Abstract
Phenotypic modification of dorsal root ganglion (DRG) neurons represents an important mechanism underlying neuropathic pain. However, the nerve injury-induced molecular changes are not fully identified. To determine the molecular alterations in a broader way, we have carried out cDNA array on the genes mainly made from the cDNA libraries of lumbar DRGs of normal rats and of rats 14 days after peripheral axotomy. Of the 7,523 examined genes and expressed sequence tags (ESTs), the expression of 122 genes and 51 expressed sequence tags is strongly changed. These genes encompass a large number of members of distinct families, including neuropeptides, receptors, ion channels, signal transduction molecules, synaptic vesicle proteins, and others. Of particular interest is the up-regulation of γ-aminobutyric acidA receptor α5 subunit, peripheral benzodiazepine receptor, nicotinic acetylcholine receptor α7 subunit, P2Y1 purinoceptor, Na+ channel β2 subunit, and L-type Ca2+ channel α2δ-1 subunit. Our findings therefore reveal dynamic and complex changes in molecular diversity among DRG neurons after axotomy.
Neuropathic pain is caused by nervous system lesions, persists long after the initiating event has healed, and may result from a pathological operation of the nervous system. Available therapies are often inadequate. Therefore, it is essential to identify the molecular changes that may lead to neuropathic pain, both for understanding underlying mechanisms and developing new therapies.
The peripherally axotomized animal represents one model to study the mechanisms of neuropathic pain (1). After the finding that vasoactive intestinal polypeptide is up-regulated after axotomy (2), many subsequent studies have reported dramatic changes in individual molecules in dorsal root ganglion (DRG) after nerve injury, and some molecules are implicated in generation and maintenance of pain (3–5). For example, down-regulation of μ-opioid receptor and up-regulation of cholecystokinin B receptor (6, 7) may contribute to the attenuated analgesic effect of opioids in neuropathic pain. Up-regulation of adrenoreceptor α2A (A-Rα2A) and neuropeptide Y (NPY) Y2-R enhances sympathetically maintained pain (8, 9). The increase in Na+ channel (Ch) III mediates ectopic activity in injured neurons (10). Moreover, hyperalgesia appearing during the course of nerve regeneration suggests a correlation between regeneration and pain. Neurotrophins play important roles in both nerve regeneration and regulation of the expression of some neuropeptides and ion channels (11, 12). Taken together, current knowledge suggests that changes in gene expression in DRGs may contribute to the generation and development of neuropathic pain. However, although in situ hybridization and other methods have been extensively used to study the changes of individual genes in DRG, a limiting factor with these methods is the lack of a comprehensive overview of the alteration of gene expression. Here, we took a broader approach, a cDNA array (13), to gain a global view of the changes in gene expression in DRGs after peripheral axotomy.
Materials and Methods
Animal Model and Tissue Preparation.
The sciatic nerves of 150 Sprague–Dawley rats (∼250 g) were transected bilaterally at mid-thigh level. Fourteen days after axotomy, lumbar (L) 4 and L5 DRGs of axotomized rats and 150 control rats were dissected for cDNA library (Lib) construction and Northern blot. The left sciatic nerves of another 100 rats were transected. The rats were allowed to survive for 2, 7, 14, and 28 days (20 rats each group). Then, L4 and L5 DRGs of these rats and of 20 normal rats were dissected for cDNA array, reverse transcription (RT)-PCR and in situ hybridization.
cDNA Lib Construction, DNA Sequencing, and Bioinformatics Analysis.
Total RNA was extracted with TRIzol reagent (Life Technologies, Rockville, MD). The mRNA was purified with an Oligotex mRNA kit (Qiagen, Hilden, Germany). cDNA Libs were constructed by using Lambda ZAP II system (Stratagene). A total of 11,910 clones was selected from the normal (5,724 clones) or the axotomized (6,186 clones) DRG cDNA Lib. The clone was sequenced from the 5′ end with T3 primer or the 3′ end with T7 primer. The qualified expressed sequence tags (ESTs) were referred to those ESTs containing less than 3% ambiguous bases and longer than 100 bp. These ESTs were subjected to blast analysis. The EST was considered as part of a known gene if the homology to the known gene was over 80%. The EST was considered as part of known EST, if it shared 95% homology with at least 100 bp of rat EST. The EST with no match to rat EST was considered a novel EST. Clustering of the EST was done with CAT3.2 (Pangea, Oakland, CA).
cDNA Array.
The cDNA clones representing 6,509 genes and ESTs were selected. All cDNA fragments were amplified by PCR with T7 and T3 primers and verified by gel electrophoresis. PCR products were purified, denatured, and spotted on 8 × 12-cm Hybond-N nylon membranes by using BioGrid 0.4-mm 384-pins-total array system arrayer (BioRotics, Cambridge, U.K.). Each cDNA fragment was placed at two different spots. Glyceraldehyde-3-phosphate dehydrogenase, pancreatic phospholipase A, α-tubulin, ornithine decarboxylase, hypoxanthine phosphoribosyltransferase, and polyubquitin were spotted on the membrane as reference genes. RT-PCR showed that the expression of these genes was not changed after axotomy. Lambda phage and pUC18 vector DNA were spotted as negative control. The array membrane was named Rat 6.5k Array. Additionally, Atlas Rat 1.2 Array membranes containing 1,176 genes were purchased from CLONTECH.
Probe labeling and hybridization were performed as described in the CLONTECH Atlas cDNA Expression Arrays User Manual (PT3140-1) with some modifications. The mRNA from each group (200 ng for Rat 6.5k Array and 1 μg for Atlas Rat 1.2 Array) were labeled in a reverse transcription reaction in the presence of 35 μCi of [α-33P ]dATP (NEN). For Rat 6.5k Array Oligo(dT) was used as a substitute for CDS Primer mix (CLONTECH), and SuperScript II RNase H− reverse transcriptase (Life Technologies) was used. The hybridization signals were scanned with a PhosphorImager (Molecular Dynamics) and analyzed with ARRAYVISION 5.1 (Imaging Research, Ontario, Canada). Each experiment was replicated at least 3 times.
The intensity of signal for reference genes was adjusted so that the mean intensities for each membrane were equal. The normalization was performed with ARRAYVISION 5.1 and presented as normalized volume (nVol). To assess the reproducibility of the normalized intensity, we compared the log2 (control/axotomy nVol ratio) of the six reference genes between different membrane sets. When the difference between normalized logarithmic ratio from three experiments was less than 1.0, we defined the data as reproducible. The reproducibility was more than 90% when the intensity of signal was above 0.01 nVol. The nVol of each spot was averaged among the tests. Only those genes with an over 2-fold change during at least one time interval were considered to show prominent differential expression.
RT-PCR and Northern Blotting.
Two hundred nanograms mRNA of normal or axotomized DRGs were reverse transcribed to cDNA. PCR products were analyzed on 2% agarose gel. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. 5′ and 3′ primers: potassium channel RCK4 (K+ Ch RCK4), 5′-catggcataattgtggcgaa3′, 5′tcccatcacggcacaattc-3′; NPY Y5-R, 5′-gctgtcgccatccagtaag-3′, 5′-ccagcttgaaatgcctattg-3′; Na+ Ch β2 subunit (Na+ Ch β2), 5′-cttcaactcctgctataccgtg-3′, 5′-ggtctggagggttggtgat3-′; P2Y1 purinoceptor (P2Y1-R), 5′-gctctggccgactttttgta-3′, 5′-agccataccagcacactgac-3′; GAPDH, 5′-atctccgccccttccgct-3′, 5′-ttgaagtcacaggagacaacct-3′.
For Northern blot, 15 μg of total RNA of normal and axotomized DRGs were separated and blotted to the nylon membrane. The probes were labeled at the 3′ end by [α-32P]dATP (NEN) by using terminal deoxynucleotidyl transferase (Amersham Pharmacia Biotech, Piscataway, NJ) and purified. The filters were hybridized at 42°C for 16–18 h and washed 2 times, and the hybridization signal was scanned with a PhosphorImager. Probes: oligonucleotide complementary to nucleotides in the code region 1367–1406 of L-type Ca2+ Ch α2δ-1 subunit (L-Ca2+ Ch α2δ-1); 134–170 of BG668901; 203–242 of BG663101.
In Situ Hybridization.
Fourteen micrometer-thick cryostat sections of DRGs of all groups were mounted on the same slide. Oligonucleotide probes were synthesized by Life Technologies. Probes: oligonucleotide complementary to nucleotides in the code region 443–481 of peripheral benzodiazepine-R (PBDZ-R); 246–284 of γ-aminobutyric acidA-R α5 subunit (GABAA-R α5); 76–114 of nicotinic acetylcholine-R α7 subunit; 402–440 of P2Y1-R; 1367–1406 of L-Ca2+ Ch α2δ-1; 315–352 of Na+ Ch β2. Probe labeling, purification, and in situ hybridization were carried out according to a published procedure (14).
The sections used for quantification were counterstained. To determine the percentage of labeled neuronal profiles (NPs), counting of both labeled and unlabeled NPs was made in all L5 DRGs. Every 15th serial section of the DRG (3 sections per ganglion) was selected for the counting. Totally, 488–503 NPs from each DRG were randomly counted, processed for t test, and presented as mean ± SEM.
Results and Discussion
Preparation of cDNA Array with the Genes and ESTs from DRG cDNA Libs.
We constructed and analyzed cDNA Libs of both normal and axotomized DRGs, because peripheral axotomy causes increases in the expression of genes that are normally expressed at low levels (3). The average size of insert in both cDNA Libs was about 1.2 Kb (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). A total of 11,229 ESTs was identified as qualified ESTs, 5,176 ESTs from the normal DRG Lib and 6,053 ESTs from the axotomized DRG Lib. All ESTs were assembled into a total of 6,509 clusters. Among them, 1,423 clusters consisted of more than one EST, the others were singletons. The high percentage of singletons indicates that the cDNA Lib contains many clones with low-copy number, suggesting a good representation of the cDNA Lib. The blast search of publicly available databases showed that about 60% of the clusters represented genes, 20% known ESTs, and 20% novel ESTs.
Identification of Differentially Expressed Genes and Verification of the Results.
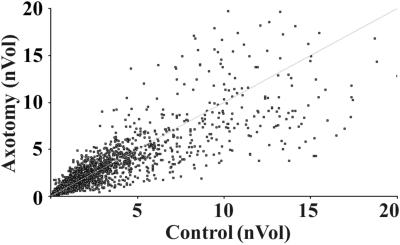
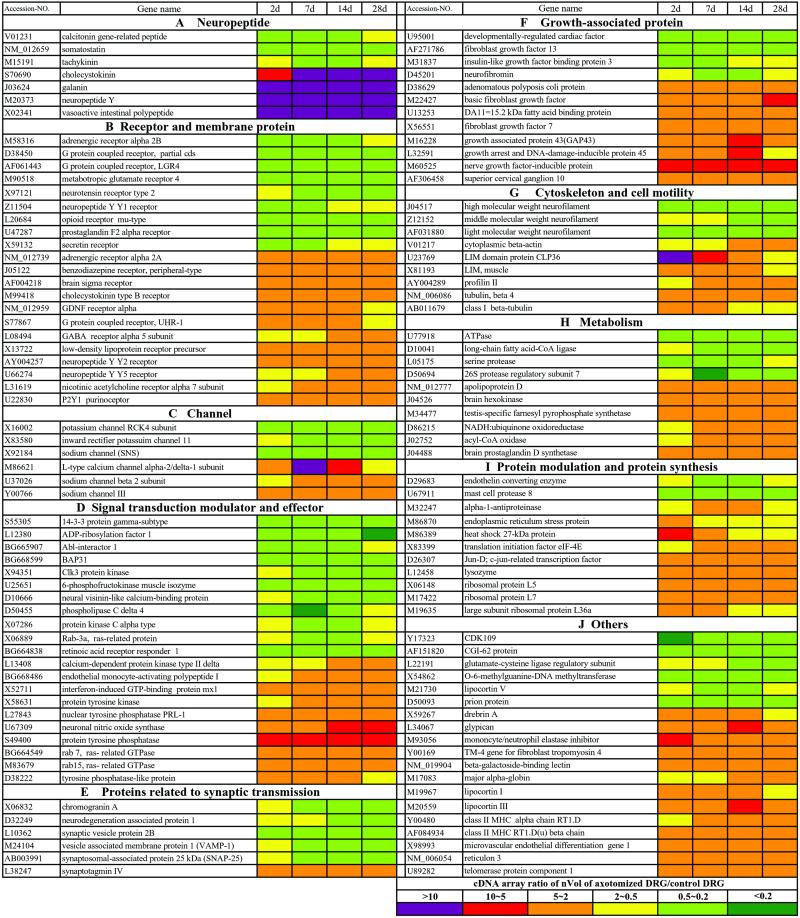
To establish gene expression profiles of normal and axotomized DRGs, the cDNA array was carried out with 7,523 genes and ESTs. The array data distribution indicated that the expression of most genes was not strongly changed (Fig. 1). The expression patterns of 162 genes on both our array and CLONTECH membranes were compatible. Based on the potential functions of the genes, we categorized the genes into 10 classes shown in Fig. 2 and Tables 2–5, which are published as supporting information on the PNAS web site. Totally, 61 distinct channels and their family members and 147 genes encoding receptors or receptor subunits were identified. Among 38 identified growth-associated proteins, 8 members of the fibroblast growth factor family were seen, indicating that fibroblast growth factors are enriched in DRGs.
Figure 1.
A scatter plot of cDNA array identifies outlying genes whose expression differs between control DRGs and the DRGs 14 days after peripheral axotomy. The line indicates that the ratio of nVol of axotomized DRG:control DRG is 1:1.
Figure 2.
The strongly regulated genes with the changes in cDNA array ratio over 2 folds are identified in the DRGs 2, 7, 14, and 28 days (d) after peripheral axotomy. The ranges of the cDNA array ratio of nVol of axotomized DRG vs. normal DRG are presented with colors indicated in the last panel.
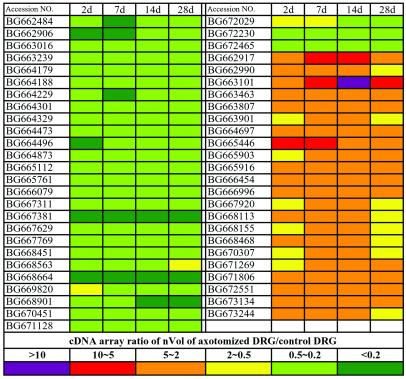
We used a 2-fold change in signal intensity, observed at least at one time interval after axotomy, as a cut-off line to consider the differential expression of a gene as significant. This criterion has also been used as a standard in other studies (15). According to this principle, we identified a total of 122 genes (Fig. 2) and 51 ESTs (Fig. 3), which represented 2.3% of total genes examined. Strikingly, 86% of these genes was not previously identified in DRGs after nerve injury. To verify the array results, we performed RT-PCR or/and Northern blot or/and in situ hybridization, and further confirmed a total of 76 of these strongly regulated genes including neuropeptides, receptors, and ion channels (Figs. 4 and 5; data not shown). These studies show that our cDNA array results accurately reflect the molecular changes. For the genes with less than 2-fold changes, the array results were included in the supporting information, because they may also contribute to the functional modification.
Figure 3.
The strongly regulated ESTs with the changes over 2 folds are identified in the DRGs after axotomy. The ranges of the cDNA array ratio of nVol of axotomized DRG vs. normal DRG are presented with colors indicated in the last panel.
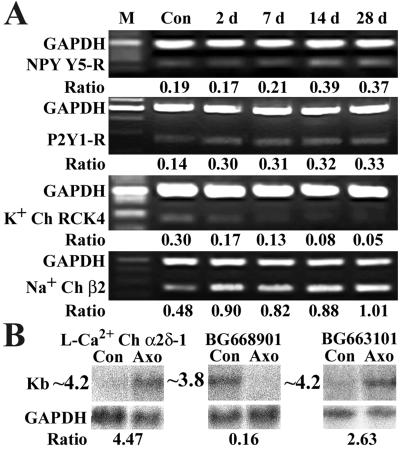
Figure 4.
Some strongly regulated genes and ESTs are confirmed with RT-PCR or Northern blot. (A) The results of the semiquantitative RT-PCR. Size of the internal control was 500 bp, and the size of the target genes is 250 bp. Ratio indicates the signal intensity of examined gene vs. that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) at different time points. (B) The results of Northern blot, with Bottom showing the internal control. Ratio indicates the signal intensity of examined gene in DRGs 14 days after axotomy vs. that in control DRGs.
Figure 5.
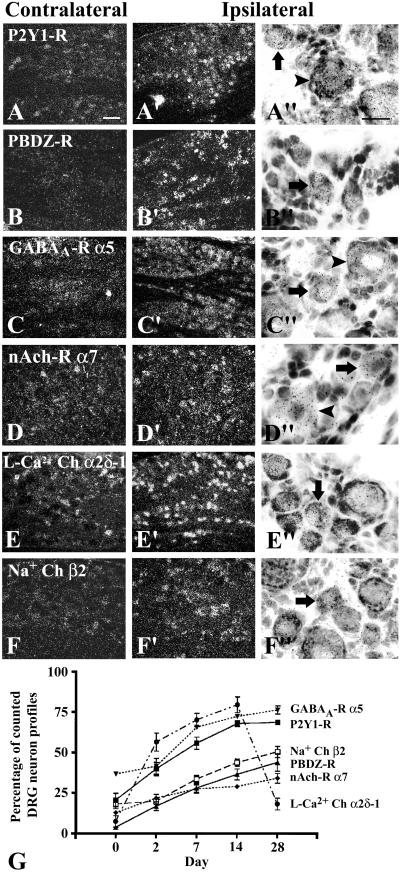
(A–F") In situ hybridization of receptors and ion channels in L5 DRG 14 days after unilateral axotomy. Arrows point to large NPs and arrowheads point to small NPs. P2Y1-R is normally expressed in some NPs (A) and up-regulated in many large and small NPs after axotomy (A′, A"). PBDZ-R is hardly seen contralaterally (B) but expressed ipsilaterally in many small NPs (B′, B"). GABAA-R α5 is normally expressed in some NPs (C) and up-regulated in many large NPs and some small NPs (C′, C"). Nicotinic acetylcholine-R α7 subunit (nAch-R α7) is expressed in some large NPs normally (D) and up-regulated in both large and small NPs (D′, D"). L-Ca2+ Ch α2δ-1 is normally seen in some small NPs (E) and increased in many small NPs after axotomy (E′, E"). Weak labeling of Na+ Ch β2 is normally seen in some NPs (F) and up-regulated ipsilaterally in many small NPs (F′, F"). [Bars = 50 μm (in A–F and A′–F′) and 20 μm (in A"–F")]. (G) The time course shows a constant increase in the percentage of NPs expressing these genes (P < 0.05 for P2Y1-R, PBDZ-R, nAch-R α7, and Na+ Ch β2 after 2 days, and for GABAA-R α5 after 7 days, compared with control), except L-Ca2+ Ch α2δ-1 showing a transient increase after 2 days (P < 0.05, compared with control) and decrease to the control level at 28 days (P > 0.05, compared with control).
In general, the most prominent change of gene expression occurred in the category of neuropeptides with 50% of the genes strongly regulated, followed by growth-associated proteins with 32% of the genes strongly regulated. The changes of these genes were dominated by up-regulation patterns. In contrast, less than 10% of the genes in the categories of receptors, channels, signal transduction, and synaptic transmission were strongly regulated, almost half of which exhibited up-regulation (Fig. 2; Table 2). Strong changes in gene expression appeared in all classes of genes, thereby revealing a surprising complexity and diversity of the gene expression profiles. The individual time course of each regulated gene could vary considerably. Several genes were transiently up-regulated at 2 days after axotomy (Fig. 2). For most genes changes started at 2 or 7 days and increased levels were maintained for the period studied, suggesting long-lasting phenotypic modifications of DRG neurons.
Molecular Adaptation to Nerve Injury.
Rat DRG neurons are able to survive and regenerate after nerve injury (16). Thus, the phenotypic switch of DRG neurons must involve a molecular adaptation to subserve this process. Such an adaptation is reflected first at transcriptional and translational levels, and then manifested by structural and functional modifications of the neuron. Transient up-regulation of heat shock protein 27, endoplasmic reticulum stress protein, and LIM domain protein CLP36 may reflect the fast adaptive responses, perhaps involved in sensory neuron survival (17). Not surprisingly, in addition to the transcription factor Jun-D (18), we show here that a translation initiation factor, eukaryotic initiation factor (eIF)-4E, was strongly up-regulated (Fig. 2). Furthermore, expression of several ribosomal proteins was also increased (Fig. 2). Nerve growth factor-mediated and ras-dependent eIF-4E phosphorylation may play a role in switching the pattern of gene expression during the differentiation of PC12 cells (19). It will be interesting to investigate whether a similar mechanism is invoked in axotomized DRG neurons. Nevertheless, our results render further support to the hypothesis that an activation of both the transcription and posttranscription machinery may constitute an integral part of mechanisms that underlie the plasticity of DRG neurons.
Axotomy up-regulates brain-derived neurotrophic factor (BDNF) (20) and glial cell line-derived neurotrophic factor (GDNF) (21), basic fibroblast growth factor, and other growth-associated proteins in DRG neurons, and these factors may not only protect neurons but also regulate the expression of some receptors and ion channels (21, 22). In the present study, the expression of both BDNF and GDNF was detected, but increases were less than 2-fold (Table 2). Unexpectedly, we observed that other growth-associated proteins were up-regulated over 2-fold (Fig. 2), such as fibroblast growth factor 7, superior cervical ganglion 10, and nerve growth factor-inducible protein.
Tyrosine kinases and tyrosine phosphorylation play important roles in virtually every function of neurons, including survival, extension of axons, and synapse formation. In agreement, we found that various tyrosine kinases and tyrosine phosphotases were up-regulated (Fig. 2). Moreover, up-regulations of actin, tubulin, and actin-associated profilin II and LIM proteins (Fig. 2) were observed. Taken together, our results reveal dynamic changes of gene expression ranging from transcriptional and translational factors to structural proteins.
Alteration of Neurotransmission.
The physiological change accompanying neuropathic pain is the spontaneous firing of primary afferent fibers, which is thought to be the result of a long-lasting alteration in neurotransmitters, receptors, ion channels, and related molecules. We observed that 50% of the strongly regulated genes were related to neurotransmission, including neuropeptides, receptors, channels, synaptic proteins, and signal transduction molecules after axotomy (Fig. 2; Tables 2–5), indicating that the physiological changes may result from the gross changes in multiple genes. The regulated expression levels of 80% of these genes were maintained over the time course, indicating their involvement in the maintenance of the pain.
Interestingly, almost all strongly regulated neuropeptides have been identified (3), which further validates our approach. Importantly, we observed that chromogranin A (CGA) was strongly down-regulated (Fig. 2; Table 4). CGA is a critical molecule for controlling dense-core secretory vesicle biogenesis (23). The decrease in CGA may result in a decrease in biogenesis of dense-core vesicles, which may be an important reason for the described sorting NPY into the constitutive secretory pathway in axotomized DRG neurons (24).
We also found a strong down-regulation of SNAP-25, VAMP-1, and Rab3A (Fig. 2; Tables 4 and 5), indicating that the docking and fusion of synaptic vesicles may be attenuated. Synaptotagmin IV was the only synaptic vesicle protein that was up-regulated (Fig. 2; Table 4). Synaptotagmin IV can form heterooligomers with synaptotagmin I, resulting in less efficiency at coupling Ca2+ to secretion that decreases evoked synaptic transmission (25). Thus, the change in the expression pattern of synaptic transmission-related proteins seems to generally work toward a decrease of synaptic transmission. However, further studies are needed to determine whether the changes of synaptic vesicle protein help to enhance or modulate synaptic transmission in a subset of neurons after axotomy.
The present study shows that G protein-coupled receptors were a major group of the regulated receptors (Fig. 2). In addition to the down-regulation of the NPY Y1-R (14) and μ-opioid receptor (6), and up-regulation of the NPY Y2-R, cholecystokinin B receptor (3), and A-R α2A (8), we further observed that the metabotropic glutamate receptor 4 was down-regulated (Fig. 2), whereas P2Y1-R (Figs. 2, 4, and 5 A–A" and G) and NPY Y5-R (Figs. 2 and 4) were up-regulated. More importantly, we found that some receptor subunits related to ion channels were strongly up-regulated (Figs. 2 and 5), such as GABAA-R α5 in both small and large NPs, PBDZ-R in many small NPs and some large NPs, and nicotinic acetylcholine-R α7 subunit in many large NPs and some small NPs. With regard to the ion channels, we observed a transient up-regulation of L-Ca2+ Ch α2δ-1 (Figs. 2, 4, and 5 E–E" and G) and an increase of Na+ Ch β2 (Figs. 2, 4, and 5 F–F" and G) in many small NPs.
It has been shown that cholecystokinin B receptor and NPY Y2-R mediate an increase in excitability of axotomized neurons (7, 9), and that A-R α2A contributes to neuropathic heat hyperalgesia (26) in addition to being involved in sympathetically maintained pain (27). P2Y1-R participates in potentiation of capsaicin receptor-mediated thermal hyperalgesia (28). The L-Ca2+ Ch is functionally involved in the exocytotic machinery and pain responses (29, 30). It contains three subunits: the α1 channel-forming subunit, the intracellular β subunit, and the α2 subunit consisting of two disulfide-lined polypeptides (α2 and δ). α2δ-1 is thought to increase the current amplitude and activation rate of Ca2+ Ch. The transient increase in L-Ca2+ Ch α2δ-1 in small neurons implies an effect on the increased excitability during the early postlesion period. A similar increase in L-Ca2+ Ch α2δ-1 is also shown with Western blot in nerve ligation and crush models (31). Up-regulation of Na+ Ch β2 may increase the cell surface Na+ Ch α subunits in small neurons, because the Na+ Ch β2 has been shown to target Na+ Ch α subunits to the cell surface (32). Thus, L-Ca2+ Ch α2δ-1, Na+ Ch III (33) and Na+ Ch β2 may also contribute to an increase in excitability. K+ currents are involved in the generation of the action potential, opposing the current flow through Na+ and Ca2+ Chs and thus decrease excitability (34). Therefore, the down-regulation of K+ Chs (Fig. 2) presumably reduces the inhibitory K+ current. These changes and decrease in inhibitory μ-opioid receptors (6) and NPY Y1-Rs (14) may contribute to a reduction in inhibition after nerve injury. These results indicate that changes in G protein-coupled receptors and ion channels generally work toward generating hypersensitivity of the neurons.
GABA is a major inhibitory neurotransmitter. GABAA-R, known as the central BDZ-R, is a GABA/drug receptor-Cl− Ch macromolecular complex including binding sites for GABA and BDZs (35). BDZs increase the affinity of the GABAA-R for GABA and the frequency of opening of the Cl− Ch. Up-regulation of GABAA-R α5 may lead to a conformation of the receptor containing α5 (36). Further studies will evaluate its role in the enhancement of GABAA-R-mediated conductances in axotomized neurons (37). Another major BDZ-R, the PBDZ-R, is normally expressed in glial cells in the brain (38). Unexpectedly, we found that PBDZ-R was expressed in axotomized small DRG neurons, which presumably significantly increases BDZ-binding sites on the neurons. However, it seems that availability of selective agonists for GABA and BDZ receptors at the spinal level after nerve injury is reduced, because GABA is decreased in the dorsal horn (39), and thus far there is no evidence for presence of an endogenous agonist for PBDZ-R at the spinal level. Thus, administration of exogenous ligands may activate an inhibitory mechanism.
Clinical evidence indicates that antidepressants such as amitriptyline, antianxiety drugs such as diazepam and midazolam, and anticonvulsants such as gabapentin and carbamazepine represent useful therapies for certain types of neuropathic pain (40, 41). Pharmalogical studies show that amitriptyline acts at Na+ Ch, diazepam and midazolam at BDZ-Rs, gabapentin at GABAA-R and L-Ca2+ Ch α2δ-1 (42), and carbamazepine at Na+ Ch (43). Our present findings show that these receptors and ion channels are strongly up-regulated. In fact, midazolam reduces C fiber-evoked firing after nerve injury (44), suggesting that agonists for these correlated receptors may be effective in reducing excitability of DRG neurons. Moreover, up-regulation of both GABAA-R α5 and L-Ca2+ Ch α2δ-1 supports a therapeutic role of gabapentin (42). Up-regulation of nicotinic acetylcholine-R α7 subunit also suggests a potential drug target, because α7 subunits are thought to form homomeric nicotinic acetylcholine-R in peripheral nervous system and may mediate antinociception (45). Our results raise the possibility that drugs designed to act specifically at these regulated subunits of the receptors and ion channels may be more effective in treating neuropathic pain with few side effects.
Conclusion
In peripherally axotomized rat DRGs, we identified marked changes in the expression of 173 genes, which encompass a large number of distinct family members including neuropeptides, receptors, ion channels, signal transduction molecules, synaptic vesicle proteins, and others. These results suggest that cascades of gene expression may be a prerequisite for the development and maintenance of neuropathic pain.
Supplementary Material
Acknowledgments
We thank W.-Q. Xu, P. Zhang, Y.-T. Xue, Y.-G. Tong, B.-F. Chen, X.-Y. Jiao, M. Zhong, X.-R. Xu, and N.-G. Li for technical assistance and Z.-F. Chen for discussion. This work was supported by 973 Program Grants G2000077800 and G1998091005 and by National Science Foundation China Grants 39840160 and 39525010.
Abbreviations
- DRG
dorsal root ganglion
- Lib
library
- L
lumbar
- L-Ca2+ Ch α2δ-1
L-type Ca2+ channel α2δ-1 subunit
- GABAA-R α5
γ-aminobutyric acidA receptor α5 subunit
- PBDZ-R
peripheral benzodiazepine receptor
- P2Y1-R
P2Y1 purinoceptor
- Na+ Ch β2
Na+ channel β2 subunit
- NPY
neuropeptide Y
- EST
expression sequence tag
- NP
neuronal profile
- RT
reverse transcription
- nVol
normalized volume
Footnotes
References
- 1.Wall P D, Waxman S, Basbaum A I. Exp Neurol. 1974;45:576–589. doi: 10.1016/0014-4886(74)90163-0. [DOI] [PubMed] [Google Scholar]
- 2.Shehab S A, Atkinson M E. Brain Res. 1986;372:37–44. doi: 10.1016/0006-8993(86)91456-3. [DOI] [PubMed] [Google Scholar]
- 3.Hökfelt T, Zhang X, Wiesenfeld-Hallin Z. Trends Neurosci. 1994;17:22–30. doi: 10.1016/0166-2236(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 4.Dray A, Urban L, Dickenson A. Trends Pharmacol Sci. 1994;15:190–197. doi: 10.1016/0165-6147(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 5.Woolf C J, Salter M W. Science. 2000;288:1765–1769. doi: 10.1126/science.288.5472.1765. [DOI] [PubMed] [Google Scholar]
- 6.deGroot J F, Coggeshall R E, Carlton S M. Neurosci Lett. 1997;233:113–116. doi: 10.1016/s0304-3940(97)00642-3. [DOI] [PubMed] [Google Scholar]
- 7.Antunes Bras J M, Laporte A M, Benoliel J J, Bourgoin S, Mauborgne A, Hamon M, Cesselin F, Pohl M. J Neurochem. 1999;72:858–867. doi: 10.1046/j.1471-4159.1999.720858.x. [DOI] [PubMed] [Google Scholar]
- 8.Shi T S, Winzer-Serhan U, Leslie F, Hökfelt T. Pain. 2000;84:319–330. doi: 10.1016/s0304-3959(99)00224-9. [DOI] [PubMed] [Google Scholar]
- 9.Abdulla F A, Smith P A. Neuroscience. 1999;89:43–60. doi: 10.1016/s0306-4522(98)00443-6. [DOI] [PubMed] [Google Scholar]
- 10.Black J A, Cummins T R, Plumpton C, Chen Y H, Hormuzdiar W, Clare J J, Waxman S G. J Neurophysiol. 1999;82:2776–2785. doi: 10.1152/jn.1999.82.5.2776. [DOI] [PubMed] [Google Scholar]
- 11.Bennett D L, Boucher T J, Armanini M P, Poulsen K T, Michael G J, Priestley J V, Phillips H S, McMahon S B, Shelton D L. J Neurosci. 2000;20:427–437. doi: 10.1523/JNEUROSCI.20-01-00427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cummins T R, Black J A, Dib-Hajj S D, Waxman S G. J Neurosci. 2000;20:8754–8761. doi: 10.1523/JNEUROSCI.20-23-08754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young R A. Cell. 2000;102:9–15. doi: 10.1016/s0092-8674(00)00005-2. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Wiesenfeld-Hallin Z, Hökfelt T. Eur J Neurosci. 1994;6:43–57. doi: 10.1111/j.1460-9568.1994.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 15.Mody M, Cao Y, Cui Z, Tay K Y, Shyong A, Shimizu E, Pham K, Schultz P, Welsh D, Tsien J Z. Proc Natl Acad Sci USA. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradbury E J, McMahon S B, Ramer M S. Trends Pharmacol Sci. 2000;21:389–394. doi: 10.1016/s0165-6147(00)01536-4. [DOI] [PubMed] [Google Scholar]
- 17.Lewis S E, Mannion R J, White F A, Coggeshall R E, Beggs S, Costigan M, Martin J L, Dillmann W H, Woolf C J. J Neurosci. 1999;19:8945–8953. doi: 10.1523/JNEUROSCI.19-20-08945.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herdegen T, Fiallos-Estrada C E, Schmid W, Bravo R, Zimmermann M. Mol Brain Res. 1992;14:155–165. doi: 10.1016/0169-328x(92)90170-g. [DOI] [PubMed] [Google Scholar]
- 19.Frederickson R M, Mushynski W E, Sonenberg N. Mol Cell Biol. 1992;12:1239–1247. doi: 10.1128/mcb.12.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tonra J R, Curtis R, Wong V, Cliffer K D, Park J S, Timmes A, Nguyen T, Lindsay R M, Acheson A, DiStefano P S. J Neurosci. 1998;18:4374–4383. doi: 10.1523/JNEUROSCI.18-11-04374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boucher T J, Okuse K, Bennett D L, Munson J B, Wood J N, McMahon S B. Science. 2000;290:124–127. doi: 10.1126/science.290.5489.124. [DOI] [PubMed] [Google Scholar]
- 22.Bennett D L, Michael G J, Ramachandran N, Munson J B, Averill S, Yan Q, McMahon S B, Priestley J V. J Neurosci. 1998;18:3059–3072. doi: 10.1523/JNEUROSCI.18-08-03059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim T, Tao-Cheng J H, Eiden L E, Loh Y P. Cell. 2001;106:499–509. doi: 10.1016/s0092-8674(01)00459-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Aman K, Hökfelt T. J Comp Neurol. 1995;352:481–500. doi: 10.1002/cne.903520402. [DOI] [PubMed] [Google Scholar]
- 25.Littleton J T, Serano T L, Rubin G M, Ganetzky B, Chapman E R. Nature (London) 1999;400:757–760. doi: 10.1038/23462. [DOI] [PubMed] [Google Scholar]
- 26.Kingery W S, Guo T Z, Davies M F, Limbird L, Maze M. Pain. 2000;85:345–358. doi: 10.1016/S0304-3959(99)00286-9. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Michaelis M, Janig W, Devor M. J Neurophysiol. 1996;76:3721–3730. doi: 10.1152/jn.1996.76.6.3721. [DOI] [PubMed] [Google Scholar]
- 28.Tominaga M, Wada M, Masu M. Proc Natl Acad Sci USA. 2001;98:6951–6956. doi: 10.1073/pnas.111025298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiser O, Trus M, Hernandez A, Renstrom E, Barg S, Rorsman P, Atlas D. Proc Natl Acad Sci USA. 1999;96:248–253. doi: 10.1073/pnas.96.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saegusa H, Kurihara T, Zong S, Minowa O, Kazuno A, Han W, Matsuda Y, Yamanaka H, Osanai M, Noda T, Tanabe T. Proc Natl Acad Sci USA. 2000;97:6132–6137. doi: 10.1073/pnas.100124197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Z D, Chaplan S R, Higuera E S, Sorkin L S, Stauderman K A, Williams M E, Yaksh T L. J Neurosci. 2001;21:1868–1875. doi: 10.1523/JNEUROSCI.21-06-01868.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catterall W A. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 33.Waxman S G, Dib-Hajj S, Cummins T R, Black J A. Proc Natl Acad Sci USA. 1999;96:7635–7639. doi: 10.1073/pnas.96.14.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudy B. Neuroscience. 1988;25:729–749. doi: 10.1016/0306-4522(88)90033-4. [DOI] [PubMed] [Google Scholar]
- 35.Smith T A. Br J Biomed Sci. 2001;58:111–121. [PubMed] [Google Scholar]
- 36.Pritchett D B, Seeburg P H. J Neurochem. 1990;54:1802–1804. doi: 10.1111/j.1471-4159.1990.tb01237.x. [DOI] [PubMed] [Google Scholar]
- 37.Oyelese A A, Eng D L, Richerson G B, Kocsis J D. J Neurophysiol. 1995;74:673–683. doi: 10.1152/jn.1995.74.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woods M J, Williams D C. Biochem Pharmacol. 1996;52:1805–1814. doi: 10.1016/s0006-2952(96)00558-8. [DOI] [PubMed] [Google Scholar]
- 39.Ibuki T, Hama A T, Wang X T, Pappas G D, Sagen J. Neuroscience. 1997;76:845–858. doi: 10.1016/s0306-4522(96)00341-7. [DOI] [PubMed] [Google Scholar]
- 40.Backonja M M. Clin J Pain. 2000;16:S67–S72. doi: 10.1097/00002508-200006001-00012. [DOI] [PubMed] [Google Scholar]
- 41.Watson C P. Clin J Pain. 2000;16:S49–S55. doi: 10.1097/00002508-200006001-00009. [DOI] [PubMed] [Google Scholar]
- 42.Gee N S, Brown J P, Dissanayake V U, Offord J, Thurlow R, Woodruff G N. J Biol Chem. 1996;271:5768–5776. doi: 10.1074/jbc.271.10.5768. [DOI] [PubMed] [Google Scholar]
- 43.Brau M E, Dreimann M, Olschewski A, Vogel W, Hempelmann G. Anesthesiology. 2001;94:137–144. doi: 10.1097/00000542-200101000-00024. [DOI] [PubMed] [Google Scholar]
- 44.Kontinen V K, Dickenson A H. Pain. 2000;85:425–431. doi: 10.1016/S0304-3959(99)00298-5. [DOI] [PubMed] [Google Scholar]
- 45.Damaj M I, Meyer E M, Martin B R. Neuropharmacology. 2000;39:2785–2791. doi: 10.1016/s0028-3908(00)00139-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.