Abstract
An earlier report has shown that herpes simplex virus 1 virions package RNA. Experiments designed to reveal the identity of the virion proteins capable of binding the RNA and to show whether the mRNA carried in the newly infected cells was expressed showed the following: (i) 32P-labeled riboprobe generated by in vitro transcription of the US8.5 ORF bound three proteins identified as the products of US11, UL47, and UL49 (VP22) genes. (ii) Viral RNA was bound to UL47 or US11 proteins immune precipitated from cells transduced with baculoviruses expressing UL47 or US11 and then superinfected with HSV-1 under conditions that blocked DNA synthesis and assembly of virions. (iii) Virions were purified from cells transduced with a baculovirus encoding a US8.5 protein fused to green fluorescent protein and superinfected with an HSV-1 mutant lacking the US8–12 genes. HEp-2 cells infected with these virions expressed the chimeric protein in ≈1% of infected cells. (iv) In mixed cultures, untreated Vero cells acquired the mRNA encoding the green fluorescent–US8.5 chimeric protein from HEp-2 cells doubly transduced with the genes encoding VP22 and the chimeric protein. The transfer was RNase sensitive and VP22 dependent, indicating that the RNA encoded by the chimeric gene was transferred to Vero cells as mRNA. We conclude that (i) three virion proteins are capable of binding RNA; (ii) the packaged RNA can be expressed in newly infected cells; and (iii) the UL47 protein was earlier reported to shuttle from nucleus to the cytoplasm and may transport RNA. VP22 thus appears to be a member of a new class of viral proteins whose major function is to bind and transport infected cell mRNA to uninfected cells to create the environment for effective initiation of infection.
Recent reports have shown that human cytomegalovirus and herpes simplex virus 1 (HSV-1) package RNA in virions (1–3). At least in the case of HSV-1, the nature of the mRNAs offers no clues as to their function. For example, both viral and cellular RNAs were detected and, whereas transcripts of some viral ORFs were invariably present in all viral preparations tested, others were detected in only some, but not all, preparations of purified virions. We also noted the packaged RNAs belong to more than one kinetic class and do not strictly correspond to the most abundant species of RNAs present in the infected cell. Furthermore, the packaged RNAs are not degradation products, because an intact virion protein was readily identified among in vitro products of translation of virion mRNAs.
To establish the physiologic role of the RNAs carried into cells during infection, it is essential to (i) identify the virion proteins that bind the mRNA, and (ii) determine whether the RNA is translated in newly infected cells.
To identify the RNA carriers, purified virions were solubilized, subjected to electrophoresis in a denaturing polyacrylamide gel, renatured, and reacted with a labeled riboprobe. This procedure, alluded to as a “Northwestern,” allowed us to identify three virion proteins that bound RNA in vitro. Further tests showed that at least two of the proteins encoded by UL47 and US11 ORFs bound RNAs in vivo.
Demonstration that the packaged mRNA is expressed presented a greater challenge, inasmuch as the experimental design required that only mRNA packaged in newly assembled virions be carried from cell to cell. In this study, we constructed a chimeric gene consisting of the US8.5 ORF fused to the ORF encoding the enhanced green fluorescent protein (EGFP). The chimeric gene was cloned into a baculovirus (Bac-US8.5-EGFP). Next, the cells were transduced with the US8.5-EGFP chimeric gene and hours later infected with a mutant virus lacking US8–12 genes. Virions produced in these cells expressed the chimeric protein in newly infected cells. In other studies, we demonstrated that the VP22 protein, a constituent of the virion tegument, mediates RNase sensitive transport and expression of the chimeric protein from transduced HEp-2 cells to untreated cocultivated Vero cells.
Material and Methods
Cells.
The sources and maintenance of HEp-2, Vero, and Sf9 cell cultures were described elsewhere (3). HSV-1(F) is the prototype strain used in this laboratory (4). The recombinant viruses R7023, lacking US8-US12, and R7105, lacking the ORFs UL46-UL47, were described elsewhere (5, 6). Virus stocks were titered on Vero cells.
Purification of Virions.
Wild-type and recombinant virus virions were purified as described elsewhere (3, 7).
In Vitro Synthesis of the Riboprobe for Binding to Virion Proteins.
A 500-bp EcoRI/NotI fragment containing the coding sequence of US8.5 was cloned into the pcDNA3.1(+) vector (Invitrogen). The radio-labeled US8.5-RNA probe was generated from 1 μg of XbaI-linearized plasmid DNA with the MAXIscrip T7 RNA polymerase kit (Ambion, Austin, TX). Briefly, the DNA was incubated for 1 h at 37°C in a final reaction volume of 20 μl, containing 1 × T7 RNA polymerase reaction buffer (Ambion), 10 mM ATP, GTP, and UTP (Ambion), 6 μl of [α32P]-CTP (800 Ci/mmol; Amersham Pharmacia, Pharmacia Biotech), and 2 μl of T7-RNA polymerase. After the addition of 1 unit of DNase I, the radiolabeled riboprobe was purified by electrophoresis on a denaturing polyacrylamide gel.
Construction of Plasmids for Recombinant Baculoviruses Production.
All recombinant baculoviruses used in this study were constructed with the aid of the shuttle vector pRB5850 (MTS1) derived from pAcSG2 baculovirus transfer vector (PharMingen). pRB5850 was generated by ligation of the XhoI-EcoRI fragment containing the human immediate early 1 promoter of human cytomegalovirus promoter/enhancer sequences. The UL47 coding sequence was amplified by PCR from cosmid cs69 (8). The forward primer, complementary to UL47 stop codon, was: UL47-F, 5′-ccgaattcccaccatgtcggctcgcgaacc-3′. The reverse primer, containing the UL47 start codon, was: UL47-R, 5′-ggaagatcttatgggcgtggcgggcctccc-3′. The entire UL49 coding sequence has been amplified by PCR starting from the plasmid pRB128, containing HSV-1 BamHI-F. The sequence of the forward primer, complementary to UL49 stop codon, was: UL49-F, 5′-ataagaatgcggccgctcactcgacgggccgtctg-3′. The reverse primer, containing the UL49 start codon, was: UL49-R, 5′-ggcgaattcatgacctctcgccgctccgtg-3′. The US11 coding sequence was amplified by PCR from pRB3910 with the forward primer 5′-ccggaattcatgagccagacccaaccc-3′ and the reverse primer 5′-ggaagatctctatacagacccgcgagccgta-3′. Each primer contained the appropriate restriction site for cloning in the shuttle vector pRB5850. The PCR products were sequenced after cloning in pRB5850. The US8.5-EGFP-pRB5850 plasmid was created as follows. An EcoRI/NotI fragment containing the coding sequence of US8.5 was amplified by PCR from cosmid cs43 (8) by using the following primers: forward, 5′-ccgaattccgccaatggatccggctttgagatc-3′; reverse, 5′-ataagaatgcggccgccataccacgcaatccc-3′. A NotI/PstI fragment containing the coding sequence of EGFP was amplified by PCR from the pCMS-EGFP vector (CLONTECH) with primers: forward, 5′-ataagaatgcggccgcatggtgagcaagggc-3′; reverse, 5′-aactgcagggcccgcctttacttctacagctcg-3′. Both fragments were ligated into pRB5850 after cleavage with EcoRI and PstI restriction enzymes. All of the recombinant baculoviruses were generated by cotransfection of Sf9 insect cells with the UL47-pRB5850, UL49-pRB5850, US11-pRB5850, or US8.5-EGFP-pRB5850 transfer plasmids along with baculoGold DNA (PharMingen), according to the manufacturer's instructions. The recombinant baculoviruses were amplified and titered in Sf9 cells. The expression of the genes cloned in baculoviruses was tested in HEp-2 cells.
Northwestern Blot Analysis.
The procedure was as described elsewhere (9, 10). Briefly, 40 μl of disrupted purified virus containing 20 μg of solubilized proteins was electrophoretically separated on a 10% denaturing polyacrylamide gel and electrically transferred to a nitrocellulose membrane at 300 mA (constant) for 4 h in Tris-glycine-methanol buffer at 4°C. The transferred proteins were renaturated overnight in PBS containing 0.01% (vol/vol) Nonidet P-40 at 40°C. Prehybridization and hybridization were performed with a single screening buffer containing 10 mM Tris (pH 7.6), 100 mM KCl, 0.1% (wt/vol) Ficoll 400-DL, 0.1% (wt/vol) polyvinylpyrrolidone, 0.01% (vol/vol) Nonidet P-40, 0.1 mM MnCl2, 0.1 mM ZnCl2, 0.1 mM EDTA, and 1 mM DTT. The prehybridization and hybridization buffers were supplemented with total yeast RNA (0.02 and 0.2 mg/ml, respectively). The membrane was prehybridized for 1 h in 25 ml of hybridization buffer and then for an additional 2 h at 25°C after addition of the [32P]-labeled riboprobe (106 cpm/ml in 5 ml of hybridization buffer). The membranes were then rinsed 4 times for 5 min each in 100 ml of hybridization buffer in the absence of yeast tRNA. The membranes were dried before autoradiography.
Antibodies.
The anti-US11 monoclonal antibody and the rabbit anti-VP13/14 polyclonal antibody R220/3 were described elsewhere (11, 12). The rabbit anti-GST-UL49 polyclonal antibody was described elsewhere (30).
Immunoblots.
After Northwestern analyses, the membranes were blocked for 1 h with 5% nonfat dry milk and reacted with the appropriate primary antibody overnight at 4°C. Polyclonal antibodies to UL47 and UL49 and monoclonal antibodies to US11 were diluted 1:500 in PBS containing 1% BSA and 0.05% Tween 20. Secondary antibodies [alkaline phosphatase (AP)-conjugated goat anti-mouse (1:3,000) and goat anti-rabbit (1:3,000) antibodies; Bio-Rad] were applied for 2 h. All rinses were done in PBS containing 0.05% Tween 20. To develop AP-conjugated secondary antibodies, the immunoblots were reacted with AP buffer [100 mM Tris (pH 9.5)/100 mM NaCl/5 mM MgCl2], followed by AP buffer containing 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium. The reactions were stopped by the addition of Tris (pH 7.6) and 10 mM EDTA.
Crosslinking Immunoprecipitation Assay.
We adapted a procedure described elsewhere (13–16). Replicates of 75-cm2 flask cultures of HEp-2 cells were exposed to 5 plaque-forming units (pfu) of baculoviruses expressing UL47 or US11 per cell. After 2 h at 37°C, the cells were rinsed and incubated for 16 h in medium containing 5 mM sodium butyrate and then exposed to HSV-1 (5 pfu per cell) in the presence of phosphonoacetate (300 μg/ml; a gift of Abbott). Formaldehyde (Fisher Scientific; 1% final concentration) was added directly to the cell culture medium 8 h after HSV-1 infection. After 20 min, fixation was arrested by the addition of glycine to a final concentration of 125 mM. The cells were then harvested, rinsed with cold PBS, resuspended in lysing buffer consisting of PBS* [PBS 1% Nonidet P-40, protease inhibitor mixture (Complete Protease mixture, Roche Diagnostics)], stored on wet ice for 30 min, and then centrifuged for 15 min at 14,000 rpm in a microcentrifuge (Eppendorf 5415C). The supernatant fluid was collected and precleared with 50% of protein-A slurry for 16 h. RNA–protein complexes were immunoprecipitated with antibodies to US11 or UL47 proteins. The precipitates, collected with protein A-beads, were rinsed three times in PBS* and eluted in 50 μl of disruption buffer [2% SDS/50 mM Tris (pH 6.8)]/3% sucrose/5% β-mercaptoethanol). All of the above procedures were done in the presence of 1 unit of RNase inhibitor (SupeRase-In, Ambion, Austin, TX) per microliter. RNA was extracted from the precipitate complexes with the aid of the RNAquose-4PCR Kit (Ambion) according to the manufacturer's instructions. The purified RNA samples were resuspended in 100 μl of elution buffer (1 mM EDTA; Ambion) and treated with 10 units of DNase I (Life Technologies, Rockville, MD) for 1 h at 37°C. Each sample was precipitated and then dissolved in 10 μl of diethyl pyrocarbonate-treated water.
Reverse Transcription and PCR Amplification of RNA Extracted from Crosslinked Immunoprecipitates.
RNAs purified from RNA–protein precipitates were reverse transcribed to yield single-stranded cDNA by using 60 units of AMV RT (Promega) in a total reaction volume of 30 μl. The reverse transcription was primed with oligo(dT)15 and random primers and performed using a pool of nucleotides containing 1 mM concentrations each of dGTP, dATP, dTTP, and dCTP (Promega). Forty units of RNasin (Promega) was added to each reaction mixture. The mixture containing only the RNA template, the oligo(dT)15, and random primers was first heated at 70°C for 10 min, chilled on wet ice, and after the addition of the other components, incubated at 42°C for 45 min, shifted at 52°C for 45 min, and then heat inactivated at 95°C for 5 min. cDNAs obtained from reverse transcription of RNA extracted from the precipitates were subjected to two cycles of amplification by using specific primer for the α22 ORF (forward: 5′-caatcagctgtttcgggtcctg-3′; reverse: 5′-atcctccgtgtcggactggga-3′). PCR conditions were: 1 min at 95°C, 45 s at 60°C, and 1 min at 72°C. Amplified products were resolved on 2% agarose gel containing ethidium bromide (0.5 μg/ml).
Detection of US8.5/EGFP Carried by Purified R7023 HSV-1 Mutant.
HEp-2 cells were exposed to 5 pfu of Bac-US8.5-EGFP per cell for 1 h at 37°C. Unabsorbed baculovirus was removed by vigorous rinsing. After 15 h of incubation in presence of 3 mM sodium butyrate, the cells were exposed to R7023 mutant (5 pfu per cell) for 1 h, vigorously rinsed, and incubated for an additional 24 h in fresh medium. The virions were then purified, titered, and used to infect HEp-2 cells grown in four-well slides. The cells were exposed to 200 or 1,000 pfu of virus per cell and examined at different times intervals. After infection, medium containing nonadherent cells was removed and the slides were mounted in 90% glycerol buffered with PBS. The slide cultures were scanned in a Zeiss confocal microscope with the aid of software provided by the manufacturer.
Detection of US8.5/EGFP Carried by VP22.
HEp-2 cells were exposed to a mixture of 25 pfu of Bac-UL49 plus 10 pfu of Bac-US8.5-EGFP or to Bac-US8.5-EGFP only and incubated for 6 h at 37°C in medium containing 5 mM sodium butyrate. In cocultivation experiments, the transduced HEp-2 cells were rinsed, trypsinized, and mixed (2:1) with untreated Vero cells. The mixtures were allowed to adhere and cocultured in 25-cm2 flasks in medium containing 1 mM sodium butyrate. After 14 h, the cells were harvested, rinsed, and resuspended at a concentration of 8 × 105 cells per ml of PBS. In some samples a mixture of RNase TI and RNase A (final concentration 80 units/ml and 2 units/ml respectively; Ambion) was present during the entire incubation time. To analyze the simultaneous expression of EGFP (FL1, green) and a surface marker (FL2, red), the cells were reacted with a monoclonal antibody against human β2 microglobulin (Becton Dickinson) for 30 min on ice and rinsed twice in PBS, reacted with a R-phycoerythrin-conjugated goat anti-mouse IgG (Molecular Probes), fixed with a paraformaldeide solution, and analyzed on a FACScan (Becton Dickinson) by using cell-quest software. Preliminary experiments showed the anti-human β2 microglobulin antibody reacted specifically with HEp-2 cells and not with Vero cells. As controls, Vero and HEp-2 cells untreated or exposed to 10 pfu of Bac-US8.5-EGFP were processed as above but were cultivated separately.
Results
Identification of Viral Protein Binding to the RNA in Vitro.
The objective of this series of experiments was to identify the virion proteins capable of binding RNA and therefore likely to be involved in the packaging of the RNAs into the HSV-1 virions. As described in Materials and Methods, protein extracts derived from purified virions were electrophoretically separated on a 10% denaturing polyacrylamide gel, electrically transferred to a nitrocellulose sheet, soaked in a protein renaturation solution, and then reacted with a 32P-riboprobe generated by in vitro transcription of US8.5 coding sequence. US8.5 is one of the viral RNAs reproducibly detected in all purified virion preparations tested (3).
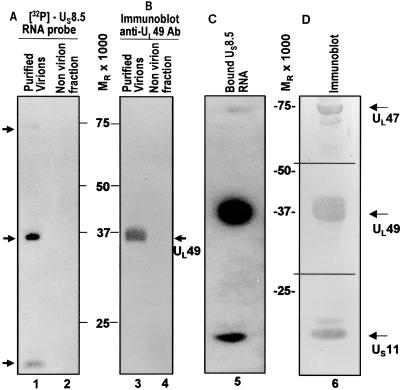
The autoradiographic images shown in Fig. 1A indicate that three protein bands bound RNA. The apparent Mr of these proteins was 75,000, 37,000, and 15,000 (Fig. 1A, lane 1). No signals were detected in the lane containing material collected from the dextran gradient immediately below the virion band (Fig. 1A, lane 2). It should be noted that mock-infected cell extracts contain numerous proteins that bind RNA (data not shown), and hence the paucity of bands binding RNA offered a measure of reassurance that they represent virion proteins.
Figure 1.
Identification of UL49, UL47, and US11 as RNA-binding proteins. Disrupted purified virions were separated on a 10% SDS/PAGE gel and transferred into a nitrocellulose membrane. After renaturation, the membrane was hybridized with US8.5 32P-labeled riboprobe (A and C) and subsequently immunoblotted with antibody to UL49 (B) and with specific antibodies to UL49, UL47, and US11 (D). Lanes 1, 3, 5, and 6 represent purified virions; lanes 2 and 4 contain no virion fraction. The positions of the proteins are indicated by arrows. Molecular weights are also shown.
The apparent molecular weights of the proteins contained in the three bands approximated very closely the apparent molecular weights of UL47, UL49, and US11 proteins, respectively. Two series of experiments were done to identify the virion proteins that bound the riboprobe. In the first, the nitrocellulose sheet containing the electrophoretically separated proteins was reacted with antibodies directed to US11, UL49, and UL47. As predicted from their apparent molecular weights, the autoradiographic images of the three bands lined up with the bands made apparent by their reactivity with the antibodies against US11, UL49, and UL47 (Fig. 1B, lane 3 and Fig. 1D, lane 6).
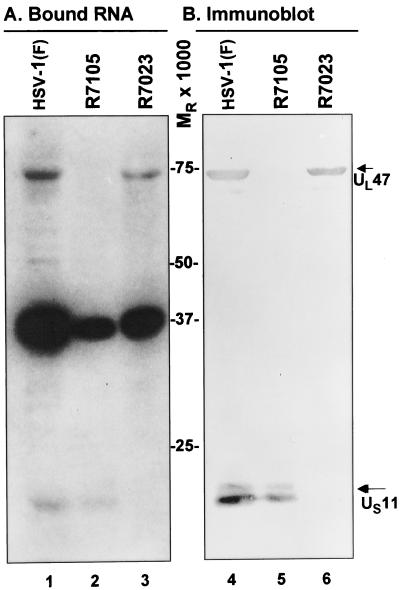
In the second series of experiments, we verified further the identity of the proteins by testing deletion mutants. In essence, the experiments described above were repeated with HSV-1(F) and mutants derived from it and lacking either UL46 and UL47 ORFs (R7105) or US8 -12 ORFs (R7023). The results (Fig. 2) were as follows: (i) neither the riboprobe nor the antibody to UL47 reacted with a Mr 75,000 protein band in electrophoretically separated virion proteins of the R7105 mutant (lane 2, Fig. 2A and lane 5, Fig. 2B). In a similar fashion, neither the riboprobe nor the antibody to US11 reacted with a Mr 15,000 protein band in lysates of electrophoretically separated virions proteins of the R7023 mutant. In contrast, all three protein bands among the resolved virion proteins of wild-type virus reacted with riboprobe, and the appropriate protein reacted with the two antibodies (lane 1, Fig. 2A and lane 4, Fig. 2B).
Figure 2.
Specificity of RNA binding to UL47 and US11 proteins. Virions produced by wild-type HSV-1(F) and mutant viruses R7105 (ΔUL46-UL47) and R7023 (ΔUS8-US12) were purified as described in Materials and Methods. Disrupted purified virions were separated on a 10% SDS/PAGE gel and transferred into a nitrocellulose membrane. After renaturation, the membrane was hybridized with US8.5 32P-labeled riboprobe (A) and subsequently immunoblotted with specific antibodies to UL47 and US11 (B). HSV-1(F), lane 1 (A) and lane 4 (B). R7105 mutant, lane 2 (A) and lane 5 (B). R7023 mutant, lane 3 (A) and lane 6 (B). Molecular weights are shown in the center. The positions of the proteins are indicated by arrows.
UL47 and US11 Proteins Bind Viral RNA in Vivo.
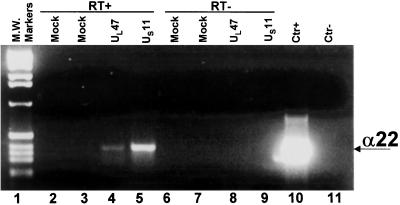
The experiments described above were done on electrophoretically separated proteins bound to nitrocellulose and renatured before exposure to labeled riboprobes. To further verify that at least UL47 and US11 proteins bind RNA in the context of infected cells, replicate 75-cm2 flask cultures of HEp-2 cells were exposed for 2 h to baculoviruses (5 pfu per cell) encoding UL47 or US11 ORFs. After 16 h of incubation at 37°C, the cells were exposed to 5 pfu of HSV-1(F) per cell in the presence of 300 μg of phosphonoacetate per ml of medium to block viral DNA synthesis. After 8 h of additional incubation, the cells were fixed with formaldehyde, lysed, and UL47 and US11 proteins were individually precipitated with the corresponding antibodies. The coprecipitated RNAs were reverse transcribed and analyzed by PCR using α22 specific primers as detailed in Materials and Methods. The α22 mRNA is one of the α (immediate early) mRNAs packaged in virions. Phosphonoacetate was added to allow for the synthesis of viral mRNA but to preclude the assembly of virus particles. The results (Fig. 3) indicate that specific signals for α22 were recovered only from RNA–protein complexes immunoprecipitated from cells transduced with UL47 and US11 and superinfected with HSV-1 (Fig. 3, lanes 4 and 5). As expected, no amplified bands were observed in the samples purified from the RNA–protein complexes immunoprecipitated from mock-infected cells (Fig. 3, lanes 2 and 3). The specificity of the signal was verified by the absence of signal from samples that were not subjected to reverse transcription before amplification of the DNA (Fig. 3, lanes 6–9). These results verified the ability of the UL47 and US11 proteins to specifically bind viral RNA in the context of infected cells.
Figure 3.
Binding of viral RNA to UL47 and US11 inside the cells. HEp-2 cells were infected with UL47- or US11-recombinant baculoviruses and superinfected with HSV-1 in the presence of phosphonoacetate. RNA–protein complexes were fixed and immunoprecipitated with specific antibodies to UL47 or US11 proteins. Lanes 2–5: reverse transcriptase was added to the reaction mixture (RT+). Lanes 6–9: reverse transcriptase was omitted from the reaction mixture (RT−). Lanes 2 and 6: mock cells immunoprecipitated with specific antibody to US11. Lanes 3 and 7: mock cells immunoprecipitated with specific antibody to UL47. Lanes 4 and 8: signal from immune precipitate obtained with anti UL47 antibody from lysates of cells infected with UL47-recombinant baculovirus and superinfected with HSV-1. Lanes 5 and 9: signal from immune precipitate obtained with anti-US11 antibody from lysates of cells infected with US11-recombinant baculovirus and superinfected with HSV-1. Lanes 10 and 11: positive and negative controls of the PCR reaction. Lane 1: 1-Kb DNA ladder. The position of the α22 amplified product is also shown.
RNA Incorporated in Virions Is Expressed in Newly Infected Cells.
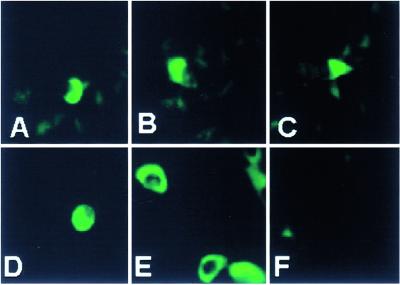
A central question arising from these studies is whether the RNA carried in virions is expressed in newly infected cells. To resolve this question, we constructed and cloned in a baculovirus a chimeric US8.5-EGFP protein, as described in Materials and Methods. In this series of experiments, HEp-2 cells were exposed to a recombinant baculovirus for 1 h at 37°C. Unabsorbed baculovirus was removed by vigorous rinsing. After 15 h of incubation at 37°C, the cells were infected with 5 pfu of R7023 (ΔUS8.5–12 mutant) and reincubated for an additional 25 h in fresh medium. The virions were purified, titered, and used to infect fresh cultures of HEp-2 cells grown in four-well slides. At different times after infection, medium containing nonadherent cells was removed, and the slides were mounted in 90% glycerol buffered with PBS. The experimental design was based on the observation that the US8.5 mRNA was reproducibly found in purified virions. Live unfixed cells expressing EGFP were tested by fluorescence microscopy at 1, 2, 4, 6, and 8 h after infection. The brightest fluorescence was observed at late time intervals. Fig. 4 shows the results obtained at 6 h after the infection in cells exposed to 200 (A–C) and 1,000 (D) pfu of purified R7023 per cell. We estimate that ≈1% of cells expressed the US8.5-EGFP chimeric protein at levels readily detected by fluorescence microscopy.
Figure 4.
Detection of chimeric US8.5-EGFP protein in HEp-2 cells. R7023 (ΔUS8-US12) mutant virions were purified from cells transduced with Bac-Us8.5-EGFP as described in Materials and Methods and used to infect fresh HEp-2 cells. The unfixed HEp-2 cells were tested for expression of EGFP-tagged US8.5 at 2, 4, 6, or 8 h after infection with the purified virions. The cells shown were examined at 6 h after infection. (A–C) Cells exposed to 200 pfu per cell; (D) cells exposed to 1,000 pfu per cell. (E) Cells transduced with baculovirus expressing the chimeric protein. (F) Mock infected cells. The images of the live cells were captured in a Zeiss confocal microscope with the aid of software provided by the manufacturer.
VP22 Encoded by the UL49 Gene Transports mRNA from Cell to Cell.
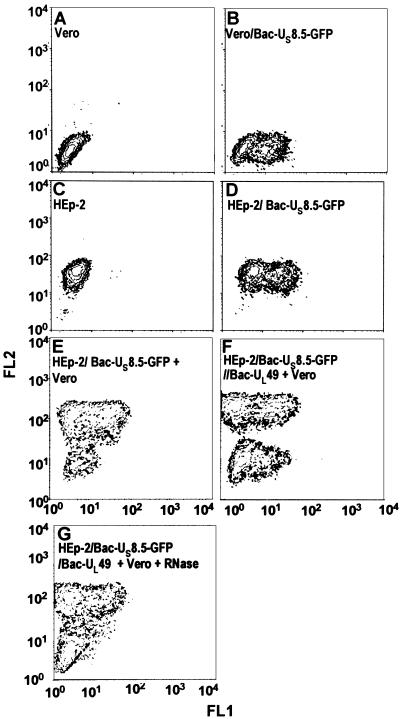
HEp-2 cells were exposed to Bac-UL49 and Bac-US8.5-EGFP or only to Bac-US8.5-EGFP and incubated for 6 h. In cocultivation experiments, at the end of infection, HEp-2 cells were rinsed, suspended with trypsin, mixed with Vero cells, and plated in 25-cm2 flasks. After 14 h of additional incubation in the presence or absence of RNase, the cells were collected, reacted with antibody, and then analyzed in a FACScan (Becton Dickinson) by using cell-quest software. The procedures were as detailed in Materials and Methods. The results (Fig. 5) were as follows: (i) Vero and HEp-2 cells, expressing or not the chimeric gene, were clearly differentiated by flow cytometry on the basis of the reactivity of the HEp-2 cells with the anti-β2 microglobulin antibody (A–D). (ii) In mixed cultures, Vero cells did not acquire the mRNA encoding the chimeric protein (US8.5-EGFP) from HEp-2 cells transduced only with the gene encoding the chimeric protein (E). (iii) Vero cells expressed the chimeric gene after incubation of cells in mixed cultures with HEp-2 cells transduced with both the gene encoding the chimeric protein and the gene encoding VP22 (F). (iv) The expression was precluded by incubation of the mixed cultures in medium containing RNase (G).
Figure 5.
Two-color flow cytometry analyses for simultaneous detection of chimeric US8.5-EGFP (FL1) protein and β2 microglobulin (FL2) in Vero and/or HEp-2 cells. (A) Untransduced Vero cells stained with anti-human β2 microglobulin. (B) Vero cells transduced with 10 pfu of Bac-US8.5-EGFP and reacted with anti-human β2 microglobulin. (C) Untransduced HEp-2 cells stained with anti-human β2 microglobulin. (D) HEp-2 cells transduced with 10 pfu of Bac-US8.5-EGFP and reacted with anti-human β2 microglobulin. (E) HEp-2 cells transduced with 10 pfu of Bac-US8.5-EGFP, cocultivated with untreated Vero cells, and then reacted with anti-human β2 microglobulin. (F) HEp-2 cells transduced with a mixture of 25 pfu of Bac-UL49 plus 10 pfu of Bac-US8.5-EGFP, cocultivated with untreated Vero cells, and then reacted with anti-human β2 microglobulin. (G) HEp-2 cells transduced and cocultivated with untreated Vero cells as in F but treated by the addition of RNase to the medium during the cocultivation.
Discussion
In this report, we show that the products of UL47, UL49, and US11 ORFs bind RNA in vitro and in the context of infected cells, and that the packaged RNAs can be expressed in infected cells. We also show that VP22, the product of the UL49 ORF, mediates the transfer of the RNA from cell to cell. Relevant to our results are the following:
(i) The procedure we have used to identify the protein capable of binding RNAs was to electrophoretically separate virion proteins in denaturing gels, renature the proteins in situ, and react them with a labeled riboprobe representing the RNA detected in all virion preparations tested. Using this procedure, we unambiguously demonstrated that three virion protein bands bind RNAs. These proteins were identified as the products of the UL47, UL49, and US11 genes. In these assays, we used as probe the most abundant RNA packaged in virions. Because we used a riboprobe representing a single viral RNA, we cannot exclude the possibility that there exist virion proteins with a high specificity for RNA sequences not present in the riboprobe. We also cannot exclude the possibility that virion proteins other than the three identified proteins bind mRNA but were not detected because they did not renature properly. Extensive studies, however, have shown that the binding of mRNA by the products of UL47, UL49, and US11 ORFs was not a spurious phenomenon caused by an incomplete or improper renaturation. Thus we have shown that RNA was associated with US11 or UL47 proteins precipitated from cells infected and maintained in the presence of phosphonoacetate in concentrations sufficient to block viral DNA synthesis and virion assembly. We have also shown that VP22, the product of the UL49, functionally interacts with mRNA inasmuch as it mediated the translocation of RNase-sensitive genetic information from cell to cell.
(ii) A key requirement for an understanding of the physiologic role of the RNA packaged in virions is that the RNA be expressed in newly infected cells. Because virions contain a finite amount of large numbers of species of RNA, it did not appear likely that we would detect the product of translation unless we chose a sensitive method for detection of the product and increased the amount of available mRNA by increasing the multiplicity of infection. To accomplish this objective, we used a recombinant baculovirus to transduce cells with a chimeric gene consisting of US8.5 fused to EGFP. The cells were then superinfected with a HSV-1 mutant lacking the US8–12 ORFs. Virions purified from these cells expressed the chimeric protein in newly infected cells. Moreover, experiments using flow cytometry demonstrated that VP22 mediated the transport of RNase-sensitive genetic information from HEp-2 cells transduced with genes expressing VP22 and the chimeric protein to cocultivated Vero cells. The evidence that virion protein-linked mRNAs is expressed is significant. Although the amount of each protein expressed by the mRNA carried into cells may be small, it may be sufficient to alter the physiologic state of the cells to render them more susceptible to infection.
(iii) Of the three proteins, US11, a protein not essential for HSV-1 replication in cultured cells, was already known to bind RNA in both a sequence- and a conformation-dependent manner (11). The domain of the protein capable of binding RNA is located at the carboxyl terminus and consists of three amino acids repeated many times. The number of repeats varies from one isolate to the next (11).
UL47 is not essential for viral replication in cells in culture (6, 17, 18). The products of the UL47 gene form two bands designated VP13 and VP14 (7). These proteins are phosphorylated and glycosylated (12). In earlier studies, UL47 was shown to enhance the transactivating activity of the α transinducing factor (α-TIF or VP16), but the ability of UL47 protein to bind RNA was not suspected (19). The protein contains a nuclear localization signal similar to that of tat, rev, and rex and has been reported to shuttle between the nucleus and cytoplasm (20). Our results indicate that this protein may act as a transporter of RNA from the nucleus to the cytoplasm and suggest that the association with mRNA constitutes the underpinning of its enhancement of the transactivating activity of α-TIF.
VP22 is an abundant tegument protein (21, 22). Full-length VP22 is necessary for viral replication and for efficient spread of the virus from cell to cell (23). VP22 is localized in the cytoplasm early in infection and accumulates in the nucleus late in infection (24). Extensive studies have shown that VP22 spreads from cell to cell by direct extension, and this function has been mapped to the protein itself (25, 26). The advantages accrued to the virus by the spread of VP22 from infected to adjacent uninfected cells have never been clear. The studies presented in this report indicate that VP22 binds RNA in the cell in which the protein was made and that it can carry the RNA to adjacent uninfected cells where it is expressed. Conceivably, a key function of VP22 is that of a carrier of mRNA from infected to uninfected cells to create the environment necessary for efficient viral replication in advance of the infection itself.
The discovery that VP22 can transport mRNA from cell to cell raises questions regarding proteins of other viruses that similarly are transported from cell to cell in advance of, or independent of, the spread of virus itself. Most notable among these is the tat protein of HIV-1. The tat protein is rapidly taken up by the cells (27, 28) and can bind RNA (reviewed in ref. 29). It is conceivable that one of the functions of the tat protein is to facilitate the initiation of infection with HIV by transport and expression of viral RNA in advance of viral infection.
Acknowledgments
We thank B. Fineschi, J. Trgovcich, P. Lopez, and M. A. Medici for invaluable discussions and T. Shenk for useful advice. These studies were initiated at the University of Chicago and completed at the University of Messina, Italy, and were aided by grants from the National Cancer Institute (CA87761, CA83939, CA71933, and CA78766) and the U.S. Public Health Service.
Abbreviations
- HSV-1
herpes simplex virus 1
- pfu
plaque-forming unit
- EGFP
enhanced green fluorescent protein
References
- 1.Brensnahan W A, Shenk T. Science. 2000;288:2373–2376. doi: 10.1126/science.288.5475.2373. [DOI] [PubMed] [Google Scholar]
- 2.Greijer A E, Dekkers C A J, Middeldorp J M. J Virol. 2000;74:9078–9082. doi: 10.1128/jvi.74.19.9078-9082.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sciortino M T, Suzuki M, Taddeo B, Roizman B. J Virol. 2001;75:8105–8116. doi: 10.1128/JVI.75.17.8105-8116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ejercito P M, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 5.Longnecker R, Roizman B. J Virol. 1986;58:583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker D E, Roizman B. Virology. 1990;177:684–691. doi: 10.1016/0042-6822(90)90534-x. [DOI] [PubMed] [Google Scholar]
- 7.Spear P G, Roizman B. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanda S K, Leibowitz J. J Virol. 2001;75:3352–3362. doi: 10.1128/JVI.75.7.3352-3362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagesser R, Martinez E, Tsagris M, Tabler M. Nucleic Acid Res. 1997;25:3816–3822. doi: 10.1093/nar/25.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roller R J, Roizman B. J Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meredith D M, Lindsay J A, Halliburton I W, Whittaker G R. J Gen Virol. 1991;72:2771–2775. doi: 10.1099/0022-1317-72-11-2771. [DOI] [PubMed] [Google Scholar]
- 13.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heise T, Guidotti L G, Chiari F V. J Virol. 2001;75:6874–6883. doi: 10.1128/JVI.75.15.6874-6883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrulis E D, Guzman E, Doring P, Werner J, Lis J T. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoborg R V, Clements J E. Virology. 1996;226:113–121. doi: 10.1006/viro.1996.0633. [DOI] [PubMed] [Google Scholar]
- 17.McLean G, Rixon F, Langeland N, Haarr L, Marsden H. J Gen Virol. 1990;71:2953–2960. doi: 10.1099/0022-1317-71-12-2953. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Sirko D A, McKnight J L. J Virol. 1991;65:829–841. doi: 10.1128/jvi.65.2.829-841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKnight J L, Doerr M, Zhang Y. J Virol. 1994;68:1750–1757. doi: 10.1128/jvi.68.3.1750-1757.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnelly M, Elliott G. J Virol. 2001;75:2566–2574. doi: 10.1128/JVI.75.6.2566-2574.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heine J W, Honess R W, Cassai E, Roizman B. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliot G D, Meredith D M. J Gen Virol. 1992;73:723–726. doi: 10.1099/0022-1317-73-3-723. [DOI] [PubMed] [Google Scholar]
- 23.Brewis N, Phelan A, Webb J, Drew J, Elliot G, O'Hare P. J Virol. 2000;74:1051–1056. doi: 10.1128/jvi.74.2.1051-1056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pomeranz L E, Blaho J A. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elliot G D, O'Hare P. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 26.Aints A, Guven H, Gahroton G, Smith C I, Dilber M S. Gene Ther. 2001;8:1051–1056. doi: 10.1038/sj.gt.3301493. [DOI] [PubMed] [Google Scholar]
- 27.Green M, Loewenstein P M. Cell. 1988;55:1179–1188. doi: 10.1016/0092-8674(88)90262-0. [DOI] [PubMed] [Google Scholar]
- 28.Frankel A, Pabo C O. Cell. 1988;55:1189–1193. doi: 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- 29.Marcello A, Zoppe M, Giacca M. IUMB Life. 2001;51:175–181. doi: 10.1080/152165401753544241. [DOI] [PubMed] [Google Scholar]
- 30.Blaho J A, Mitchell C, Roizman B. J Biol Chem. 1994;269:17401–17410. [PubMed] [Google Scholar]