Abstract
Cowpox virus (Brighton Red strain) possesses one of the largest genomes in the Orthopoxvirus genus. Sequence analysis of a region of the genome that is type-specific for cowpox virus identified a gene, vCD30, encoding a soluble, secreted protein that is the fifth member of the tumor necrosis factor receptor family known to be encoded by cowpox virus. The vCD30 protein contains 110 aa, including a 21-residue signal peptide, a potential O-linked glycosylation site, and a 58-aa sequence sharing 51–59% identity with highly conserved extracellular segments of both mouse and human CD30. A vCD30Fc fusion protein binds CD153 (CD30 ligand) specifically, and it completely inhibits CD153/CD30 interactions. Although the functions of CD30 are not well understood, the existence of vCD30 suggests that the cellular receptor plays a significant role in normal immune responses. Viral inhibition of CD30 also lends support to the potential therapeutic value of targeting CD30 in human inflammatory and autoimmune diseases.
As the immune system has evolved mechanisms to counter virus infection, viruses have evolved mechanisms to counter the immune system. These viral countermeasures not only illuminate processes important in restricting viral replication but also often provide new insights into regulatory mechanisms within the immune system.
Viruses with large genomes, such as the poxviruses, herpesviruses, and adenoviruses, are especially capable at interfering with immune defenses (reviewed in ref. 1). These viruses use a wide range of countermeasures to immune defenses, with viruses of different types often targeting different processes, reflecting virus host-ranges and modes of replication. For example, many of the poxviruses, particularly those in the orthopoxvirus genus, including cowpox, variola, monkeypox, ectromelia, raccoonpox, and vaccinia viruses, employ a variety of countermeasures primarily targeting innate immune responses (reviewed in refs. 2 and 3).
Cowpox virus is one of the most adept at these countermeasures, possessing more known cytokine-response modifiers than other poxviruses, as illustrated by its use of soluble secreted receptors to interfere with immune responses. In common with many poxviruses, it encodes receptors for cytokines such as tumor necrosis factor (TNF) and lymphotoxin (4, 5) IL-1β (6, 7), IFN-γ (8, 9), IFN I (10, 11), β-chemokines (12–14), and IL-18 (15–17). However, whereas most poxviruses express only a subset of these receptors, cowpox viruses typically express each of them, including up to four different receptors for TNF, the CrmB, CrmC, CrmD, and CrmE proteins (18–21).
The full complement of cytokine receptors encoded by cowpox virus has not been determined. The largest group of homologous receptors encoded by cowpox virus are the four targeting TNF and lymphotoxin, prototypic members of the TNF superfamily, which includes at least 18 members, many of which have crucial roles in immune responses (reviewed in ref. 22). The similarities among the poxviral and cellular TNF receptor (TNFR) suggest that the viral TNFR genes originated from their cellular counterparts (4, 5), raising the possibility that cowpox virus may have acquired other members of the TNFR family. In this study, we show cowpox virus encodes one additional member, a soluble, secreted form of CD30, the receptor for CD153.
Methods
Cells and Viruses.
Cowpox virus, Brighton Red strain (CPV-BR), vaccinia virus, Western Reserve strain (VV-WR), and recombinant viruses were grown in human osteosarcoma 143B cells. VTF7–3, vaccinia virus expressing the T7 RNA polymerase (23), was provided by B. Moss (National Institutes of Health, Bethesda, MD).
Recombinant CPV-BR (A624) containing an inactivated vCD30 gene was constructed as follows. Plasmid p1890 was generated containing the Escherichia coli gpt gene encoding a xanthine-guanine phosphoribosyltransferase under the control of the vaccinia virus p7.5 promoter flanked by XmaI sites within a pGem7zf vector. The latter insert was derived by PCR from the template of pTK61-gpt (24), provided by B. Moss. A SmaI fragment of p1890, containing the gpt gene cassette, was inserted into the repaired BspmI site in p1923, a pGEM7zf vector containing a 1.8-kb XbaI–ClaI fragment of CPV-BR DNA spanning the vCD30 gene, to create plasmid p1926. This SmaI fragment insertion disrupted the vCD30 gene by placing the gpt cassette about 40 bp downstream of the initiation codon. The intact vCD30 gene in the CPV-BR genome was replaced by the disrupted version in p1926 as described (24).
Recombinant VV-WR (A632) containing the CPV-BR vCD30 gene was constructed as follows. Plasmid p2110 was constructed containing the vCD30 gene, including its predicted promoter and the ORF downstream of the vCD30 gene. The insert was derived by PCR from the template of p1649, a pGEM7zf vector containing a 4.3-kb EcoRI–ClaI fragment of CPV-BR DNA spanning the vCD30 gene. After cleavage with EcoRI and ClaI, the PCR product was inserted into EcoRI–ClaI-cleaved plasmid p1378, which is a pUC19 vector lacking the polylinker region except the HindIII site, where the HindIII J fragment of the VV-WR DNA is inserted. Plasmid p2110 was used to generate virus A632 by standard methods as described (25).
Recombinant VV-WR (A593) was constructed to express the vCD30 protein fused to the Fc portion of human (hu) IgG1 (26). The intact vCD30 gene was amplified from p1649 by PCR, cleaved with BspH1 and EcoRI, and inserted into NcoI–EcoRI-cleaved pTM1 vector (27) to create plasmid p1784. The region encoding the Fc portion of huIgG1 was inserted in-frame with the 3′ end of the coding region of the vCD30 gene by insertion of an SpeI–BglII fragment of p1783 (the Fc portion of huIgG1 as an Asp718–SpeI fragment in a pBluescript SK vector) into BglII–EcoRI cut p1784, to generate plasmid p1812. This plasmid was used to generate virus A593 as described (25).
Recombinant VV-WR (A608) expressing a secreted version of the Fc portion of huIgG1 was constructed as described above, with plasmid p1882. The latter was generated by PCR from p1812 to delete all of the region encoding the vCD30 protein except for the signal sequence, which was retained upstream of the Fc portion of huIgG1 with an amino-terminal Flag epitope tag.
Sequence Analyses.
DNAs of cowpox virus strain OPV 90/2 (28) and cowpox viruses designated cat poxviruses 3 and 5, isolated from a cheetah and a cat (29, 30), were provided by H. Meyer (Institute of Microbiology, Federal Armed Forces Medical Academy, Munich, Germany). Raccoonpox virus (strain V71-I-85A) DNA and the latter cowpox virus DNAs were used as templates for PCR to generate copies of the regions predicted to contain the vCD30 genes. The nucleotide sequence of the vCD30 gene of CPV-BR was determined from p1649.
Purification and Biotinylation of Recombinant Proteins.
To prepare vCD30Fc fusion protein, 143B cells were coinfected with A593 and VTF7–3 viruses. After 16 hr the medium containing vCD30Fc was removed, clarified by centrifugation, and passed through a column of staphylococcal protein-A-Sepharose equilibrated with PBS containing 0.05% Tween-20. After washing with PBS containing 0.05% Tween-20, the vCD30Fc protein was eluted with 0.1 M glycine, pH 3.0. The eluate was neutralized with 1.0 M Tris, pH 9.0, and dialyzed overnight against PBS. huIgG1Fc was similarly purified after expression from virus A608. For use in flow cytometric analysis, each protein was biotinylated in 100 mM sodium bicarbonate buffer (pH 8.5) with sulfo-NHS-LC-biotin (Pierce) for 4 hr at room temperature. Excess biotin was removed by using a Centricon-10 centrifugal filter device (Millipore).
Immunoprecipitations of CD153 from Chinese Hamster Ovary (CHO) Cell Lines.
huCD153 was immunoprecipitated from CHO-CD153 cells surface-biotinylated as described (31). Briefly, cell lysates were precleared with murine or human IgG (Sigma) and protein-A-Sepharose, then immunoprecipitated overnight with 2.0 μg/ml P3 (mouse IgG1 control antibody), anti-huCD153 mAbs M80 and M82, huCD30Fc (Immunex), huIgG1Fc, and vCD30Fc. After incubation with protein-A-Sepharose, proteins were washed at 4°C with lysis buffer containing, in succession, 600 mM NaCl, 300 mM NaCl, and 150 mM NaCl. Portions of samples were resuspended in 90 mM sodium phosphate buffer, pH 7.5, containing 1% SDS and 1% 2-mercaptoethanol, boiled for 5 min, diluted with a 200 mM sodium phosphate buffer containing 1.0% Nonidet P-40, and incubated 16 hr at 37°C, with or without 0.5 unit of N-glycosidase F (Boehringer Mannheim). Proteins from the equivalent of 0.5 × 108 cells were resolved by SDS/PAGE in 10% polyacrylamide gels, and transferred to Immobilon-P membranes (Millipore). Biotinylated proteins were detected by using streptavidin-conjugated horseradish peroxidase (HRP) and the ECL detection system (Amersham Pharmacia).
Flow Cytometry.
CHO and CHO-CD153 cells were harvested by using 0.5 mM EDTA in PBS and resuspended in PBS containing 1% BSA and 20% normal rabbit serum. Peripheral blood mononuclear cells (PBMCs) were purified from heparin-treated blood by using a Ficoll–Hypaque gradient (Organon-Teknika). PBMCs were plated at a density of 2 × 106 cells per ml in RPMI medium 1640 (GIBCO/BRL) containing 2% heat-inactivated FBS for 1 hr. Adherent cells were harvested and activated for 22 hr with 10 μg/ml Escherichia coli O111:B4 lipopolysaccharide (Sigma) in RPMI medium 1640 plus 10% heat-inactivated FBS. Cells were stained with 5 μg/ml anti-huCD153 (M80 or M82) or 2.5 μg/ml biotinylated vCD30Fc and phycoerythrin (PE)-conjugated goat-anti-mouse reagent (Caltag, Burlingame, CA) or PE-conjugated streptavidin (PharMingen). Isotype-matched antibodies (Dako) or huIgGFc were used as controls. Cells were analyzed by using a FACStarPlus flow cytometer and CELL QUEST software (Becton Dickinson).
Ligand-Binding Assays.
A huCD30Fc fusion protein containing the entire extracellular portion of CD30 fused to truncated IgG1 heavy chain was constructed, expressed, and purified as described (32). Transient expression of membrane-bound murine CD153 (CV1-muCD153) involved transfection of full-length muCD153 cDNA into CV1/EBNA cells by using a pDC409 expression vector and standard procedures (32). Equilibrium binding isotherms between vCD30Fc, huCD30Fc, and recombinant, full-length, cell surface-expressed muCD153 were determined by competitive inhibition with 125I-vCD30Fc as described (4). vCD30Fc was radioiodinated to a specific activity of 3 × 1015 cpm/mmol with Iodo-Gen (Pierce) without significant loss of binding activity. Briefly, CV1-muCD153 cells were diluted 50-fold into carrier cells (Daudi) lacking CD153 and were incubated with a constant amount of 125I-vCD30Fc in medium for 2 hr at 4°C in the presence or absence of increasing concentrations of specific inhibitors. Duplicate aliquots of each concentration were centrifuged through oil to separate bound and free 125I-vCD30Fc, and data were plotted using the assumption of single-site competitive inhibition.
Results
Cowpox Virus Encodes a Soluble Secreted Protein Similar to Part of the Ligand-Binding Portion of CD30.
Sequence analysis of the genome of CPV-BR revealed a gene, vCD30, encoding a protein whose amino acid sequence shares 51–59% identity with conserved segments of the ligand-binding portions of both mouse and human CD30 (Fig. 1). The sequence suggests that the primary product of the vCD30 gene is a 110-aa protein containing a signal peptide sequence and one potential O-linked glycosylation site (36), both features consistent with secretion of the protein. The nucleotide sequence also suggests that the vCD30 gene possesses a promoter corresponding to the consensus sequence of viral late promoters (37).
Figure 1.
The cowpox virus genome contains a gene, vCD30, encoding a protein similar to part of the extracellular portion of CD30. (A) The predicted amino acid sequence of the vCD30 protein. The arrow indicates the predicted cleavage site of the signal peptide sequence (33). (B) The cowpox virus vCD30 protein shares a high degree of identity with portions of both human and mouse CD30 (34, 35). Boxed areas show regions of identity among the three proteins. In the region shown, the vCD30 protein shares 51% (31/60) identity with human CD30, and 59% (35/59) identity with mouse CD30.
The vCD30 corresponds to only a short portion of the predicted ligand-binding region of mouse and human CD30, raising the possibility that the ORF identified in CPV-BR might be a truncated form of longer vCD30 genes present in other poxviruses. To address this possibility we examined the sequences of the vCD30 genes in three additional different natural isolates of cowpox virus and also one strain of raccoonpox virus, whose genome is comparable in size to that of cowpox virus (38). Each of the cowpox virus strains examined has almost identical vCD30 ORFs, as does the GRI strain of cowpox virus (39), whereas the raccoonpox virus lacks an intact vCD30 gene (GenBank accession nos. AF419543–419547). Thus, we have not found evidence of a viral gene encoding a longer version of vCD30.
To characterize the protein encoded by the vCD30 gene, the following two recombinant viruses were constructed: a cowpox virus (A624) containing a vCD30 gene inactivated by the insertion of the gpt gene; and a vaccinia virus (A632) containing the cowpox virus vCD30 gene under the control of its own promoter. Radiolabeled proteins secreted from cells infected with either one of these viruses, cowpox virus, or vaccinia virus, are shown in Fig. 2. Insertion of the vCD30 gene into the vaccinia virus genome results in the production of secreted proteins of two types (≈13 kDa and 14.3 kDa) that are not encoded by the wild-type virus. Proteins of both types are secreted from cells infected with the wild-type cowpox virus, but these are not secreted from cells infected with the cowpox virus A624, containing the interrupted vCD30 gene. Similar proteins were produced when epitope-tagged versions of the vCD30 gene were expressed by using vaccinia virus vectors (data not shown). Consistent with the nucleotide sequence data, these results suggest that the vCD30 gene encodes a late, secreted 13-kDa protein that may be modified posttranslationally to a form with a molecular mass of 14.3 kDa.
Figure 2.
The cowpox virus vCD30 gene encodes soluble proteins secreted from virus-infected cells during the late phase of virus replication. Virus-infected human 143 cells (3 × 106) were labeled with 100 μCi of [35S]cysteine (37 TBq/mmol) in 1.5 ml of cysteine-free medium without serum, from 11 to 17 hr after infection. Labeled secreted proteins were concentrated 10-fold, resolved by SDS/PAGE in a 16% polyacrylamide gel, and visualized by autoradiography of the dried gel. Proteins were secreted from cells infected with wild-type (wt) CPV-BR (lane 1); cowpox virus A624 containing an inactivated vCD30 gene (lane 2); vaccinia virus A632, containing the cowpox virus vCD30 gene (lane 3); and wt VV-WR, which lacks the vCD30 gene (lane 4). Arrows indicate the proteins encoded by the vCD30 gene. The positions of 14C-labeled standards (Amersham Pharmacia) are indicated.
vCD30 Protein Binds Specifically to CD153.
To facilitate analysis of the binding properties of the vCD30 protein, a fusion protein (vCD30Fc) was constructed by addition of the Fc region of huIgG1 to the COOH terminus of vCD30 protein. To determine whether vCD30Fc protein could bind specifically to human CD153, three approaches were used.
First, the ability of vCD30Fc to bind either CD153 or other TNF family ligands was tested by use of cultures of CV-1/EBNA cells on sterile glass slides transiently expressing recombinant type II membrane forms of each ligand. After incubation of these cells (30 min at room temperature) with either vCD30Fc or control cognate receptor/Fc protein, the cells were washed twice with PBS, and then incubated with 125I-labeled goat anti-huFc F(ab′)2 fragments. Bound radiolabeled antibodies were detected by slide autoradiography as described (32). This assay showed that vCD30Fc binds specifically to cells expressing either human or mouse CD153, but none of 17 other TNF superfamily members tested, including human and murine forms of TNF, lymphotoxin-α (LTα), lymphotoxin-β (LTβ), LTα/LTβ complexes, and the ligands for CD40, 4–1BB, OX40, Fas, and CD27 (data not shown).
Second, the ability of vCD30Fc to bind CD153 expressed on the surface of CHO cells was examined by immunoprecipitation of surface-biotinylated CHO cells or CHO cells stably expressing high levels of huCD153 (CHO-CD153). M82 (anti-huCD153) mAb, huCD30Fc protein, and vCD30Fc protein each bound to biotinylated proteins of the expected molecular masses of CD153 (40–58 kDa) only in extracts of the CHO cells expressing CD153 (Fig. 3A). These biotinylated proteins were not bound either by isotype control antibody or by huIgG1Fc. The heterogeneity in the electrophoretic mobilities of CD153 proteins is produced by different degrees of glycosylation of a 28-kDa polypeptide (32). When immunoprecipitated proteins were deglycosylated with N-glycosidase F, the molecular mass of each of the biotinylated proteins precipitated by the anti-CD153 mAb (M82), the huCD30Fc, or the vCD30Fc protein was reduced to 28 kDa (Fig. 3A), consistent with the interpretation that each of these proteins can specifically bind to CD153 on the surface of the CHO-CD153 cells.
Figure 3.
The vCD30Fc protein binds to CD153 expressed on the surface of CHO cells. (A) Proteins on the surfaces of CHO cells, or CHO cells stably expressing human CD153, were biotinylated. Lysates of the cells were immunoprecipitated with mouse mAb against human CD153 (M82), huCD30Fc fusion protein, vCD30Fc fusion protein, isotype-matched P3 antibodies, or huIgGFc. Selected samples were deglycosylated with N-glycosidase F, then samples were resolved by electrophoresis through SDS/PAGE gels and transferred to Immobilon-P membranes. Biotinylated proteins were detected with streptavidin-conjugated horseradish peroxidase and the ECL detection system. The bracket indicates the region containing the glycosylated CD153 proteins, and the arrow indicates the position of the deglycosylated CD153. The positions of standards are shown on the right. (B) CHO or CHO-CD153 cells were stained with either mouse mAb against human CD153 (M82) or the vCD30Fc fusion protein. (C) CD153 on activated human monocytes was detected by using either anti-CD153 (M80) or vCD30Fc. Isotype-matched murine IgG2b and human IgG1Fc were used as negative controls.
Third, flow cytometry was used to test the ability of vCD30Fc to bind CD153 on the surface of CHO cells expressing CD153 and on primary monocytes. Anti-huCD153 mAb M82 and vCD30Fc bound equally well to the surface of CHO-CD153 cells, but not to the surface of control CHO cells (Fig. 3B). vCD30Fc also bound specifically to the surface of human monocytes which express low levels of CD153 (Fig. 3C).
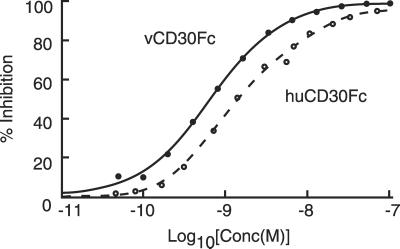
To quantify the equilibrium binding of vCD30 and huCD30 to murine CD153, we performed competitive inhibition assays using transiently expressed recombinant murine CD153 in CV1/EBNA cells and radioiodinated vCD30 (125I-vCD30Fc) protein (Fig. 4). Unlabeled vCD30Fc and huCD30Fc each specifically and completely inhibited binding of 125I-vCD30Fc to mouse CD153 in a dose-dependent manner, with the viral receptor showing reproducibly higher affinity for the murine ligand (Ki for vCD30Fc = 0.5 nM; Ki for huCD30Fc = 2.5 nM), consistent with the fact that rodents are the natural hosts for cowpox virus.
Figure 4.
The vCD30Fc competes with huCD30Fc for binding to membrane-bound murine CD153. Unlabeled vCD30Fc (solid line) and huCD30Fc (broken line) were incubated with a constant amount of 125I-vCD30Fc (0.2 nM) and CV1-muCD153 cells. The ratio of bound/free 125I-vCD30 was determined and the data were plotted assuming single-site competitive inhibition. The viral receptor showed reproducibly higher affinity for the murine ligand (Ki for vCD30Fc = 0.5 nM; Ki for huCD30Fc = 2.5 nM).
Discussion
This study has shown that cowpox virus encodes a soluble secreted version of CD30, which binds specifically and with high affinity to CD153. This protein is the fifth member of the TNFR family encoded by cowpox virus (Fig. 5), the only virus known to encode this many TNF receptors. The conservation of these different receptors suggests that cowpox virus derives an advantage from the expression of each of these five receptors during infections in vivo.
Figure 5.
Cowpox virus encodes five members of the TNFR superfamily. The diagram shows the viral members of the TNF family of receptors and their homologues. The structural modules of the ligand-binding regions are depicted according to the model of Naismith and Sprang (40) as follows: A1, open circle; B2, shaded oval; B1, shaded circle; A2, open oval; and D2, hexagon. Potential N-linked glycosylation sites are indicated by stars. Open rectangles represent the conserved COOH-terminal tails of the CrmB and CrmD proteins.
The structure of the vCD30 protein is similar to that of the other viral TNFR family members, except that the latter resemble almost all of the cysteine-rich domains (CRD) in both their sequences and the organization of component structural modules (40), whereas the vCD30 resembles only a small portion of either mouse or human CD30 (Fig. 5). Our results show that the vCD30 specifically binds human and mouse CD153, suggesting that additional components of the CRDs similar to those present in mouse and human CD30 are not required for CD153 binding. This interpretation is consistent with crystallographic studies of TNF ligand/receptor complexes, which show two main contact regions involving receptor residues corresponding to those in the three structural modules (A1, B2, A1) in the second and third CRDs (reviewed in ref. 41). These three modules correspond to those present in vCD30 (Fig. 5). Thus, vCD30 may be viewed as a structurally minimized CD153-binding protein. The vCD30 and the viral TNFRs also each contain regions similar to the pre-ligand-binding assembly domains (PLADs) of cellular receptors (42–44), suggesting that these viral TNFR members may be capable of forming both heterotypic and homotypic receptor complexes with themselves and cell-surface receptors. Whether this capacity is important for the function of viral TNFR family members remains to be determined.
CD30, originally identified as a cell-surface antigen and clinical marker of Reed–Sternberg cells of Hodgkin's disease (HD), is also expressed at low levels on 3–31% of human peripheral blood T cells (mostly CD8+), as well as on resting murine B cells, macrophages, and natural killer (NK) cells (45–49). Activation or viral transformation increases CD30 expression on B and T cells, including γδ T cells (50, 51). CD153 is expressed on activated T cells, monocytes and macrophages (32), eosinophils (52), neutrophils (53), and both normal and malignant B cells (54). Therefore, both CD30 and CD153 are expressed on cells expected to be present at the site of a poxvirus infection. Presumably, vCD30 interferes with CD30–CD153 interactions among some of these cells.
The biological functions of CD30 and CD153 are not fully understood. One important role is in costimulation of B and T cell responses. CD153 is costimulatory with IL-4 and IL-5 for mouse B cell proliferation and immunoglobulin secretion, and with IL-2 and IL-5 for antigen-specific responses (48). Signaling through CD30 enhances IL-5 production in CD30+ cytotoxic T lymphocytes (35), and it costimulates T cell proliferation in response to CD3 crosslinking (55). Conversely, CD153 inhibits constitutive and CD40L-induced Iɛ gene transcription by human B cell lines (56), and B cell CD153 engagement by CD30+ T cells inhibits isotype class switching and antibody production (57–59). Despite the lack of death-effector domains, CD30 may play a role in apoptosis of activated CD8 T cells after cessation of TCR signaling (60). However, while these in vitro studies indicate a role for CD30–CD153 interactions in both B and T cell activities, the CD30-deficient mouse model has provided controversial evidence for defects in B or T cell function (61). Although T cell negative selection appeared to be impaired in CD30-deficient mice (61), subsequent studies suggest that negative selection can proceed normally in the absence of CD30 (62). Meanwhile, other in vivo studies, involving mouse models of autoimmunity, have shown that blocking CD30 ligand/receptor interactions appears to reduce disease severity (K. Mohler, personal communication).
The presence and conservation of the poxvirus vCD30 gene strongly suggests that vCD30 provides some advantage to cowpox virus in vivo. As noted, cowpox virus is known to use soluble secreted receptors to target a specific subset of cytokines, including IL-1, TNF, LT-α, IL-18, β-chemokines, and both type I and type II IFNs, all of which are major contributors to antiviral immune responses. The inclusion of CD153 within this highly select group suggests that CD153-mediated processes play a significant role in antiviral processes.
The exact role of vCD30 in countering antiviral processes has yet to be determined. One potential antiviral process involving CD30 is the interaction between CD153 and CD30 on γδ T cells, which may be providers of an important link between innate and acquired immune responses (reviewed in refs. 63 and 64). γδ T cells play a significant role in protection against infection with vaccinia virus, in either the presence or the absence of an adaptive immune response (65–67). Although γδ T cells constitute less than 10% of peripheral lymphoid T cells, they are the major T cell components of the skin, intestinal epithelium, and pulmonary epithelium (reviewed in ref. 64). Consequently, γδ T cells are likely to be one of the most abundant CD30-expressing cells in the immediate vicinity of virus-infected cells during the initial phase of the infection. Importantly, CD30 enhanced CD3-induced expression of a variety of cytokines, including IL-4, IFN-γ, IL-8, and the β-chemokines I-309 and MDC, by γδ T cells (51). Thus, CD30–CD153 signaling between γδ T cells and neutrophils or activated macrophages may be an important factor in the development of immune responses to virus infection, and consequently one of the targets for vCD30. A second potential antiviral process that may be targeted by vCD30 is the ligation of CD153 on neutrophils, which, by reverse signaling, can initiate a rapid oxidative burst and the induction of IL-8 production (53).
In addition to the implications for virus–host interactions, the existence of vCD30 has broader implications for the role of CD30–CD153 interactions in health and disease. Elevated levels of cell surface expression of CD30 and shed soluble CD30 are often symptomatic of a variety of diseases, including Hodgkin's lymphoma (45), several non-Hodgkin's lymphomas (46), embryonal carcinoma (68), rheumatoid arthritis (69, 70), systemic lupus erythematosus (71), systemic sclerosis (72), atopic dermatitis (73), Wegener's granulomatosis (74), and Omenn's syndrome (75). Elevated levels of CD153 are found on various hematopoietic malignancies (76); and infections with Epstein–Barr virus (77), hepatitis B virus (78), and HIV (79). Signaling through CD30 activates HIV gene expression in latently infected CD4+ T cells (80, 81). However, in many of these instances it has been unclear whether CD30–CD153 interactions contribute to the disease. The finding that a virus may gain an advantage from secretion of a soluble version of CD30 from virus-infected cells suggests that inhibition of cellular CD30 signaling has a significant impact on the immune response.
Thus, viral genomics again illuminates crucial molecular components of the immune system, adding new significance to CD30/CD153 interactions. These results also suggest that CD30 agonism may be useful for reducing severity of viral infections, while CD30 antagonism may be of therapeutic value in the treatment of autoimmune disease. In fact, the early discovery of poxvirus-encoded soluble TNFRs (4) presaged the use of soluble cellular TNFRs in the treatment of rheumatoid arthritis.
Acknowledgments
This work was supported by Grants AI32982 and T32CA09111 (to J.F.P.) from the National Institutes of Health. D.J.P. is a member of the Duke University Comprehensive Cancer Center, whose shared core facilities were used in this study.
Abbreviations
- CPV-BR
cowpox virus, Brighton Red strain
- CHO
Chinese hamster ovary
- hu
human
- LT
lymphotoxin
- mu
murine
- TNF
tumor necrosis factor
- TNFR
TNF receptor
- v
viral
- VV-WR
vaccinia virus, Western Reserve strain
Footnotes
References
- 1.Tortorella D, Gewurz B E, Furman M H, Schust D J, Ploegh H L. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 2.Alcami A, Koszinowski U H. Immunol Today. 2000;21:447–455. doi: 10.1016/S0167-5699(00)01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moss B, Shisler J L. Semin Immunol. 2001;13:59–66. doi: 10.1006/smim.2000.0296. [DOI] [PubMed] [Google Scholar]
- 4.Smith C A, Davis T, Anderson D, Solam L, Beckmann M P, Jerzy R, Dower S K, Cosman D, Goodwin R G. Science. 1990;248:1019–1023. doi: 10.1126/science.2160731. [DOI] [PubMed] [Google Scholar]
- 5.Smith C A, Davis T, Wignall J M, Din W S, Farrah T, Upton C, McFadden G, Goodwin R G. Biochem Biophys Res Commun. 1991;176:335–342. doi: 10.1016/0006-291x(91)90929-2. [DOI] [PubMed] [Google Scholar]
- 6.Alcami A, Smith G L. Cell. 1992;71:153–167. doi: 10.1016/0092-8674(92)90274-g. [DOI] [PubMed] [Google Scholar]
- 7.Spriggs M K, Hruby D E, Maliszewski C R, Pickup D J, Sims J E, Buller R M, VanSlyke J. Cell. 1992;71:145–152. doi: 10.1016/0092-8674(92)90273-f. [DOI] [PubMed] [Google Scholar]
- 8.Upton C, Mossman K, McFadden G. Science. 1992;258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- 9.Alcami A, Smith G L. J Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colamonici O R, Domanski P, Sweitzer S M, Larner A, Buller R M. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 11.Symons J A, Alcami A, Smith G L. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 12.Graham K A, Lalani A S, Macen J L, Ness T L, Barry M, Liu L Y, Lucas A, Clark-Lewis I, Moyer R W, McFadden G. Virology. 1997;229:12–24. doi: 10.1006/viro.1996.8423. [DOI] [PubMed] [Google Scholar]
- 13.Smith C A, Smith T D, Smolak P J, Friend D, Hagen H, Gerhart M, Park L, Pickup D J, Torrance D, Mohler K, et al. Virology. 1997;236:316–327. doi: 10.1006/viro.1997.8730. [DOI] [PubMed] [Google Scholar]
- 14.Alcami A, Symons J A, Collins P D, Williams T J, Smith G L. J Immunol. 1998;160:624–633. [PubMed] [Google Scholar]
- 15.Novick D, Kim S H, Fantuzzi G, Reznikov L L, Dinarello C A, Rubinstein M. Immunity. 1999;10:127–136. doi: 10.1016/s1074-7613(00)80013-8. [DOI] [PubMed] [Google Scholar]
- 16.Xiang Y, Moss B. Proc Natl Acad Sci USA. 1999;96:11537–11542. doi: 10.1073/pnas.96.20.11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith V P, Bryant N A, Alcami A. J Gen Virol. 2000;81:1223–1230. doi: 10.1099/0022-1317-81-5-1223. [DOI] [PubMed] [Google Scholar]
- 18.Hu F Q, Smith C A, Pickup D J. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- 19.Smith C A, Hu F Q, Smith T D, Richards C L, Smolak P, Goodwin R G, Pickup D J. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- 20.Loparev V N, Parsons J M, Knight J C, Fanelli Panus J, Ray C A, Buller R H L, Pickup D J, Esposito J J. Proc Natl Acad Sci USA. 1998;95:3786–3791. doi: 10.1073/pnas.95.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saraiva M, Alcami A. J Virol. 2001;75:226–233. doi: 10.1128/JVI.75.1.226-233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locksley R M, Killeen N, Lenardo M J. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 23.Fuerst T R, Earl P L, Moss B. Mol Cell Biol. 1987;7:2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falkner F G, Moss B. J Virol. 1988;62:1849–1854. doi: 10.1128/jvi.62.6.1849-1854.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackett M, Smith G L, Moss B. J Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fanslow W C, Anderson D M, Grabstein K H, Clark E A, Cosman D, Armitage R J. J Immunol. 1992;149:655–660. [PubMed] [Google Scholar]
- 27.Moss B, Elroy Stein O, Mizukami T, Alexander W A, Fuerst T R. Nature (London) 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 28.Czerny C P, Zeller-Lue C, Eis-Hubinger A M, Kaaden O R, Meyer H. Arch Virol Suppl. 1997;13:13–24. doi: 10.1007/978-3-7091-6534-8_2. [DOI] [PubMed] [Google Scholar]
- 29.Baxby D, Ashton D G, Jones D M, Thomsett L R. J Hyg (London) 1982;89:365–372. doi: 10.1017/s0022172400070935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baxby D. Vet Rec. 1984;115:91. doi: 10.1136/vr.115.4.91. (lett.). [DOI] [PubMed] [Google Scholar]
- 31.Lantz L M, Holmes K L. BioTechniques. 1995;18:56–60. [PubMed] [Google Scholar]
- 32.Smith C A, Gruss H J, Davis T, Anderson D, Farrah T, Baker E, Sutherland G R, Brannan C I, Copeland N G, Jenkins N A. Cell. 1993;73:1349–1360. doi: 10.1016/0092-8674(93)90361-s. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Durkop H, Latza U, Hummel M, Eitelbach F, Seed B, Stein H. Cell. 1992;68:421–427. doi: 10.1016/0092-8674(92)90180-k. [DOI] [PubMed] [Google Scholar]
- 35.Bowen M A, Lee R K, Miragliotta G, Nam S Y, Podack E R. J Immunol. 1996;156:442–449. [PubMed] [Google Scholar]
- 36.Hansen J E, Lund O, Tolstrup N, Gooley A A, Williams K L, Brunak S. Glycoconjugate J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 37.Davison A J, Moss B. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 38.Knight J C, Goldsmith C S, Tamin A, Regnery R L, Regnery D C, Esposito J J. Virology. 1992;190:423–433. doi: 10.1016/0042-6822(92)91228-m. [DOI] [PubMed] [Google Scholar]
- 39.Shchelkunov S N, Safronov P F, Totmenin A V, Petrov N A, Ryazankina O I, Gutorov V V, Kotwal G J. Virology. 1998;243:432–460. doi: 10.1006/viro.1998.9039. [DOI] [PubMed] [Google Scholar]
- 40.Naismith J H, Sprang S R. Trends Biochem Sci. 1998;23:74–79. doi: 10.1016/s0968-0004(97)01164-x. [DOI] [PubMed] [Google Scholar]
- 41.Bodmer J L, Schneider P, Tschopp J. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 42.Naismith J H, Devine T Q, Brandhuber B J, Sprang S R. J Biol Chem. 1995;270:13303–13307. doi: 10.1074/jbc.270.22.13303. [DOI] [PubMed] [Google Scholar]
- 43.Chan F K, Chun H J, Zheng L, Siegel R M, Bui K L, Lenardo M J. Science. 2000;288:2351–2354. doi: 10.1126/science.288.5475.2351. [DOI] [PubMed] [Google Scholar]
- 44.Siegel R M, Frederiksen J K, Zacharias D A, Chan F K, Johnson M, Lynch D, Tsien R Y, Lenardo M J. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 45.Schwab U, Stein H, Gerdes J, Lemke H, Kirchner H, Schaadt M, Diehl V. Nature (London) 1982;299:65–67. doi: 10.1038/299065a0. [DOI] [PubMed] [Google Scholar]
- 46.Stein H, Gerdes J, Schwab U, Lemke H, Mason D Y, Ziegler A, Schienle W, Diehl V. Int J Cancer. 1982;30:445–459. doi: 10.1002/ijc.2910300411. [DOI] [PubMed] [Google Scholar]
- 47.Cambiaggi A, Cantoni C, Marciano S, De Totero D, Pileri S, Tazzari P L, Stein H, Ferrini S. Br J Haematol. 1993;85:270–276. doi: 10.1111/j.1365-2141.1993.tb03166.x. [DOI] [PubMed] [Google Scholar]
- 48.Shanebeck K D, Maliszewski C R, Kennedy M K, Picha K S, Smith C A, Goodwin R G, Grabstein K H. Eur J Immunol. 1995;25:2147–2153. doi: 10.1002/eji.1830250805. [DOI] [PubMed] [Google Scholar]
- 49.Agrawal B, Reddish M, Longenecker B M. J Immunol. 1996;157:3229–3234. [PubMed] [Google Scholar]
- 50.Stein H, Mason D Y, Gerdes J, O'Connor N, Wainscoat J, Pallesen G, Gatter K, Falini B, Delsol G, Lemke H. Blood. 1985;66:848–858. [PubMed] [Google Scholar]
- 51.Biswas P, Rovere P, De Filippi C, Heltai S, Smith C, Dagna L, Poli G, Manfredi A A, Ferrarini M. Eur J Immunol. 2000;30:2172–2180. doi: 10.1002/1521-4141(2000)30:8<2172::AID-IMMU2172>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 52.Pinto A, Aldinucci D, Gloghini A, Zagonel V, Degan M, Improta S, Juzbasic S, Todesco M, Perin V, Gattei V, et al. Blood. 1996;88:3299–3305. [PubMed] [Google Scholar]
- 53.Wiley S R, Goodwin R G, Smith C A. J Immunol. 1996;157:3635–3639. [PubMed] [Google Scholar]
- 54.Younes A, Consoli U, Zhao S, Snell V, Thomas E, Gruss H J, Cabanillas F, Andreeff M. Br J Haematol. 1996;93:569–571. doi: 10.1046/j.1365-2141.1996.d01-1686.x. [DOI] [PubMed] [Google Scholar]
- 55.Gilfillan M C, Noel P J, Podack E R, Reiner S L, Thompson C B. J Immunol. 1998;160:2180–2187. [PubMed] [Google Scholar]
- 56.Jumper M D, Nishioka Y, Davis L S, Lipsky P E, Meek K. J Immunol. 1995;155:2369–2378. [PubMed] [Google Scholar]
- 57.Cerutti A, Schaffer A, Shah S, Zan H, Liou H C, Goodwin R G, Casali P. Immunity. 1998;9:247–256. doi: 10.1016/s1074-7613(00)80607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerutti A, Schaffer A, Goodwin R G, Shah S, Zan H, Ely S, Casali P. J Immunol. 2000;165:786–794. doi: 10.4049/jimmunol.165.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cerutti A, Kim E C, Shah S, Schattner E J, Zan H, Schaffer A, Casali P. Nat Immunol. 2001;2:150–156. doi: 10.1038/84254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Telford W G, Nam S Y, Podack E R, Miller R A. Cell Immunol. 1997;182:125–136. doi: 10.1006/cimm.1997.1228. [DOI] [PubMed] [Google Scholar]
- 61.Amakawa R, Hakem A, Kundig T M, Matsuyama T, Simard J J, Timms E, Wakeham A, Mittruecker H W, Griesser H, Takimoto H, et al. Cell. 1996;84:551–562. doi: 10.1016/s0092-8674(00)81031-4. [DOI] [PubMed] [Google Scholar]
- 62.DeYoung A L, Duramad O, Winoto A. J Immunol. 2000;165:6170–6173. doi: 10.4049/jimmunol.165.11.6170. [DOI] [PubMed] [Google Scholar]
- 63.Mak T W, Ferrick D A. Nat Med. 1998;4:764–765. doi: 10.1038/nm0798-764. [DOI] [PubMed] [Google Scholar]
- 64.Hayday A C. Annu Rev Immunol. 2000;18:975–1026. doi: 10.1146/annurev.immunol.18.1.975. [DOI] [PubMed] [Google Scholar]
- 65.Bukowski J F, Morita C T, Brenner M B. J Immunol. 1994;153:5133–5140. [PubMed] [Google Scholar]
- 66.Welsh R M, Lin M Y, Lohman B L, Varga S M, Zarozinski C C, Selin L K. Immunol Rev. 1997;159:79–93. doi: 10.1111/j.1600-065x.1997.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 67.Selin L K, Santolucito P A, Pinto A K, Szomolanyi-Tsuda E, Welsh R M. J Immunol. 2001;166:6784–6794. doi: 10.4049/jimmunol.166.11.6784. [DOI] [PubMed] [Google Scholar]
- 68.Pallesen G, Hamilton-Dutoit S J. Am J Pathol. 1988;133:446–450. [PMC free article] [PubMed] [Google Scholar]
- 69.Gerli R, Muscat C, Bistoni O, Falini B, Tomassini C, Agea E, Tognellini R, Biagini P, Bertotto A. Clin Exp Immunol. 1995;102:547–550. doi: 10.1111/j.1365-2249.1995.tb03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerli R, Pitzalis C, Bistoni O, Falini B, Costantini V, Russano A, Lunardi C. J Immunol. 2000;164:4399–4407. doi: 10.4049/jimmunol.164.8.4399. [DOI] [PubMed] [Google Scholar]
- 71.Caligaris-Cappio F, Bertero M T, Converso M, Stacchini A, Vinante F, Romagnani S, Pizzolo G. Clin Exp Rheumatol. 1995;13:339–343. [PubMed] [Google Scholar]
- 72.Mavalia C, Scaletti C, Romagnani P, Carossino A M, Pignone A, Emmi L, Pupilli C, Pizzolo G, Maggi E, Romagnani S. Am J Pathol. 1997;151:1751–1758. [PMC free article] [PubMed] [Google Scholar]
- 73.Dummer W, Brocker E B, Bastian B C. Br J Dermatol. 1997;137:185–187. doi: 10.1046/j.1365-2133.1997.18031887.x. [DOI] [PubMed] [Google Scholar]
- 74.Wang G, Hansen H, Tatsis E, Csernok E, Lemke H, Gross W L. Am J Med. 1997;102:517–523. doi: 10.1016/s0002-9343(97)00049-1. [DOI] [PubMed] [Google Scholar]
- 75.Chilosi M, Facchetti F, Notarangelo L D, Romagnani S, Del Prete G, Almerigogna F, De Carli M, Pizzolo G. Eur J Immunol. 1996;26:329–334. doi: 10.1002/eji.1830260209. [DOI] [PubMed] [Google Scholar]
- 76.Gattei V, Degan M, Gloghini A, De Iuliis A, Improta S, Rossi F M, Aldinucci D, Perin V, Serraino D, Babare R, et al. Blood. 1997;89:2048–2059. [PubMed] [Google Scholar]
- 77.Vinante F, Morosato L, Siviero F, Nadali G, Rigo A, Veneri D, de Sabata D, Vincenzi C, Chilosi M, Semenzato G. Haematologica. 1994;79:413–419. [PubMed] [Google Scholar]
- 78.Fattovich G, Vinante F, Giustina G, Morosato L, Alberti A, Ruol A, Pizzolo G. Clin Exp Immunol. 1996;103:105–110. doi: 10.1046/j.1365-2249.1996.915607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pizzolo G, Vinante F, Morosato L, Nadali G, Chilosi M, Gandini G, Sinicco A, Raiteri R, Semenzato G, Stein H. AIDS. 1994;8:741–745. doi: 10.1097/00002030-199406000-00003. [DOI] [PubMed] [Google Scholar]
- 80.Biswas P, Smith C A, Goletti D, Hardy E C, Jackson R W, Fauci A S. Immunity. 1995;2:587–596. doi: 10.1016/1074-7613(95)90003-9. [DOI] [PubMed] [Google Scholar]
- 81.Maggi E, Annunziato F, Manetti R, Biagiotti R, Giudizi M G, Ravina A, Almerigogna F, Boiani N, Alderson M, Romagnani S. Immunity. 1995;3:251–255. doi: 10.1016/1074-7613(95)90094-2. [DOI] [PubMed] [Google Scholar]