blood for 8–12 h, and then enter the tissues where they function for 2–5 days before dying. They kill bacteria in blood or interstitial fluid by phagocytosing them, thereby exposing the ingested bacteria to a variety of potent bactericidal proteins and oxidizing agents.
Neutrophils are the predominant white blood cells in blood, accounting for ≈60% of the total leukocyte pool. The concentration of neutrophils in the blood of healthy humans ranges from ≈3 to 6 million cells per ml. In humans experiencing a bacterial infection (e.g., appendicitis), the concentration of neutrophils in blood may rise acutely to 15–40 million per ml. Conversely, under conditions of bone-marrow aplasia, the concentration of neutrophils in blood may fall more than 10-fold to 0.1–0.5 million per ml and show little or no increase in response to bacterial infections.
Maintaining neutrophil concentration at physiological level is of primary importance to host defense. The host becomes predisposed to life-threatening bacterial and fungal infections when the blood neutrophil concentration falls below 5 × 105 per ml, a condition called neutropenia (1–3). Neutropenia occurs spontaneously in newborn infants and is a commonly recognized consequence of HIV infection (4) or treatment with immunosuppressive or cytotoxic drugs. Restoring the neutrophil concentration such as through neutrophil transfusion has been used successfully in the treatment of neutropenic patients. For example, over 70% of neutropenic patients who developed bacterial infections while undergoing stem-cell transplantation resolved these infections after neutrophil transfusions that restored their blood neutrophil concentration to ≈2 × 106 per ml (5).
These observations indicate that microbicidal activities of neutrophils are compromised when blood neutrophil concentrations fall below ≈5 × 105 per ml. The reason, however, has remained unknown.
The efficiency of neutrophil microbicidal activity has been studied primarily (6–8) at physiological neutrophil concentrations (3–6 × 106 per ml) and has been reported to depend on the ratio of neutrophils to bacteria (6, 8). At neutrophil concentrations of 3–6 × 106 per ml and in the presence of appropriate opsonins (e.g., IgG and complement), each neutrophil can ingest up to 100 Staphylococcus aureus or Escherichia coli in 30 min (6, 8). During bacteremia, the concentration of bacteria in blood rarely exceeds 103 per ml (9, 10). At this concentration of bacteria, even if the blood contains only 1–5 × 105 neutrophils per ml, the ratio of neutrophils to bacteria would be 100:1–500:1. Thus, if the ratio of neutrophils to bacteria were the principal factor determining the efficiency of killing, a blood neutrophil concentration of 1–5 × 105 per ml still would be sufficient to control growth of ≈ 103 bacteria per ml; but clinical evidence indicates that it is not.
This paper address the questions of why host defense against bacterial and fungal infections is compromised when neutrophil concentrations fall below physiological level and why ≈5 × 105 per ml is a critical threshold value. We hypothesize that the rate of bacterial killing by neutrophils is determined by the neutrophil concentration and not by the ratio of neutrophils to bacteria. To test this hypothesis, we measured neutrophil killing of a clinical isolate of Staphylococcus epidermidis (strain H753) at concentrations varying from 103 colony-forming units (cfu)/ml to 108 cfu/ml in suspension with neutrophil concentrations varying from 105 to 107 per ml.
Methods
S. Epidermidis.
S. epidermidis H753, a clinical isolate from the cerebrospinal fluid of a patient with an infected cerebrospinal fluid shunt, was provided by the Diagnostic Microbiology Laboratory at New York-Presbyterian Hospital (New York). For experiments, S. epidermidis H753 was streaked onto 3% trypticase soy broth (TSB) agar plates, and the plates were incubated at 37°C for 10 h. Colonies were harvested, suspended, and washed three times in PBS (Dulbecco's PBS with Ca2+ and Mg2+), and the absorbance at 600 nm of the final suspension was measured. The number of viable bacteria (in cfu) in this suspension was determined by reference to a previously determined growth curve relating the cfu of S. epidermidis H753 to A600.
Human Serum.
Human serum was derived from AB plasma provided by Columbia Presbyterian Medical Center Transfusion Service (New York). In brief, citrated plasma was restored to physiological Ca2+ concentration by the addition of CaCl2 and clotted at room temperature by the addition of thrombin to a final concentration of 1 unit/ml. Clots were removed by centrifugation. Serum was sterilized by filtration through a 0.22-μm filter and stored at −80°C.
Human Neutrophils.
Neutrophils were prepared as described (11). The purity of neutrophils was >95% as determined by Wright–Giemsa staining. Purified neutrophils were resuspended in PBS supplemented with 5 mM glucose and 0.1% human serum albumin (PBS-G-HSA).
Neutrophil Bacterial Killing in Suspension.
Killing of S. epidermidis was assayed as described (7) with modifications. In brief, 500 μl of PBS-G-HSA containing neutrophils (105–107 per ml), S. epidermidis (103–108 cfu/ml), and human serum [10, 40, or 75% (vol/vol)] was placed in sterile 1.5-ml Eppendorf tubes (Continental Lab Products, San Diego). The tubes were capped and incubated at 37°C on an Orbit Environ-shaker (Lab-Line Instruments) rotating at 200 rpm. Control samples containing 103–108 cfu/ml S. epidermidis and human serum (10, 40, or 75%) were incubated in parallel. Samples were prepared in duplicate. The number of viable bacteria in these suspensions was determined using a pour-plate method as follows. After 90 min of incubation, a 350-μl aliquot of each sample was diluted in sterile distilled water at pH 11, incubated for 5 min at 37°C to lyse neutrophils (12), and serially diluted in PBS. The dilutions were mixed in Petri dishes with 12 ml of liquid TSB agar (3% TSB/1.5% Bacto-agar) at 45°C. The plates were incubated overnight at 37°C, and the colonies were counted manually. We obtained identical results by plating serial dilutions on TSB agar at 37°C.
To determine the time course of killing, 500-μl aliquots of PBS-G-HSA containing 1, 2, or 4 × 106 neutrophils per ml, ≈1 × 105 S. epidermidis per ml, and 10% human serum (vol/vol) were incubated as described above. Controls containing 10% human serum and 1 × 105 per ml S. epidermidis were incubated in parallel. A separate tube was used for each time point. At the indicated times, the number of cfu was determined as described above.
Results and Discussion
The Effect of Neutrophil Concentration on Bacterial Killing in Suspension.
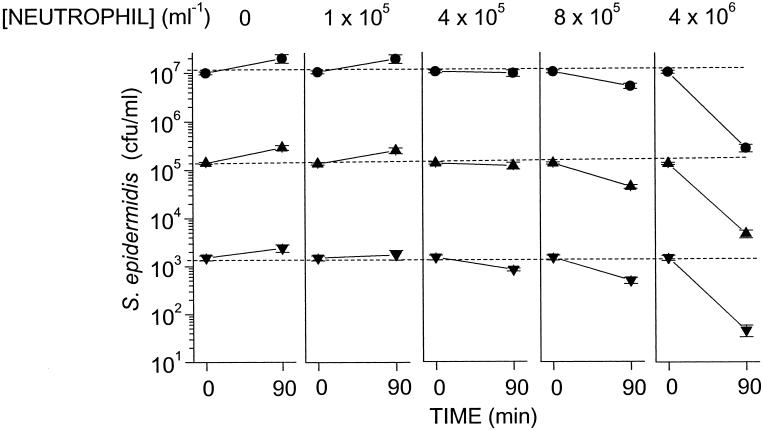
Previous studies (13) showed that incubation of S. epidermidis H753 with human serum opsonized these bacteria with IgG and the C3 component of complement, and that opsonization with both IgG and C3 is required for killing in suspension by neutrophils. We used this system to assess whether killing of S. epidermidis by neutrophils depends on the neutrophil concentration or the ratio of neutrophils to bacteria. We incubated neutrophils at concentrations varying from ≈1 × 105 per ml to ≈1 × 107 per ml with concentrations of S. epidermidis varying from ≈1 × 103 per ml to ≈1 × 107 per ml. These mixtures yielded ratios of neutrophils to S. epidermidis varying from 1:100 to 10,000:1. We found that after 90 min of incubation of neutrophils at concentrations of 8 × 105 per ml and higher with any of these concentrations of bacteria, there was a significant reduction in bacterial concentration (Fig. 1). For example, at 4 × 106 per ml neutrophils, bacterial concentrations were reduced by ≈90% after a 90-min incubation. In contrast, at neutrophil concentrations of 4 × 105 per ml or less, there was no significant reduction in bacterial concentration even though the ratio of neutrophils to bacteria was as high as 100:1 (Fig. 1, 1 × 105 neutrophils per ml vs. 103 cfu/ml bacteria).
Figure 1.
Effects of neutrophil concentration and the ratio of neutrophils/bacteria on S. epidermidis. Human neutrophils were mixed continuously in suspension with S. epidermidis in PBS-G-HSA buffer containing 75% human serum at 37°C. Shown are the concentrations of viable bacteria at time 0 and at 90 min in suspensions containing the indicated concentrations of neutrophils. Not shown are data for neutrophils per ml. The data represent the means and SEM of four independent experiments.
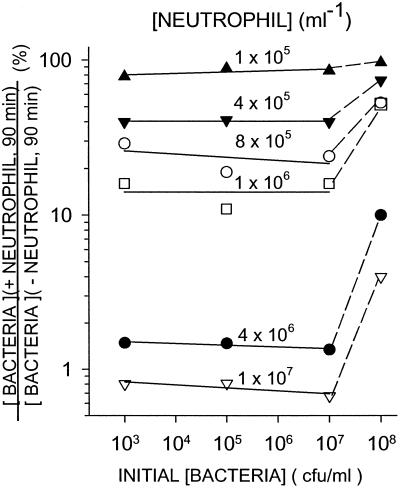
In the absence of neutrophils, S. epidermidis H753 grew in the medium (Fig. 1). Thus, in the presence of neutrophils, the bacterial concentration after 90 min of incubation is the net result of bacterial growth and neutrophil killing. To assess the extent of neutrophil killing of bacteria during the 90-min incubation, bacterial growth must be taken into account. The bacterial concentration after 90 min in the presence of neutrophils divided by the bacterial concentration after 90 min in the absence of neutrophils depends only on neutrophil concentration and is independent of initial bacterial concentrations between 103 and 107 cfu/ml (Fig. 2). For example, neutrophils at a concentration of 4 × 105 per ml killed ≈60% of the bacteria at all bacterial concentrations between 103 to 107 cfu/ml tested (Fig. 2). Nonetheless, they were incapable of killing the bacteria faster than the bacteria were replicating. For this reason, neutrophils at 4 × 105 per ml or less did not reduce significantly the concentration of bacteria in the mixture after 90 min of incubation below that present at time 0 (Fig. 1).
Figure 2.
Viable S. epidermidis recovered after 90 min of incubation with varying concentrations of neutrophils as a fraction of viable S. epidermidis recovered after a 90-min incubation without neutrophils. Human neutrophils were mixed continuously in suspension with S. epidermidis in buffer containing 75% human serum at 37°C. The data represent the means of four independent experiments.
In contrast, at a bacterial concentration in excess of 107 per ml, the killing efficiency of the neutrophils decreased, which is likely because of a combination of the saturation of neutrophil-killing capacity and toxic factors released by the bacteria.
Killing of S. epidermidis at all neutrophil concentrations was blocked by cytochalasin D (data not shown), confirming (14) that killing depends on phagocytosis and not on microbicidal substances secreted into the medium by neutrophils.
A Mathematical Model That Describes Bacterial Killing as a Function of Neutrophil Concentration and Time.
Bacterial killing in suspension has been described as an exponential function of time by Leijh et al. (15) and Hampton and Winterbourn (16). In neither of their models, however, was neutrophil concentration a variable. In contrast, we find that neutrophil concentration is a critical variable in determining the number of bacteria killed (Figs. 1 and 2). A model incorporating the role of neutrophil concentration is as follows.
We assume that neutrophils kill bacteria in a second-order collisional process, in which the neutrophils are not consumed, i.e.,
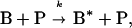 |
1 |
where k is second-order rate constant, B is a bacterium, B* is a killed bacterium, and P is a neutrophil. At the same time, the bacteria are replicating in a first-order reaction characterized by the first-order rate constant, g, i.e.,
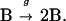 |
2 |
The change in the concentration of viable bacteria (b) with time (t) is
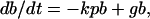 |
3 |
where p (neutrophil concentration) is assumed not to change.
We obtain
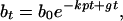 |
4 |
where bt is the concentration of viable bacteria after incubation time t, and b0 is the initial concentration of viable bacteria.
Eq. 4 can be expressed also with t factored out,
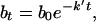 |
5 |
where
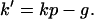 |
6 |
Similarly to equations reported by others (15, 16), Eq. 5 describes bacterial concentration as a function of time. The rate constant k′, however, is a pseudo first-order rate constant that depends on neutrophil concentration, p (Eq. 6).
The Rate Constant k for Neutrophil Killing of S. epidermidis in Suspension.
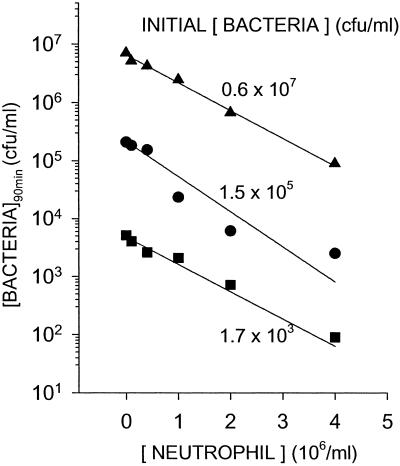
We mixed neutrophils at different concentrations with S. epidermidis in medium containing 10% human serum and determined the concentration of viable bacteria after 90 min. We found that the bacterial concentration decreased exponentially as a function of neutrophil concentration (Fig. 3). Fitting Eq. 4 to these data yields values for k and g. As shown in Table 1, k was nearly constant over a 10,000-fold range in the initial bacterial concentration. The maximum values of k (≈2.5 × 10−8 ml per neutrophil per min) were obtained in the presence of a 40% or higher concentration of serum. The value of g varied with bacterial concentration only in 10% serum but was constant at ≈8 × 10−3 per min in 40 and 75% serum (Table 1).
Figure 3.
Determination of the rate constants of bacterial killing (k) and growth (g) at varying neutrophil concentrations. Human neutrophils were mixed continuously in suspension with S. epidermidis in buffer containing 10% human serum at 37°C. The symbols refer to the number of viable bacteria recovered after a 90-min incubation with the indicated concentration of neutrophils (b90 min). Solid lines indicate the curves fitted to these data by using Eq. 4 (R2 = 0.96–0.98). Nonlinear least-squares fits were carried out in SIGMA PLOT by using Eq. 4 with the indicated b0, t = 90 min, p (neutrophil concentrations) as the independent variable, and b90 min as the dependent variable. Shown are the data of one representative experiment for each initial bacterial concentration. The average values of k and g are shown in Table 1 (under the heading of “10% Human serum”).
Table 1.
Effect of serum concentration on rate constants characterizing the killing of S. epidermidis by human neutrophils in suspension
| Human serum, % (v/v) | b0, cfu/ml | k, ml per neutrophil per min | g/min | CNC = g/k, neutrophils per ml |
|---|---|---|---|---|
| 10 | 103 | 1.8 ± 0.4 × 10−8 | 8.6 ± 1.0 × 10−3 | 5.2 ± 1.0 × 105 |
| 105 | 2.2 ± 0.3 × 10−8 | 6.8 ± 1.3 × 10−3 | 3.1 ± 0.3 × 105 | |
| 107 | 1.4 ± 0.2 × 10−8 | 3.0 ± 0.8 × 10−3 | 2.1 ± 0.3 × 105 | |
| 1.8 ± 0.2 × 10−8 | ||||
| 40 | 103 | 2.6 ± 0.5 × 10−8 | 8.5 ± 1.3 × 10−3 | 3.4 ± 0.6 × 105 |
| 105* | 2.7 ± 0.1 × 10−8 | 8.6 ± 0.2 × 10−3 | 3.2 ± 0.1 × 105 | |
| 107 | 2.1 ± 0.1 × 10−8 | 8.0 ± 0.7 × 10−3 | 3.7 ± 0.3 × 105 | |
| 2.5 ± 0.2 × 10−8 | ||||
| 75 | 103* | 2.5 ± 0.6 × 10−8 | 7.8 ± 2.0 × 10−3 | 3.0 ± 0.3 × 105 |
| 105* | 2.4 ± 0.1 × 10−8 | 8.6 ± 1.3 × 10−3 | 3.5 ± 0.7 × 105 | |
| 107 | 2.2 ± 0.2 × 10−8 | 8.3 ± 2.0 × 10−3 | 3.7 ± 0.8 × 105 | |
| 2.4 ± 0.1 × 10−8 |
Experiments were carried out as described for Fig. 1 in the presence of various serum concentrations. For each experiment, k and g were obtained by fitting Eq. 4 with t = 90 min and p as the independent variable. The correlation coefficient, R2, for these experiments was 0.98–0.99. Shown are means and SEM for k, g, and CNC, and in bold means and SD for k at the indicated concentration of serum.
For each condition indicated, three independent experiments were performed. For all other conditions, four independent experiments were performed.
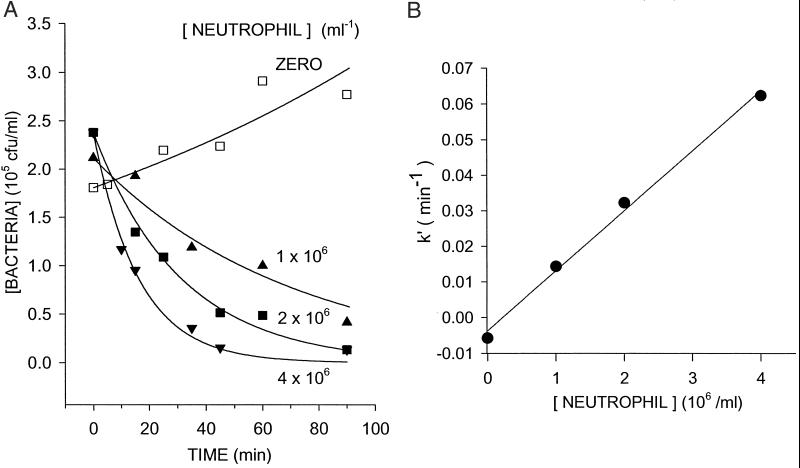
We also measured bacterial concentration as a function of time and neutrophil concentration (Fig. 4A). The fit of Eq. 5 to the data yields pseudo first-order rate constants, k′, that increased linearly with neutrophil concentration (Fig. 4B). Fitting Eq. 6 to the values of k′ yielded values for k and g. The means for three determinations with neutrophils from different donors were k = (1.7 ± 0.06) × 10−8 ml per neutrophil per min and g = (7.4 ± 0.1) × 10−3 per min. These values are similar to those obtained by measuring bacterial concentration after a fixed period of incubation (90 min) in the presence of varying concentrations of neutrophils (e.g., Table 1, 10% human serum/105 cfu/ml inoculum).
Figure 4.
Determination of rate constants for k and g with varying neutrophil concentrations and time. (A) Human neutrophils were mixed continuously in suspension with S. epidermidis in buffer containing 10% human serum at 37°C. The symbols indicate the concentration of viable bacteria (bt) recovered at 5- or 15-min intervals in samples incubated without (open squares) or with (solid symbols) neutrophils from 0 to 90 min. Solid lines indicate the curves fitted to these data by using Eq. 5 (R2 = 0.92–0.99). (B) The pseudo first-order rate constant, k′, is plotted versus p, the neutrophil concentration. A linear least-squares fit of Eq. 6 yielded k (slope) and −g (intercept), and R2 = 0.99. The data shown in A and B are from one representative experiment. The mean and SEM of k and g from three independent experiments are: k = (1.7 ± 0 06) × 10−8 ml per neutrophil per min and g = (7.4 ± 0.1)× 10−3 per min.
Eq. 4 fits our data with initial bacterial concentrations from 103 to 107 per ml. At 1 × 108 bacteria per ml, however, a bacterial concentration in excess of that found in most clinical settings, k decreased ≈3-fold (data not shown).
Analysis of Published Data on Neutrophil Killing of S. aureus, E. coli, and Pseudomonas aeruginosa.
Leijh et al. (8) and Hammer et al. (6) reported that neutrophil killing of S. aureus, E. coli, and P. aeruginosa in suspension depended on the ratio of these bacteria to neutrophils. They used 10% human serum and varied the ratio of bacteria to neutrophils over a 100-fold range. We found that values of k derived from their data (Table 2) are similar to the ones we obtained for neutrophil killing of S. epidermidis (Table 1, 10% human serum). These findings support our hypothesis that the rate of bacterial killing by neutrophils depends on the concentration of neutrophils and not on the ratio of bacteria to neutrophils, at least up to bacterial concentrations of 107 cfu/ml.
Table 2.
Rate constants characterizing the killing of bacteria in suspension by human neutrophils
| Bacteria (ref. no.) | Ratio, p/b0 | b0, cfu/ml | k, ml per neutrophil per min | R2 |
|---|---|---|---|---|
| S. aureus (8) | ≈10:1 | 4 × 105 | 0.9–1.2 × 10−8 | 0.92 |
| 1:1 | 4 × 106 | 1.7–2.0 × 10−8 | 0.99 | |
| 1:10 | 4 × 107 | 1.8–2.1 × 10−8 | 0.99 | |
| E. coli − K− (7) | ≈10:1 | 5 × 105 | 1.2–1.6 × 10−8 | 0.99 |
| E. coli (8) | ≈10:1 | 7 × 105 | 1.9–2.2 × 10−8 | 0.99 |
| 1:1 | 6 × 106 | 1.8–2.1 × 10−8 | 0.99 | |
| 1:10 | 6 × 107 | 1.0–1.3 × 10−8 | 0.90 | |
| Pseudomonas (6) | ≈1:1 | 4 × 106 | 1.0–1.5 × 10−8 | 0.90 |
Values of k were obtained first by fitting Eq. 5 with values of b0, bt, and t in the indicated references to obtain k′ and then by applying Eq. 6 to the values of k′, p = 5 × 106 per ml for S. aureus and E. coli and 3 × 106 per ml for Pseudomonas as used in the indicated references and growth-rate constants (g) corresponding to bacterial doubling times of 30, 60, and 90 min. Shown are the range of the values of k and the correlation coefficient R2 from fitting Eq. 5 to the data.
The Critical Neutrophil Concentration (CNC).
In a suspension of bacteria and neutrophils, the bacterial concentration changes as a result of two processes, bacterial growth, characterized by a rate constant g, and bacterial killing by neutrophils, characterized by a rate constant k. When the rate of bacterial growth equals the rate of bacterial killing, the bacterial concentration will remain unchanged, which occurs when p = g/k (Eqs. 4 and 5). We call the concentration of p = g/k the CNC. In this study, we found that the CNC ranges from 3 to 4 × 105 neutrophils per ml (Table 1). These values are similar to the blood neutrophil concentration (i.e., 5 × 105 per ml) widely recognized to predispose humans to lethal bacterial infection (17). Thus our mathematical model provides one explanation for the reduced capacity of neutropenic patients to clear bacteria from the blood.
Neutropenia also is likely to limit the delivery of neutrophils to tissues (17). Low delivery of neutrophils to tissues would enable the small number of bacteria that gains access to extravascular sites via the airways and mucous membranes to grow to sufficient numbers to seed the blood, thereby leading to bacteremia. Evidence supporting this concept has been obtained in neutropenic animals, in which both mucosal and systemic defenses against bacterial and fungal infections were seriously compromised (18–22). We have found that bacterial killing in fibrin gels, a tissue-like environment, obeys Eq. 4, albeit, the CNC is approximately three times larger in these gels than in suspension (unpublished data). This finding suggests that a higher neutrophil concentration is required in tissues than in blood to combat bacterial invasion and growth. A requirement for a higher neutrophil concentration in tissues and tissue-like environments than in blood to control bacterial growth would further reduce the capacity of a neutropenic host to defend against bacterial invasion and spread.
In summary, we have developed an equation (Eq. 4) that fits killing of Gram-positive and Gram-negative bacteria, opsonized by IgG and C3, by neutrophils in suspension. The killing rate constant, k, is an essential feature of the equation. We have used the concepts embodied in Eq. 4 to define a parameter that we have termed the CNC. It is the concentration of neutrophils at which the rate of bacterial killing by the neutrophils matches the rate of bacterial growth. The values we have obtained for the CNC [(3–4) ×105 neutrophils per ml] correspond closely with that (≤5 × 105 per ml) known to predispose humans to lethal bacterial infections, thereby providing a quantitative explanation for the relationship between blood neutrophil concentrations of ≤5 × 105 per ml and susceptibility to bacterial sepsis.
Acknowledgments
We thank Dr. Marcus Horwitz for suggesting the term CNC and Ms. Emily Lu for technical assistance. We are grateful to the blood donors who, after informed consent, provided the neutrophils used in this study. This work was supported by National Institutes of Health Grants R37-AI20516-18 (to S.C.S.), T32-AI07525 (to Y.L.), and NS07065 (to A.K.).
Abbreviations
- cfu
colony-forming unit(s)
- CNC
critical neutrophil concentration
References
- 1.Lane T A. Transfus Med Rev. 1990;4:23–34. doi: 10.1016/s0887-7963(90)70245-3. [DOI] [PubMed] [Google Scholar]
- 2.Schiffer C A. Transfus Med Rev. 1990;4:2–7. doi: 10.1016/s0887-7963(90)70242-8. [DOI] [PubMed] [Google Scholar]
- 3.Pizzo P A. N Engl J Med. 1993;328:1323–1332. doi: 10.1056/NEJM199305063281808. [DOI] [PubMed] [Google Scholar]
- 4.Aboulafia D M, Mitsuyasu R T. Hematol Oncol Clin North Am. 1991;5:195–214. [PubMed] [Google Scholar]
- 5.Price T H, Bowden R A, Boeckh M, Bux J, Nelson K, Liles W C, Dale D C. Blood. 2000;95:3302–3309. [PubMed] [Google Scholar]
- 6.Hammer M C, Baltch A L, Sutphen N T, Smith R P, Conroy J V. J Lab Clin Med. 1981;98:938–948. [PubMed] [Google Scholar]
- 7.Horwitz M A, Silverstein S C. J Clin Invest. 1980;65:82–94. doi: 10.1172/JCI109663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leijh P C, van den Barselaar M T, van Zwet T L, Dubbeldeman-Rempt I, van Furth R. Immunology. 1979;37:453–465. [PMC free article] [PubMed] [Google Scholar]
- 9.Wain J, Diep T S, Ho V A, Walsh A M, Nguyen T T, Parry C M, White N J. J Clin Microbiol. 1998;36:1683–1687. doi: 10.1128/jcm.36.6.1683-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner A S, Cobbs C G, Kaye D, Hook E W. J Am Med Assoc. 1967;202:199–203. [PubMed] [Google Scholar]
- 11.Loike J D, el Khoury J, Cao L, Richards C P, Rascoff H, Mandeville J T, Maxfield F R, Silverstein S C. J Exp Med. 1995;181:1763–1772. doi: 10.1084/jem.181.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gargan R A, Brumfitt W, Hamilton-Miller J M. J Immunol Methods. 1989;124:289–291. doi: 10.1016/0022-1759(89)90368-2. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Loike J D, Ember J A, Cleary P P, Lu E, Budhu S, Cao L, Silverstein S C. J Immunol. 2002;168:816–824. doi: 10.4049/jimmunol.168.2.816. [DOI] [PubMed] [Google Scholar]
- 14.Barkalow K, Hartwig J H. Biochem Soc Trans. 1995;23:451–456. doi: 10.1042/bst0230451. [DOI] [PubMed] [Google Scholar]
- 15.Leijh P C, van den Barselaar M T, Dubbeldeman-Rempt I, van Furth R. Eur J Immunol. 1980;10:750–757. doi: 10.1002/eji.1830101005. [DOI] [PubMed] [Google Scholar]
- 16.Hampton M B, Winterbourn C C. J Immunol Methods. 1999;232:15–22. doi: 10.1016/s0022-1759(99)00147-7. [DOI] [PubMed] [Google Scholar]
- 17.Dale D C, Liles C. Lancet. 1998;351:1752–1753. doi: 10.1016/S0140-6736(05)78742-0. [DOI] [PubMed] [Google Scholar]
- 18.Ernst J D, Decazes J M, Sande M A. Infect Immun. 1983;41:275–279. doi: 10.1128/iai.41.1.275-279.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haraoka M, Hang L, Frendeus B, Godaly G, Burdick M, Strieter R, Svanborg C. J Infect Dis. 1999;180:1220–1229. doi: 10.1086/315006. [DOI] [PubMed] [Google Scholar]
- 20.Fulurija A, Ashman R B, Papadimitriou J M. Microbiology. 1996;142:3487–3496. doi: 10.1099/13500872-142-12-3487. [DOI] [PubMed] [Google Scholar]
- 21.Czuprynski C J, Brown J F, Maroushek N, Wagner R D, Steinberg H. J Immunol. 1994;152:1836–1846. [PubMed] [Google Scholar]
- 22.Fitzgeorge R B, Featherstone A S, Baskerville A. Br J Exp Pathol. 1988;69:105–112. [PMC free article] [PubMed] [Google Scholar]