Summary
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), has caused >770 million infections since 2020. SARS-CoV-2 outcomes are largely influenced by host immune responses. Among key mediators of innate immunity, high-mobility group box 1 (HMGB1) has gained attention for its role in inflammation during SARS-CoV-2 infection, and its levels are significantly elevated in acute and post-COVID-19 cases, correlating with disease severity. This study investigated the role of HMGB1 in COVID-19 pathogenesis. Our findings demonstrate that the SARS-CoV-2 spike protein directly interacts with HMGB1, forming an HMGB1–SARS-CoV-2 complex. This complex interacts with the receptor for advanced glycation end-products (RAGE), facilitating clathrin-mediated endocytosis and enhancing SARS-CoV-2 infection in human lung cells in vitro and in mouse models of infection. Overall, this study demonstrates the role of HMGB1 in promoting viral entry via RAGE, emphasizing its potential as a therapeutic target in severe COVID-19 cases.
Subject areas: Immunology, Virology, Cell biology
Graphical abstract

Highlights
-
•
HMGB1 directly binds to SARS-CoV-2 spike protein
-
•
The HMGB1-SARS-CoV-2 complex interacts with RAGE
-
•
SARS-CoV-2 can infect host cells via RAGE-mediated endocytosis
Immunology; Virology; Cell biology
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19),1,2 has caused more than 770 million infections since 2020.3 SARS-CoV-2 infection results in broad-spectrum outcomes ranging from mild symptoms to severe respiratory failure and even death.4 Similar to other viral infections, a key factor influencing the variability in COVID-19 severity is the host immune response. Immune-related cytokines, including IL-6 and TNF-α, have gained significant attention.5 Among these, high-mobility group box 1 (HMGB1) has emerged as one of the most extensively studied mediators of the inflammatory response during SARS-CoV-2 infection,6,7 and its serum levels are reported to be significantly higher in patients with severe symptoms and fatal outcomes.8
HMGB1 is a well-characterized damage-associated molecular pattern molecule released during cellular stress and is involved in regulating inflammation and immune responses.9 The structure of HMGB1 is highly conserved and comprises 215 amino acid (aa) residues forming two homologous DNA-binding domains—box A (aa 1–79) and box B (aa 89–162)—as well as a C-terminal acidic tail domain (aa 186–215).10,11 Box B plays a crucial role in pro-inflammatory activities and interacts with Toll-like receptors (TLRs),12 whereas box A interacts with box B and the C-terminal acidic tail to inhibit their inflammatory functions.13 Residues 150–183 are known to interact with the receptor for advanced glycation end-products (RAGE)14; further, post-translational modifications of HMGB1 are critical for regulating its roles in nuclear-cytoplasmic translocation and extracellular secretion.9,15,16,17,18 HMGB1 endocytosis via RAGE has been observed in macrophages, where it induces pyroptosis during endotoxemia.19 Furthermore, extracellular HMGB1 binds to pro-inflammatory mediators such as lipopolysaccharide (LPS),20 and the resulting HMGB1–LPS complexes bind to RAGE on the cell surface, facilitating endocytosis into the endolysosomal compartment.19 Within this compartment, HMGB1 permeabilizes the lysosomal membrane under acidic conditions, ultimately mediating pyroptosis.21 SARS-CoV-2 infection of host cells occurs primarily through interaction with specific cell surface receptors, such as angiotensin-converting enzyme 2 (ACE2).22,23 Although ACE2 is the most extensively studied receptor in SARS-CoV-2 infection,24,25,26 recent studies highlight that the virus can also infect cells and organs that do not express ACE2.27,28
Beyond its role in pathogen sensing and inflammatory signaling, HMGB1 has been implicated in the pathogenesis of various diseases, including sepsis,29 autoimmune disorders,30,31,32 and acute lung injury.33,34 Severe COVID-19 shares key pathological features with sepsis, including cytokine storm and endothelial dysfunction,7,35,36 suggesting a potential role for HMGB1 in COVID-19 progression. However, the specific role of HMGB1 following its release during SARS-CoV-2 infection remains unclear.
This study aimed to deepen our understanding of the role of HMGB1 in COVID-19 pathogenesis and its involvement in viral entry through its interaction with RAGE. HMGB1 binds to the spike protein of SARS-CoV-2, and the SARS-CoV-2–HMGB1 complex interacts with RAGE to promote infection via endocytosis. This study sought to determine the potential of HMGB1 as a therapeutic target and contribute to the discovery and development of effective treatment strategies for severe COVID-19.
Results
HMGB1 directly interacts with the S1 subunit of the SARS-CoV-2 spike protein
Our previous report showed that HMGB1 is passively released from cells undergoing PANoptosis during SARS-CoV-2 infection,37 prompting us to investigate its physiological role in the context of SARS-CoV-2 infection. Therefore, we aimed to determine whether HMGB1 binds to the SARS-CoV-2 spike protein. We extracted and purified full-length HMGB1 (HMGB1-WT), A box (HMGB1-A box), B box (HMGB1-B box), as well as acidic C-tail-deleted HMGB1 (HMGB1-ΔC) from E. coli (Figure 1A) and performed surface plasmon resonance (SPR) analysis. For this assay, spike protein was immobilized and various concentrations of HMGB1 domains were passed over the surface. The analysis showed that HMGB1 domains directly bound to the spike protein in a concentration-dependent manner, with HMGB1-WT showing an equilibrium dissociation constant (KD) of 3.553 μM. HMGB1-A box, HMGB1-B box, and HMGB1-ΔC also bound to the spike protein with KD values of 6.099 μM, 1.025 μM, and 1.381 μM, respectively (Figure 1B). The corresponding association (Ka) and dissociation (Kd) rate constants are provided (Table S1). Next, to determine whether HMGB1 binds specifically to the S1 subunit of the spike protein, we performed an immunoprecipitation (IP) assay. HMGB1-WT and HMGB1-ΔC, both containing the RAGE-binding site at aa 150–183, exhibited strong interactions with the S1 subunit compared to those of the other domains. The acidic C-tail region appeared to have no significant influence on the interaction between HMGB1 and the S1 subunit. The HMGB1-A and -B boxes showed weak binding with S1 (Figure 1C).
Figure 1.
HMGB1 directly interacts with the S1 subunit of the SARS-CoV-2 spike protein
(A) Various HMGB1 domains were purified and (B) direct binding of HMGB1 domains to the spike protein was detected using SPR. Spike protein was immobilized on a CM5 chip, and each HMGB1 analyte was added. KD values were measured using the Biacore T200 evaluation software. Also see Table S1. HMGB1 domains were incubated with the S1-subunit of spike protein for 1 h at 37°C. Recombinant protein concentration was calculated according to its molecular mass. Immunoprecipitation (C) between the S1 subunit and HMGB1 domains was performed (left panel), and non-specific binding of the HMGB1 protein molecules to the beads was observed as a control (right panel). HMGB1, high-mobility group box 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SPR, surface plasmon resonance.
High mobility group box 1 enhances SARS-CoV-2 infectivity
Next, we investigated the effects of HMGB1 on viral infectivity using A549, a lung-derived adenocarcinoma cell line that expresses ACE2 at low levels.38 After excluding the possibility of HMGB1 binding to ACE2 (Figure 2A), HMGB1 was incubated with SARS-CoV-2 in vitro before infection. The addition of high concentrations of HMGB1 significantly increased the SARS-CoV-2 nucleocapsid protein (NP) levels and viral load 3 h post-infection (hpi) (Figures 2B and 2C). To determine whether HMGB1 functions during viral entry rather than post-entry, we assessed NP levels at 1 hpi (Figure S1). The results were consistent with those observed at 3 hpi, indicating that HMGB1 likely acts during the early stages of infection. Furthermore, immunofluorescence staining of A549 cells showed a significant increase in NP levels and in the total number of infected cells in the presence of high HMGB1 concentrations (Figure 2D). Early detection of NP by western blotting and immunofluorescence staining following SARS-CoV-2 infection is consistent with previous reports.39,40
Figure 2.
HMGB1 enhances SARS-CoV-2 Infectivity
(A) hACE2 and HMGB1 were incubated for 1 h at 37°C. hACE2 was immunoprecipitated in vitro, and the presence of HMGB1 was assessed via western blotting.
(B and C) A549 cells were infected for 1 h with 1 MOI SARS-CoV-2, which was preincubated with HMGB1 for 1 h at 37°C, cultured further for 3 h, and subjected to western blotting (B) and qRT-PCR for viral RNA measurement (C) n = 3. S.E., short exposure; L.E., long exposure. Also see Figure S1.
(D) A549 cells were infected with 5 MOI SARS-CoV-2 as above, fixed, and stained with anti-NP for confocal imaging. Representative images are shown. The percentage of infected cells and NP intensity were measured by counting at least 200 visible cells. n = 4.
(E and F) A549 cells were infected with 1 MOI SARS-CoV-2 preincubated with various HMGB1 domains and were harvested for western blotting, quantification (E) n = 3, and qRT-PCR for viral RNA measurement (F) n = 2.
(G) A549 cells were infected with 5 MOI SARS-CoV-2 preincubated with various HMGB1 domains. Cells were fixed, permeabilized, and stained with anti-NP for confocal imaging. Representative images are shown. The percentage of infected cells and NP intensity were measured by counting at least 200 visible cells. n = 4. Scale bars represent 5 μm. Also see Figure S2. Data are presented as mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant, using one-way ANOVA with Tukey’s multiple comparison test. HMGB1, high-mobility group box 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; hACE2, human angiotensin-converting enzyme 2; MOI, multiplicity of infection; qRT-PCR, quantitative reverse transcription polymerase chain reaction; NP, nucleocapsid protein; SEM, standard error of the mean; ANOVA, analysis of variance.
To exclude the possibility that HMGB1 induces SARS-CoV-2 replication independently of SARS-CoV-2–HMGB1 complex formation, we pretreated NCI-H1975 cells—a lung-derived adenocarcinoma cell line with no detectable ACE2 expression38,40—with HMGB1, followed by extensive washing before infection. Concurrently, cells infected with the virus preincubated with HMGB1 showed increased NP levels, whereas HMGB1-pretreated cells exhibited NP levels comparable to those in untreated infected controls (Figure S2). These results indicate that the increased infectivity is specifically dependent on the formation of SARS-CoV-2–HMGB1 complexes.
To further assess the effects of different HMGB1 mutant domains on SARS-CoV-2 infectivity, we preincubated SARS-CoV-2 with the HMGB1-A box, -B box, and -ΔC domains. These recombinant proteins were used at an equivalent molarity and were preincubated with SARS-CoV-2 under identical conditions. Interestingly, the mutant HMGB1 proteins exhibited varying effects on SARS-CoV-2 infectivity. HMGB1-WT and HMGB1-ΔC, both of which contain the known RAGE-binding site (Figure 1A), enhanced viral infectivity, whereas the HMGB1-A box and -B box mutants had a negative effect on infectivity (Figures 2E–2G). These findings suggest that the HMGB1-mediated increase in SARS-CoV-2 infection is dependent on the presence of the RAGE-binding site, suggesting the potential involvement of the HMGB1–RAGE interaction during SARS-CoV-2 infection.
High mobility group box 1 enhances SARS-CoV-2 infection in a advanced glycation end-product-dependent manner
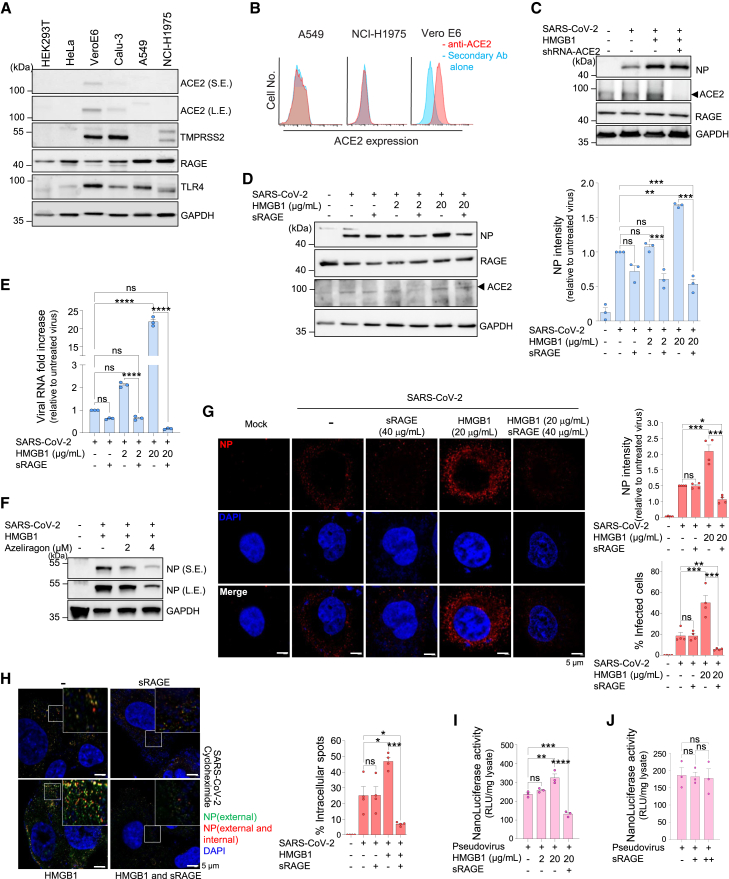
To investigate the mechanism by which the SARS-CoV-2–HMGB1 complex enhances infectivity, we focused on RAGE. For this purpose, cells with high RAGE expression but low or no ACE2 expression were selected. Vero E6 and Calu-3 cells expressed moderate levels of ACE2 and TMPRSS2, while HEK293T and HeLa cells did not. Consistent with previous reports,38,40 A549 and NCI-H1975 cells exhibited low and undetectable ACE2 expression levels, respectively. (Figure 3A). Surface expression was evaluated using flow cytometry, which confirmed that ACE2 expression in A549 and NCI-H1975 cells was undetectable (Figure 3B). Moreover, A549 and NCI-H1975 cells expressed high levels of RAGE, as previously reported,41,42 and were therefore selected for our study. To verify whether the increased SARS-CoV-2 infectivity in the presence of HMGB1 was ACE2-independent, ACE2 expression was knocked down using shRNA-ACE2 transfection. Incubation with HMGB1 before infection significantly enhanced SARS-CoV-2 infectivity, while successful ACE2 knockdown in A549 cells did not result in a significant change in infectivity (Figure 3C). Collectively, these results indicate that HMGB1-mediated SARS-CoV-2 infection occurs independently of ACE2 expression.
Figure 3.
HMGB1 induces SARS-CoV-2 infection in an ACE2-independent RAGE-dependent manner
(A) Western blot analysis of receptors responsible for SARS-CoV-2 infection and HMGB1 binding. S.E., short exposure; L.E., long exposure.
(B) Flow cytometry analysis of ectodomain ACE2 in A549 and NCI-H1975 cells used in this study. Vero E6 cells were used as control.
(C) A549 cells were transfected with shRNA-ACE2 for 48 h prior to infection with 1 MOI SARS-CoV-2, which was preincubated with HMGB1 for 1 h at 37°C, cultured further for 3 h, and subjected to western blotting.
(D and E) A549 cells were infected for 1 h with 1 MOI SARS-CoV-2, which was preincubated with HMGB1 for 1 h, in the presence of 40 μg/mL sRAGE at 37°C as indicated. Cells were harvested at 3 hpi and subjected to western blotting (D) n = 3, and qRT-PCR for viral RNA measurement (E) n = 3.
(F) NCI-H1975 cells were infected with SARS-CoV-2 preincubated with 20 μg/mL HMGB1 in the presence of azeliragon. Cells were harvested at 3 hpi and subjected to western blotting.
(G) A549 cells were infected with 5 MOI SARS-CoV-2, which was treated as above to observe NP. Representative confocal images are shown. The percentage of infected cells and NP intensity were measured by counting at least 700 visible cells. n = 4.
(H) Cycloheximide pretreated NCI-H1975 cells were infected with 5 MOI SARS-CoV-2 (preincubated with HMGB1). Cells were stained for NP before permeabilization (green; external) and post-permeabilization (red; external and internal). Representative images and their magnifications are shown. The percentage of intracellular spots was measured by counting at least 200 visible cells. n = 4.
(I and J) SARS2pp was preincubated with or without HMGB1 in the presence of sRAGE before transduction in NCI-H1975 cells. NanoLuc luciferase activity was measured 72 h post-transduction. n = 3 Scale bars represent 5 μm. Data are presented as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant, using one-way ANOVA with Tukey’s multiple comparison test and Student’s unpaired t-test. HMGB1, high-mobility group box 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MOI, multiplicity of infection; qRT-PCR, quantitative reverse transcription polymerase chain reaction; NP, nucleocapsid protein; SEM, standard error of the mean; ANOVA, analysis of variance; hpi, hours post-infection; RAGE, receptor for advanced glycation end-products; ACE2, angiotensin-converting enzyme 2; SARS2pp, SARS-CoV-2 spike protein (S)-pseudotyped retrovirus.
Soluble RAGE (sRAGE) binds RAGE ligands, preventing the ligands from binding to membrane-bound RAGE, thereby blocking RAGE signaling and inhibiting endocytosis.43,44 Infection of A549 cells with HMGB1-bound SARS-CoV-2 in the presence of sRAGE significantly hindered infectivity (Figures 3D and 3E), and treatment with azeliragon, a well-established RAGE antagonist,45,46,47 produced similar results to sRAGE in a dose-dependent manner (Figure 3F). Correspondingly, the increased SARS-CoV-2 NP levels and the percentage of cells infected with SARS-CoV-2–HMGB1 were significantly decreased in the presence of sRAGE, as determined using confocal imaging (Figure 3G).
To confirm these observations, we treated NCI-H1975 cells with cycloheximide, a eukaryotic translation inhibitor,48 followed by consecutive NP staining procedures to differentiate between internalized and extracellular SARS-CoV-2 viral particles. At 3 hpi, we observed an increase in the number of intracellular viral particles (stained red) in infected cells in the presence of HMGB1, whereas additional sRAGE treatment significantly decreased SARS-CoV-2 infectivity (Figure 3H). For verification, we used an MLV-based SARS-CoV-2 spike protein (S)-pseudotyped retrovirus (pseudovirus, SARS2pp) and compared its viral internalization patterns in NCI-H1975 cells. NanoLuc luciferase activity, indicative of viral entry, was greatly increased when SARS2pp was incubated with HMGB1 before transduction, whereas it was decreased with additional sRAGE treatment (Figure 3I). sRAGE alone did not affect SARS2pp internalization in the absence of HMGB1 (Figure 3J). Collectively, these findings suggest that HMGB1 binding to SARS-CoV-2 enhances viral infectivity via RAGE.
The SARS-CoV-2–high-mobility group box 1–receptor for advanced glycation end-products complex is required for endocytic viral entry
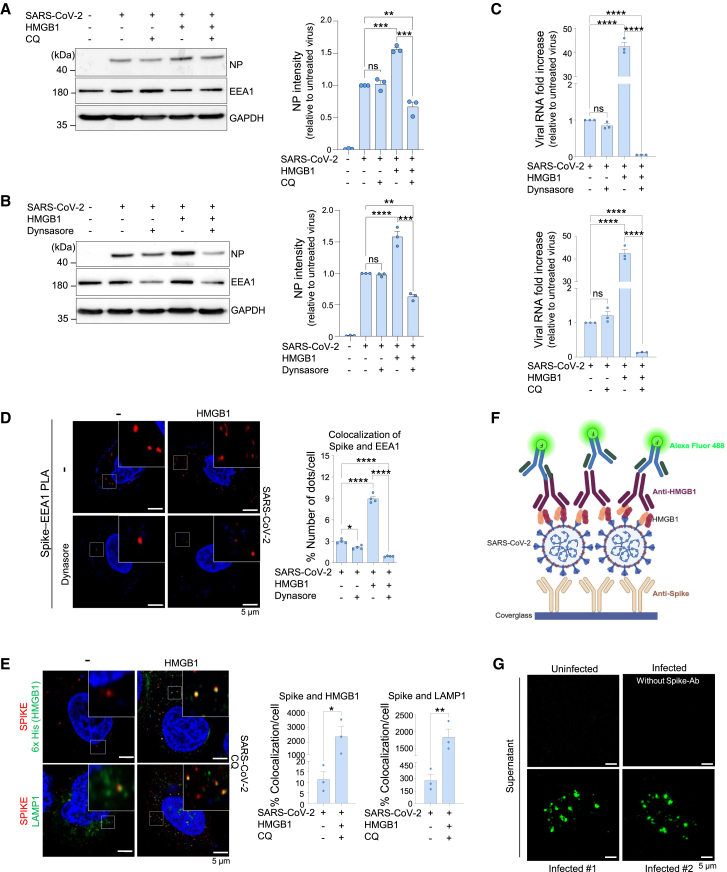
Extracellular HMGB1–LPS complexes bind to RAGE expressed on cell surfaces, enabling endocytosis in the endolysosomal compartment.19 To investigate whether HMGB1- and RAGE-dependent viral entry mechanisms were involved in endocytosis, we pretreated NCI-H1975 cells with chloroquine (CQ) and dynasore, which inhibit endosomal acidification and endocytosis, respectively. NCI-H1975 cells infected with SARS-CoV-2 in the presence of HMGB1, along with CQ and dynasore treatments, showed reduced viral NP levels in western blotting (Figures 4A and 4B) as well as reduced viral load (Figure 4C) compared to that in the untreated controls. TLRs (TLR2/4) are also known to interact with HMGB1; however, this interaction predominantly drives inflammatory responses rather than endocytosis.49
Figure 4.
The SARS-CoV-2–HMGB1–RAGE complex is required for endocytic viral entry
(A–C) NCI-H1975 cells were pretreated with either dynasore or chloroquine (CQ) and infected with 1 MOI SARS-CoV-2, which was preincubated with HMGB1 for 1 h at 37°C, cultured further for 3 h, and subjected to western blotting (A and B) n = 3, and qRT-PCR for viral RNA measurement (C) n = 3.
(D) NCI-H1975 cells were pretreated with dynasore and infected with 5 MOI SARS-CoV-2, preincubated with HMGB1, and analyzed using a proximity ligation assay. Representative images and their magnifications are shown with red dots indicating the colocalization of spike protein and EEA1. The percentage of the number of dots per cell was determined by counting at least 10 visible cells. n = 4.
(E) NCI-H1975 cells were pretreated with CQ and infected with 5 MOI SARS-CoV-2 (preincubated with HMGB1). Cells were stained for spike (SARS-CoV-2) and either for 6x His for HMGB1 or LAMP1. Representative images and their magnifications are shown. The percentage of colocalization per cell was determined by counting at least 30 visible cells. n = 3.
(F) Scheme of virus pull-down assay.
(G) Supernatants from infected cells were incubated on spike antibody-coated cover glass and subsequently stained for HMGB1. Green signals indicate SARS-CoV-2–HMGB1 complexes. Also see Figure S3n = 2. Scale bars represent 5 μm. Data are presented as the mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant, using one-way ANOVA with Tukey’s multiple comparison test and Student’s unpaired t-test. HMGB1, high-mobility group box 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; MOI, multiplicity of infection; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SEM, standard error of the mean; ANOVA, analysis of variance; RAGE, receptor for advanced glycation end-products.
To further study the role of HMGB1 in viral internalization, we performed a proximity ligase assay (PLA). Red signals represented the colocalization of EEA1 and the SARS-CoV-2 spike protein. The increased red dots in the presence of HMGB1 and the significant reduction in red dots with dynasore treatment further supported the increased endocytosis (Figure 4D). These results suggest that HMGB1-bound SARS-CoV-2 can be endocytosed via the RAGE-mediated pathway. An immunofluorescence assay was then performed to visualize the colocalization of HMGB1 and the SARS-CoV-2 spike protein within cells after CQ treatment. We observed yellow dots indicating the colocalization of HMGB1 and the spike protein, while the lysosomal marker LAMP1 showed increased colocalization with the spike protein in the presence of HMGB1 (Figure 4E). These results suggest that HMGB1 promotes viral internalization via endocytosis.
While virus internalization was detected in cells, to determine whether SARS-CoV-2–HMGB1 complexes form under biologically relevant conditions, we used supernatants from SARS-CoV-2-infected NCI-H1975 cells. These supernatants were incubated on spike antibody-coated coverglass, followed by incubation with an HMGB1-specific antibody for detection by confocal microscopy (Figure 4F). Green signals indicating the presence of SARS-CoV-2-HMGB1 complexes were detected in infected samples, whereas the signal was undetectable in the uninfected samples (Figure 4G). The corresponding controls have been added (Figure S3).
High-mobility group box 1 enhances SARS-CoV-2 infectivity in a murine model
We then assessed SARS-CoV-2 infectivity in the presence of HMGB1 in vivo. BALB/c mice, which are susceptible to infection with the B.1.351 variant,50,51 were selected for the experiment. Mice were infected intranasally with 2 × 105 plaque-forming units (PFUs) of the B.1.351 variant, which was incubated with 2 and 20 μg/mL of HMGB1 before infection. The lungs and trachea were collected at 24 and 48 hpi for plaque assays and immunohistochemistry (IHC) analyses (Figure 5A). In the presence of high-concentration (20 μg/mL) HMGB1, a significant increase in PFU was noted at 24 and 48 hpi (Figure 5B). IHC was performed on the left lobe and trachea to detect SARS-CoV-2 NP at 48 hpi. Tracheal respiratory epithelia and alveolar epithelia stained for SARS-CoV-2 NP showed a significant increase in the percentage of stained cells when the mice were infected with the virus incubated with a high concentration of HMGB1 (20 μg/mL) (Figures 5C and 5D). Both plaque assays and IHC yielded comparable results, demonstrating that HMGB1 presence is strongly associated with SARS-CoV-2 infection in vivo.
Figure 5.
HMGB1 enhances SARS-CoV-2 infectivity in a murine model
(A) Schematic overview of the timeline of the experiment. BALB/c mice were infected with SARS-CoV-2 (preincubated with HMGB1). The lungs and trachea were harvested at 24 and 48 hpi for (B) the plaque assay n = 4, and (C and D) immunohistochemistry analysis using anti-NP, anti-RAGE, anti-ACE2, and anti-HMGB1 antibodies. n = 3. Scale bars represent 100 μm. Data are presented as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, not significant, using one-way ANOVA with Tukey’s multiple comparison test. HMGB1, high-mobility group box 1; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SEM, standard error of the mean; ANOVA, analysis of variance; RAGE, receptor for advanced glycation end-products; ACE2, angiotensin-converting enzyme 2; hpi, hours post-infection.
Discussion
Our study revealed the molecular role of extracellular HMGB1 in SARS-CoV-2 infection. We observed a direct interaction between HMGB1 and the SARS-CoV-2 spike protein, which significantly increased viral infectivity in RAGE-expressing host cells with low to undetectable expression of ACE2. SARS-CoV-2 infection induces HMGB1 secretion via post-translational modifications and PANoptosis, resulting in its translocation from the nucleus to the cytoplasm, as observed in postmortem lung tissues.37 The elevated levels of HMGB1 and RAGE in the serum of severely infected individuals,52,53 along with the presence of SARS-CoV-2–HMGB1 complexes in infected supernatants, support their significant involvement in the microenvironment during infection. It is well known that HMGB1 plays a central role in multiple inflammatory and immune-mediated conditions. High levels of HMGB1 in these pathological states may predispose patients to enhanced SARS-CoV-2 susceptibility and severity. Elevated HMGB1 levels have also been reported in patients with severe COVID-19,52,54,55,56 and our findings raise the possibility that circulating HMGB1 may facilitate SARS-CoV-2 entry through RAGE-mediated endocytosis.
RAGE is one of the receptors for HMGB1. HMGB1 released from cells during infection binds to RAGE, and this receptor–ligand complex is internalized through clathrin-coated pits.19,57 While the A- and B-box domains of HMGB1 bind to the SARS-CoV-2 spike protein, their inability to interact and be endocytosed by RAGE results in reduced viral infectivity compared to full-form HMGB1. Although TLRs (such as TLR2 and TLR4) are also known to interact with HMGB1, these interactions primarily drive inflammatory responses rather than endocytosis.49 The colocalization of spike protein with the early endosomal marker EEA1 indicates that the SARS-CoV-2–HMGB1 complex utilizes clathrin-mediated endocytosis for cellular entry. Furthermore, treatment with sRAGE and azeliragon or dynasore, which blocks RAGE and inhibits endocytosis, respectively, effectively suppressed infectivity. These observations support the conclusion that HMGB1-mediated viral internalization occurs via RAGE. Following endocytosis, the virus may escape from the endosome through acidic lysis,58 a process potentially facilitated by the HMGB1-mediated permeabilization of the lysosomal membrane.21 This hypothesis is supported by the increased colocalization of the lysosomal marker LAMP1 with spike protein following CQ treatment. Notably, even in the absence of HMGB1, some colocalization of spike protein with EEA1 and LAMP1 was observed, suggesting that alternative endocytosis-mediated pathways for SARS-CoV-2 entry exist. Although direct binding of SARS-CoV-2 with RAGE has been reported to drive monocyte infection,59 our results demonstrate that viral infection via RAGE is significantly promoted in the presence of HMGB1 compared to the absence of HMGB1.
SARS-CoV-2 primarily utilizes two major routes of host cell entry: either through the cell-surface protease TMPRSS2 or through endocytosis triggered by cathepsin proteases.22,23 There have been reports of SARS-CoV-2 infecting cells that do not express ACE2.27,40,60 While SARS-CoV-2 may engage alternative receptors such as KREMEN1 and ASGR161 to facilitate infection, ACE2 knockdown does not affect HMGB1-mediated SARS-CoV-2 infection in human lung carcinoma cells, suggesting that this infection is independent of ACE2. While our findings demonstrate that HMGB1 binding to SARS-CoV-2 facilitates virus internalization through RAGE, this does not indicate the identification of a new primary receptor for SARS-CoV-2. Rather, HMGB1 appears to function as an adaptor molecule, particularly under conditions of low ACE2 expression, providing an accessory pathway that complements primary receptors. Targeting accessory pathways such as HMGB1–RAGE interactions could represent a complementary therapeutic strategy to mitigate viral entry and disease severity, especially in populations characterized by high systemic HMGB1 levels.
The HMGB1–RAGE signaling pathway disrupts the renin-angiotensin system by sustaining inflammation and oxidative stress, both key contributors to COVID-19-associated multi-organ injury,62 and the increasing prevalence of post-acute sequelae of SARS-CoV-2 (PASC or long COVID). Despite extensive research on the mechanism of SARS-CoV-2 infection and a decline in new COVID-19 cases, the increasing incidence of PASC among recovering patients has become a major concern. Baseline plasma HMGB1 levels at the final follow-up were reported to be significantly elevated in patients presenting with PASC symptoms.63 This finding indicates that HMGB1 may serve as a potential predictor of PASC risk and a target for therapeutic interventions.
Our study highlights the critical role of HMGB1 in SARS-CoV-2 infection and its potential as a therapeutic target to mitigate severe COVID-19 outcomes and long-term complications.
Limitations of the study
While in vitro immunoprecipitation shows a direct interaction between the spike protein and HMGB1, the specific binding site remains undefined and requires further investigation. In vivo models may not fully capture the complexity of COVID-19 in patients with diverse inflammatory symptoms. Further research is needed to evaluate viral strain variations, genetic backgrounds, and therapeutic implications, including the use of RAGE inhibitors in mouse models. While sex-specific effects were not the primary focus of this study, we acknowledge that the exclusive use of female subjects may limit the generalizability of our findings.
Resource availability
Lead contact
Requests for further information and resources should be directed to and will be fulfilled by the lead contact Jeon-Soo Shin (jsshin6203@yuhs.ac).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Data reported in this article will be shared by the lead contact upon request. This article does not report any original code. Additional information required to reanalyze the data reported in this article is available from the lead contact upon request.
Acknowledgments
This study was supported by grants from the National Research Foundation of Korea (NRF), funded by the Korean government (RS-2019-NR040072, RS-2022-NR068972, and RS-2021-NR057630), and the Research Center Program of the Institute for Basic Science (IBS) in Korea (IBS-R026-D1). We thank Professor Hye Ryun Kim from Yonsei University College of Medicine, Seoul, South Korea, for providing NCI-H1975 cells and Professor Hyun Soo Cho at Yonsei University, Seoul, South Korea, for the recombinant SARS-CoV-2 spike protein and S1 subunit. The diagrams were created in BioRender. Jung, S. (2025) https://BioRender.com/g8gedfu.
Author contributions
Conceptualization, S.F.J., M.S.K., and J.S.S.; Methodology, S.F.J. and M.S.K.; investigation, S.F.J., I.H.P., M.G.H., and S.Y.C.; formal analysis, S.F.J.; resources, S.F.J., M.S.K., I.H.P., H.Y.S., and J.W.O.; writing-original draft, S.F.J.; writing-review and editing, S.F.J., M.S.K., I.H.P., M.G.H., and J.S.S.; visualization, S.F.J.; supervision, J.S.S.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-SARS-CoV-2 NP | Sino Biological | Cat # 40143-R001; RRID: AB_2827974 Cat # 40143-MM08; RRID: AB_2827978 |
| Anti-ACE2 | Proteintech | Cat # 21115-1-AP; RRID: AB_10732845 |
| Anti-ACE2 | R&D Systems | Cat # MAB9331; RRID: AB_2242330 |
| Anti-RAGE | Santa Cruz | Cat # sc-365154; RRID: AB_10707685 |
| Anti-TMPRSS2 | Santa Cruz | Cat # sc-515727; RRID: AB_2892118 |
| Anti-HMGB1 | Abcam | Cat # ab18256; RRID: AB_444360 |
| Anti-EEA1 | CST | Cat # 3288; RRID: AB_2096811 |
| Anti-6x His | Abcam | Cat # ab9108; RRID: AB_307016 |
| Anti-GAPDH | Abfrontier | Cat # LF-PA0212; RRID: N/A |
| Anti-GAPDH | Abclonal | Cat # A19056; RRID: AB_2862549 |
| Anti-SARS-CoV-2 spike antibody | GeneTex | Cat # GTX632604; RRID: AB_2864418 |
| Anti-LAMP1 | Abcam | Cat # ab24170; RRID: AB_775978 |
| Alexa Fluor 594 | Invitrogen™ | Cat # A-11005; AB_2534073/A-11012; RRID: AB_2534079 |
| Alexa Fluor 488 | Invitrogen™ | Cat # A-11008; RRID: AB_143165/A-11001; RRID: AB_2534069 |
| horseradish peroxidase (HRP)-conjugated secondary antibodies | Jackson Immunoresearch | Cat # 111-035-003; RRID: AB_2313567 Cat # 115-035-003; RRID: AB_10015289 |
| Bacterial and virus strains | ||
| SARS-CoV-2 | NCCP | Cat # NCCP43326 |
| SARS2pp | Department of Biotechnology, Yonsei University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Spike protein and S1 subunit | Gift from Prof. Hyun Soo Cho | N/A |
| HMGB1-WT, -A and B box, ΔC domain | This study | N/A |
| hACE2 | Sigma-Aldrich | Cat # SAE0064 |
| Dynasore | Sigma-Aldrich | Cat # D7693 |
| Chloroquine | Sigma-Aldrich | Cat # C6628 |
| sRAGE | Sino Biological | Cat # 11629-HCCH |
| Azeliragon | Selleckchem | Cat # S6415 |
| BS3 | Sigma-Aldrich | Cat # S5799 |
| Critical commercial assays | ||
| Duolink in situ fluorescence kit | Sigma-Aldrich | Cat # DUO92101 |
| Nano-Glo® Luciferase Assay System | Promega | Cat #N1110 |
| Experimental models: Cell lines | ||
| A549 | ATCC | ATCC CCL-185 |
| NCI-H1975 | Gift from Prof. Hye Ryun Kim | N/A |
| HeLa | Korean Cell Line Bank | KLCB No. 10002 |
| HEK293T | ATCC | ATCC CRL-1573 |
| Calu-3 | Korean Cell Line Bank | KLCB No. 30055 |
| Vero E6 | ATCC | ATCC CRL-1586 |
| Experimental models: Organisms/strains | ||
| BALB/c | Orientbio Inc | N/A |
| Oligonucleotides | ||
| RdRp SARS-CoV-2 5′-GTGARATGGTCATGTGTGGCGG-3′ and 5′-CARATGTTAAASACACTATTAGCATA-3′ |
Macrogen | N/A |
| GAPDH 5′-TCT-TCTTGGGCTACACTGAGGAC-3′ and 5′-TGTTGCTGTAGCCGTATTCA-3′ |
Macrogen | N/A |
| Recombinant DNA | ||
| HMGB1- expressing vectors | Previous study from Ohet al.64 | N/A |
| shRNA-hACE2 | Yonsei Genomics Center System Biology Core | TRCN0000046693 |
| Software and algorithms | ||
| ImageQuant 800 | LAS | N/A |
| QuantStudio™ Real-Time PCR System | Applied Biosystems | N/A |
| FlowJo | BD Bioscience | V10.10 |
| ZEN Blue edition | Carl Zeiss | V3.0. |
| Prism 10 | GraphPad | V10.4.0 |
| Biacore™ T200 | Cytiva | N/A |
| BIA evaluation software | Cytiva | VT.200 |
| Confocal microscope | Carl Zeiss | LSM780, 980 |
| ImageJ | ImageJ | V1.54k |
| BD FACS Verse II system | BD | N/A |
Experimental model and study participant details
Animal model and cell lines
Seven-week-old female BALB/c mice were purchased from Orientbio Inc. (Seongnam-si, South Korea) and housed in groups of 4 in an animal BSL-3 facility, with controlled temperature and humidity. The animals were allowed to acclimate for 2 weeks before any treatment. Animal studies were approved and strictly conducted according to the protocols of the Institutional Animal Care and Use Committee of Yonsei University College of Medicine (2020-0227).
A549, Vero E6, HEK293T (CCL-185, CRL-1586, CRL-1573, ATCC), HeLa, Calu-3 (Korean Cell Line Bank, Seoul, South Korea), and NCI-H1975 (a gift from Professor Hye Ryun Kim at Yonsei University College of Medicine, Seoul, South Korea) cells were maintained at 37°C in a humidified atmosphere with 5% CO2. Vero E6, HEK293T, HeLa, and Calu-3 cells were cultured in Dulbecco’s modified Eagle’s medium with high glucose (DMEM; LM0001-05, Welgene, Gyeongsan-si, South Korea); NCI-H1975 cells were cultured in RPMI-1640 medium (10-040-CV, Corning, Glendale, AZ, USA); and A549 cells were cultured in HAM’s/F-12K nutrient mixture (LM010-03, Welgene). All media were supplemented with 10% heat-inactivated fetal bovine serum (FBS; 35-015-CV, Corning) and 100 U/mL penicillin-streptomycin (PS; 15140122, Gibco, Grand Island, NY, USA). All cell lines used in this study were routinely tested for mycoplasma contamination and were confirmed to be negative.
Method details
Cell transfection
Lipofectamine 2000 transfection reagent (52887, Invitrogen, Carlsbad, CA, USA) was used to perform shRNA transfection according to the manufacturer’s instructions. Human ACE2 shRNA plasmid (TRCN0000046693) produced at the BROAD Institute (Cambridge, MA, USA) was purchased from Yonsei Genomics Center System Biology Core (Yonsei University, Seoul, South Korea).
SARS-CoV-2 virus production and titration
All experiments involving SARS-CoV-2 were conducted in a BSL-3 facility in accordance with the institutional guidelines. SARS-CoV-2 B.1.351 strain BetaCoV/Korea/KCDC03/2020 NCCP43326 was obtained from the National Culture Collection for Pathogens (NCCP), Korea Disease Control and Preventive Agency (KDCA, Osong, South Korea). The virus was propagated in Vero E6 cells and titrated using a plaque assay before all experiments.
Virus infection
BALB/c mice were randomly divided into four groups (four mice/group). The treatment group mice were intranasally infected with 2 × 105 plaque-forming units (PFUs) of the SARS-CoV-2 B.1.351 strain, with or without HMGB1, and the control group was administered PBS. A total volume of 50 μL was administered intranasally under isoflurane anesthesia. Body weight was measured twice: before infection and at sacrifice. All groups were euthanized with CO2 at 24- or 48-h post-infection (hpi) to collect the lungs and trachea for plaque assays and IHC analyses. The left lung lobes were fixed in 4% paraformaldehyde and embedded in paraffin blocks to prepare formalin-fixed, paraffin-embedded (FFPE) slices.
For cell infection, A549 or NCI-H1975 cells were seeded at a density of 1.8 × 105 cells per well in 12-well plates and allowed to adhere overnight at 37°C. SARS-CoV-2 at a multiplicity of infection (MOI) of 1 was preincubated with HMGB1 at various concentrations (2, 20 μg/mL) for 1 h at 37°C. Cells were then infected with SARS-CoV-2 (control or pre-incubated) for 1 h at 37°C in serum-free media, with gentle rocking every 15 min, followed by removal of the virus and brief washing with warm Dulbecco’s phosphate-buffered saline (DPBS; LB001-02; Welgene). Fresh medium containing 10% FBS was added, and the cells were cultured for an additional 3 h unless otherwise specified, at 37°C before being harvested for western blotting and viral RNA quantification. 40 μg/mL sRAGE (11629-HCCH, Sino Biological, Oklahoma City, OK, USA), azeliragon (S6415, Selleckchem, Houston, TX, USA), dynasore (D7693, Sigma-Aldrich, St. Louis, MO, USA) and chloroquine (C6628, Sigma-Aldrich) were pretreated for 2 h at 37°C, prior to the infection procedure. For pretreatment with HMGB1, cells were incubated with 20 μg/mL HMGB1 for 1 h, followed by thorough washing with DPBS prior to infection with SARS-CoV-2.
Recombinant proteins
HMGB1 recombinant protein was expressed with vectors,64 extracted and purified as previously described.16 6 x His-tagged HMGB1-WT, HMGB1-A box (aa 1–79), HMGB1-B box (aa 89–162), and HMGB1-ΔC (aa 1–186) proteins were produced in Escherichia coli (E. coli) SoluBL21. Transformed SoluBL21 E. coli was treated with 0.5 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) and incubated for 18 h at 37°C. The recombinant proteins were purified using Ni2+-NTA and heparin columns, followed by gel filtration chromatography. Endotoxin detoxification was performed using the Triton X-114 method and verified using the Limulus amebocyte lysate (LAL) test. Recombinant human ACE2 protein was purchased (SAE0064, Sigma-Aldrich), and SARS-CoV-2 spike protein and S1 subunit were a gift from Professor Hyun Soo Cho at Yonsei University, Seoul, South Korea.
SDS-PAGE and western blot analysis
Cells were washed twice with DPBS, collected via centrifugation at 7,000 rpm for 5 min at 4°C, and lysed in RIPA buffer supplemented with a protease inhibitor cocktail (R4100-010, P3100, GenDEPOT, Baker, TX, USA) for 1 h with vigorous vortexing. Quantified samples in reducing sample buffer (100 mM Tris-HCl pH 6.8, 2% SDS, 25% glycerol, 0.1% bromophenol blue, and 5% β-mercaptoethanol) were separated via SDS-PAGE, transferred onto a nitrocellulose (NC) membrane (10600001, Cytiva, Wilmington, DE, USA), and blocked with 5% skim milk in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBST). The NC membranes were then incubated overnight at 4°C with primary antibodies. Antibodies against SARS-CoV-2 nucleocapsid (SARS-CoV-2 NP; 40143-R001, Sino Biological), ACE2 (21115-1 AP, Proteintech, St. Louis, MO, USA), RAGE (sc-365154, Santa Cruz, Dallas, TX, USA), TMPRSS2 (sc-515727, Santa Cruz), TLR4 (MA5-16216, Invitrogen), 6x His (ab9108; Abcam, Cambridge, UK), EEA1 (3288S, CST, Danvers, MA, USA), and GAPDH (LF-PA0212, Abfrontier, Seoul, South Korea/A19056, Abclonal, Woburn, MA, USA) were used at dilutions of 1:2,000–3,000. After incubation, the membranes were washed with 0.1% TBST and incubated for 1 h at room temperature (RT) with horseradish peroxidase (HRP)-conjugated secondary antibodies (111-035-003, 115-035-003, Jackson Immunoresearch, West Grove, PA, USA) at a dilution of 1:3,000. West-Q Pico ECL Solution (W3652-020, GenDEPOT) was used to visualize the membranes with ImageQuant 800 (LAS). Membrane stripping was performed using stripping buffer (BWS-0500; Biomax, Guri-si, South Korea).
Quantitative PCR
The level of viral replication was assessed using quantitative reverse transcription PCR (qRT-PCR). RNA was extracted using TRIzol (15596018, Thermo Fisher Scientific, Waltham, MA, USA). cDNA was synthesized using cDNA EcoDry Premix (639543, Takara Bio, San Jose, CA, USA) and subjected to qRT-PCR using Power SYBR Green Master Mix (4367659, Thermo Fisher Scientific). Reactions were performed in triplicate using the following primer sequences: RdRp SARS-CoV-2 5′-GTGARATGGTCATGTGTGGCGG-3′ and 5′-CARATGTTAAASACACTATTAGCATA-3′ (Macrogen, Seoul, South Korea); GAPDH, 5′-TCT-TCTTGGGCTACACTGAGGAC-3′ and 5′-TGTTGCTGTAGCCGTATTCA-3′. Amplification and quantitation were performed on the QuantStudio Real-Time PCR System (Applied Biosystems). GAPDH mRNA levels were used to correct and convert the Ct values to relative RNA levels.
In vitro immunoprecipitation
For immunoprecipitation, recombinant proteins were incubated for 1 h at 37°C. Simultaneously, Dynabeads protein G magnetic beads (10004D, Invitrogen) were incubated with an anti-SARS-CoV-2 spike antibody (GTX632604, GeneTex, San Antonio, TX, USA) for 1 h at RT. The antibody-bound beads were then incubated overnight at 4°C with the S1 subunit of SARS-CoV-2 spike protein and HMGB1-WT, -A box, -B box, and -ΔC. Following incubation, the beads were thoroughly washed with PBS containing 0.05% Tween 20 (PBST) and subsequently eluted with a reducing protein sample buffer for SDS-PAGE analysis.
Surface plasmon resonance
Direct binding of HMGB1 domains to the spike protein was analyzed using Biacore T200 (Cytiva). For this, a CM5 dextran sensor chip was used to immobilize spike protein at 10 μg/mL in 10 mM sodium acetate buffer (pH 4.0). To derive Rligand values, a coupling process was performed based on the molecular weights of the spike (MW ligand) and HMGB1 domains (MW analyte). MW analyte was diluted in HBS-EP buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, 0.05% Tween 20, pH 7.4) and passed over the sensor chip at a flow rate of 20 μL/min for 3 min. Sensorgrams were recorded and analyzed in real-time using the control software on a Biacore T200 system. All data were processed using the Biacore Evaluation Software (Cytiva).
Confocal microscopy
A549 or NCI-H1975 cells were seeded (6.0 × 104 and 3.0 × 104) onto 4-chamber and 8-chamber slides, respectively, and allowed to adhere overnight before treatment or infection. After infection, the cells were fixed with 4% paraformaldehyde for 30 min at RT and permeabilized with 1% Triton X-100 for 10 min at RT, followed by blocking with 1% BSA in PBST for 1 h at 37°C. The cells were incubated overnight at 4°C with antibodies against SARS-CoV-2 NP (40143-R001/40143-MM08, Sino Biological), SARS-CoV-2 spike (GTX632604, GeneTex), LAMP1 (ab24170, Abcam), and 6x His (ab9108, Abcam). Cells were then washed and incubated with the secondary antibodies tagged with Alexa Fluor 594 (A11005/A11012, Invitrogen) or Alexa Fluor 488 (A11008/A11001, Invitrogen) for 45 min at 37°C. DAPI (Vector Laboratories, Burlingame, CA, USA) was added, and the images were analyzed using a confocal microscope (LSM 780, 980, ZEISS, Oberkochen, Germany).
Immunofluorescence staining
SARS-CoV-2 internalization was observed using a previously described assay.40 Briefly, 6.0 x 104 NCI-H1975 cells were seeded onto 4-chamber slides and allowed to adhere overnight before treatment or infection. After infection, the cells were fixed with 4% paraformaldehyde, blocked with 1% BSA in PBST, and sequentially stained with an antibody against SARS-CoV-2 NP (40143-R001, Sino Biological) and an Alexa Fluor 488-conjugated secondary antibody (A11008, Invitrogen). On the next day, the cells were permeabilized with 1% Triton X-100, followed by repeated staining with the SARS-CoV-2 NP antibody (40143-R001, Sino Biological) overnight and then with an Alexa Fluor 594-conjugated antibody (A11005, Invitrogen). DAPI (Vector Laboratories) was added, and the images were analyzed using a confocal microscope (LSM 780, ZEISS). The percentage of intracellular spots was determined by counting four individual images with at least 200 visible cells. The ImageJ software was used to process the images and quantify the internalized red spots.
Visualization of SARS-CoV-2-HMGB1 complex was performed using an adapted assay.65 NCI-H1975 cells were infected with SARS-CoV-2, and supernatants were collected at 72 hpi. Crosslinking was done by adding 2 mM BS3 (S5799, Sigma-Aldrich) to the supernatants, followed by concentration using Amicon Ultra Centrifugal Filters (10 kDa MWCO; UFC5010, Sigma-Aldrich). The concentrated supernatants were incubated on anti-SARS-CoV-2 spike antibody (GTX632604, GeneTex)-coated coverglasses for 2 h at 37°C. After incubation and washing, immunofluorescence staining was performed as described above, using antibody against HMGB1 (ab18256, Abcam) and Alexa Fluor 488-conjugated secondary antibody (A11008, Invitrogen). Samples were analyzed by confocal microscopy (LSM 780, ZEISS).
Proximity ligation assay (PLA)
PLA was performed using the Duolink in situ fluorescence kit (DUO92101; Sigma-Aldrich). A total of 6.0 x 104 NCI-H1975 cells were seeded onto 4-chamber slides and pretreated with 30 μM dynasore before viral infection. After fixation and permeabilization, the cells were blocked with the blocking solution provided in the kit, followed by incubation overnight at 4°C with antibodies against EEA1 (3288S, CST) and SARS-CoV-2 spike (GTX632604, GeneTex), diluted in antibody diluent. Next, the cells were washed and incubated with the PLA probe mixture for 1 h and with the ligation solution for 30 min at 37°C. After a brief wash under gentle agitation, the cells were incubated in the amplification-polymerase solution for 100 min at 37°C. After a final round of washing, the cells were mounted with DAPI for imaging using a confocal microscope (LSM 980, ZEISS).
Pseudovirus assays
An MLV-based SARS-CoV-2 spike protein (S)-pseudotyped retrovirus (SARS2pp) was used in this study. For the virus internalization assay, NCI-H1975 cells were seeded at a density of 1 × 105 cells. SARS2pp was preincubated with HMGB1 in the presence or absence of 40 μg/mL sRAGE for 1 h at 37°C before transduction in serum-free RPMI 1640 medium. The culture medium was exchanged with 10% FBS-RPMI 1640 after 24 h of transduction, and the cells were further incubated for 48 h at 37°C. Cells were lysed directly with Glo Lysis Buffer (E2661, Promega, Madison, WI, USA) for the luciferase assay using Nano-Glo Luciferase Assay System (N1110, Promega).
Flow cytometry analysis
A549, NCI-H1975, and Vero E6 cells were subjected to live cell surface staining. Cells were gently washed twice with PBS and resuspended in ice-cold 5% FBS-PBS followed by blocking in 0.5% BSA 2% FBS-PBS at 4°C. The cells were then incubated with an antibody against ACE2 (MAB9331, R&D Systems, Minneapolis, MN, USA) in 5% FBS-PBS for 30 min at 4°C in the dark, followed by washing. A secondary antibody conjugated with Alexa Fluor 488 was added, and the cells were incubated for 30 min at 4°C in the dark. Cells were then resuspended in 5% FBS-PBS and transferred into polystyrene round-bottom Falcon tubes with a strainer cap before analysis on a BD FACS Verse II system (BD, Franklin Lakes, NJ, USA).
IHC
FFPE lung and tracheal sections were deparaffinized for IHC analysis through a series of xylene and ethanol washes, followed by a brief rinse in distilled water. Antigen retrieval was performed by heating the slides at 95°C for 20 min in pH 6.0 FLEX target retrieval solution (k8005, Dako, Carpinteria, CA, USA) for SARS-CoV-2 NP and HMGB1 and in pH 9.0 buffer for RAGE and ACE2. Slides were incubated with 3% hydrogen peroxide (H2O2; 3059, Duksan, Ansan-si, South Korea) for 10 min to block endogenous peroxidase activity. Primary antibodies against SARS-CoV-2 NP (40143-MM08, Sino Biological), ACE2 (21115-1 AP, Proteintech), RAGE (sc-365154, Santa Cruz), and HMGB1 (ab18256, Abcam) were applied at dilutions of 1:500–1,000 for 1 h at RT. Mouse or rabbit Envision Plus system-HRP-labeled polymer (k4003/k4001, DAKO) secondary antibodies were used. After development with DAB (k3468, DAKO) for 5 min, the slides were counterstained with hematoxylin, dehydrated using ethanol and xylene, and mounted for scanning.
Plaque-forming assay
For the plaque-forming assay, 3 × 105 Vero E6 cells were seeded onto 12-well plates and allowed to adhere overnight. The medium was removed, and the cells were washed with DPBS. The right lobes of the lungs from infected mice were weighed and lysed with metal beads to obtain viral suspensions. A 200 μL aliquot of these suspensions was diluted 10-fold and applied to each well to initiate infection. The cells were incubated for 1 h at 37°C, with gentle rocking every 15 min. After incubation, the supernatants were removed, and a 1:1 mixture of 2x DMEM and 2% low-melting-point agar solution was added to each well, which is a critical step for viral immobilization. The cells with agar overlay were incubated for an additional 2–4 days, which was the optimal period for the development of detectable viral plaques. Once the plaques formed, the cells were stained with a 0.5% crystal violet solution in 20% methanol. Quantitative analysis of viral infectivity was performed by counting the plaques in each well and calculating the PFU per gram of lung tissue.
Quantification and statistical analysis
All data are presented as mean ± standard error of mean (SEM) of at least three individual measures (n > 2), unless otherwise stated. Variable n denotes the number of biologically independent experiments. Confocal images were analyzed using the ImageJ software (NIH). Statistical significance was determined using one-way analysis of variance with Tukey’s multiple comparison test, and Student’s unpaired t-test was used for comparisons between two groups. The tests were performed using GraphPad Prism 10 software, with ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 considered to indicate statistical significance. Details of statistical analyses and statistical significance are included in the corresponding figure legends.
Published: July 5, 2025
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2025.113063.
Supplemental information
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuai H., Chan J.F.W., Hu B., Chai Y., Yuen T.T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization 2023 data.who.int, WHO Coronavirus (COVID-19) dashboard > More resources [Dashboard] https://data.who.int/dashboards/covid19/more-resources
- 4.Omolo C.A., Soni N., Fasiku V.O., Mackraj I., Govender T. Update on therapeutic approaches and emerging therapies for SARS-CoV-2 virus. Eur. J. Pharmacol. 2020;883 doi: 10.1016/j.ejphar.2020.173348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson U., Ottestad W., Tracey K.J. Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19? Mol. Med. 2020;26:42. doi: 10.1186/s10020-020-00172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wulandari S., Wibawa T., Hartono, Wibawa T. The role of HMGB1 in COVID-19-induced cytokine storm and its potential therapeutic targets: A review. Immunology. 2023;169:117–131. doi: 10.1111/imm.13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vicentino A.R.R., Fraga-Junior V.D.S., Palazzo M., Tasmo N.R.A., Rodrigues D.A.S., Barroso S.P.C., Ferreira S.N., Neves-Borges A.C., Allonso D., Fantappié M.R., et al. High mobility group box 1, ATP, lipid mediators, and tissue factor are elevated in COVID-19 patients: HMGB1 as a biomarker of worst prognosis. Clin. Transl. Sci. 2023;16:631–646. doi: 10.1111/cts.13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwak M.S., Kim H.S., Lee B., Kim Y.H., Son M., Shin J.S. Immunological Significance of HMGB1 Post-Translational Modification and Redox Biology. Front. Immunol. 2020;11:1189. doi: 10.3389/fimmu.2020.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi M.E., Falciola L., Ferrari S., Lilley D.M. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992;11:1055–1063. doi: 10.1002/j.1460-2075.1992.tb05144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S.A., Kwak M.S., Kim S., Shin J.S. The role of high mobility group box 1 in innate immunity. Yonsei Med. J. 2014;55:1165–1176. doi: 10.3349/ymj.2014.55.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Qiang X., Harris H.E., Czura C.J., Wang H., Ulloa L., Kokkola R., Tabibzadeh S., Yang R., Ochani M., et al. Structural Basis for the Proinflammatory Cytokine Activity of High Mobility Group Box 1. Mol. Med. 2003;9:37–45. [PMC free article] [PubMed] [Google Scholar]
- 13.Gong Q., Xu J.F., Yin H., Liu S.F., Duan L.H., Bian Z.L. Protective effect of antagonist of high-mobility group box 1 on lipopolysaccharide-induced acute lung injury in mice. Scand. J. Immunol. 2009;69:29–35. doi: 10.1111/j.1365-3083.2008.02194.x. [DOI] [PubMed] [Google Scholar]
- 14.Huttunen H.J., Fages C., Kuja-Panula J., Ridley A.J., Rauvala H. Receptor for advanced glycation end products-binding COOH-terminal motif of amphoterin inhibits invasive migration and metastasis. Cancer Res. 2002;62:4805–4811. [PubMed] [Google Scholar]
- 15.Kim Y.H., Kwak M.S., Park J.B., Lee S.A., Choi J.E., Cho H.S., Shin J.S. N-linked glycosylation plays a crucial role in the secretion of HMGB1. J. Cell Sci. 2016;129:29–38. doi: 10.1242/jcs.176412. [DOI] [PubMed] [Google Scholar]
- 16.Youn J.H., Shin J.S. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 2006;177:7889–7897. doi: 10.4049/jimmunol.177.11.7889. [DOI] [PubMed] [Google Scholar]
- 17.Ito I., Fukazawa J., Yoshida M. Post-translational methylation of high mobility group box 1 (HMGB1) causes its cytoplasmic localization in neutrophils. J. Biol. Chem. 2007;282:16336–16344. doi: 10.1074/jbc.M608467200. [DOI] [PubMed] [Google Scholar]
- 18.Kwak M.S., Kim H.S., Lkhamsuren K., Kim Y.H., Han M.G., Shin J.M., Park I.H., Rhee W.J., Lee S.K., Rhee S.G., Shin J.S. Peroxiredoxin-mediated disulfide bond formation is required for nucleocytoplasmic translocation and secretion of HMGB1 in response to inflammatory stimuli. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J., Jiang Y., Wang J., Shi X., Liu Q., Liu Z., Li Y., Scott M.J., Xiao G., Li S., et al. Macrophage endocytosis of high-mobility group box 1 triggers pyroptosis. Cell Death Differ. 2014;21:1229–1239. doi: 10.1038/cdd.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youn J.H., Oh Y.J., Kim E.S., Choi J.E., Shin J.S. High mobility group box 1 protein binding to lipopolysaccharide facilitates transfer of lipopolysaccharide to CD14 and enhances lipopolysaccharide-mediated TNF-alpha production in human monocytes. J. Immunol. 2008;180:5067–5074. doi: 10.4049/jimmunol.180.7.5067. [DOI] [PubMed] [Google Scholar]
- 21.Deng M., Tang Y., Li W., Wang X., Zhang R., Zhang X., Zhao X., Liu J., Tang C., Liu Z., et al. The Endotoxin Delivery Protein HMGB1 Mediates Caspase-11-Dependent Lethality in Sepsis. Immunity. 2018;49:740–753. doi: 10.1016/j.immuni.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayati A., Kumar R., Francis V., McPherson P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Bio. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J., Li Y., Liu Q., Yao Q., Wang X., Zhang H., Chen R., Ren L., Min J., Deng F., et al. SARS-CoV-2 cell tropism and multiorgan infection. Cell Discov. 2021;7:17. doi: 10.1038/s41421-021-00249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin S., Wang H., Yuan R., Li H., Ochani M., Ochani K., Rosas-Ballina M., Czura C.J., Huston J.M., Miller E., et al. Role of HMGB1 in apoptosis-mediated sepsis lethality. J. Exp. Med. 2006;203:1637–1642. doi: 10.1084/jem.20052203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magna M., Pisetsky D.S. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Mol. Med. 2014;20:138–146. doi: 10.2119/molmed.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris H.E., Andersson U., Pisetsky D.S. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat. Rev. Rheumatol. 2012;8:195–202. doi: 10.1038/nrrheum.2011.222. [DOI] [PubMed] [Google Scholar]
- 32.Kim S.Y., Son M., Lee S.E., Park I.H., Kwak M.S., Han M., Lee H.S., Kim E.S., Kim J.Y., Lee J.E., et al. High-Mobility Group Box 1-Induced Complement Activation Causes Sterile Inflammation. Front. Immunol. 2018;9:705. doi: 10.3389/fimmu.2018.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu L., Chen C., Chen Y., Li Y., Tang F., Huang H., He W., Zhang R., Shen L. High-Mobility Group Box 1 (HMGB1) and Autophagy in Acute Lung Injury (ALI): A Review. Med. Sci. Monit. 2019;25:1828–1837. doi: 10.12659/msm.912867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li R., Zhang J., Pan S., Yuan Y., Qi H., Shu H., Hu Y., Ren L., Jiang Y., Yuan S. HMGB1 aggravates lipopolysaccharide-induced acute lung injury through suppressing the activity and function of Tregs. Cell. Immunol. 2020;356 doi: 10.1016/j.cellimm.2020.104192. [DOI] [PubMed] [Google Scholar]
- 35.Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghaffarpour S., Ghazanfari T., Ardestani S.K., Naghizadeh M.M., Vaez Mahdavi M.R., Salehi M., Majd A.M.M., Rashidi A., Chenary M.R., Mostafazadeh A., et al. Cytokine profiles dynamics in COVID-19 patients: a longitudinal analysis of disease severity and outcomes. Sci. Rep. 2025;15 doi: 10.1038/s41598-025-98505-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwak M.S., Choi S., Kim J., Lee H., Park I.H., Oh J., Mai D.N., Cho N.H., Nam K.T., Shin J.S. SARS-CoV-2 Infection Induces HMGB1 Secretion Through Post-Translational Modification and PANoptosis. Immune Netw. 2023;23 doi: 10.4110/in.2023.23.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang C.W., Parsi K., Somasundaran M., Vanderleeden E., Liu P., Cruz J., Cousineau A., Finberg R., Kurt-Jones E. A Newly Engineered A549 Cell Line Expressing ACE2 and TMPRSS2 Is Highly Permissive to SARS-CoV-2, Including the Delta and Omicron Variants. Viruses. 2022;14:1369. doi: 10.3390/v14071369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gioia U., Tavella S., Martínez-Orellana P., Cicio G., Colliva A., Ceccon M., Cabrini M., Henriques A.C., Fumagalli V., Paldino A., et al. SARS-CoV-2 infection induces DNA damage, through CHK1 degradation and impaired 53BP1 recruitment, and cellular senescence. Nat. Cell Biol. 2023;25:550–564. doi: 10.1038/s41556-023-01096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baggen J., Jacquemyn M., Persoons L., Vanstreels E., Pye V.E., Wrobel A.G., Calvaresi V., Martin S.R., Roustan C., Cronin N.B., et al. TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry. Cell. 2023;186:3427–3442. doi: 10.1016/j.cell.2023.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiong X., Dou J., Shi J., Ren Y., Wang C., Zhang Y., Cui Y. RAGE inhibition alleviates lipopolysaccharides-induced lung injury via directly suppressing autophagic apoptosis of type II alveolar epithelial cells. Respir. Res. 2023;24:24. doi: 10.1186/s12931-023-02332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Xia Yu W.C.P., Cheng Y.F. Silencing of advanced glycosylation and glycosylation and product-specific receptor (RAGE) inhibits the metastasis and growth of non-small cell lung cancer. Am J Transl Res. 2017;9:2760–2774. [PMC free article] [PubMed] [Google Scholar]
- 43.Hofmann M.A., Drury S., Hudson B.I., Gleason M.R., Qu W., Lu Y., Lalla E., Chitnis S., Monteiro J., Stickland M.H., et al. RAGE and arthritis: the G82S polymorphism amplifies the inflammatory response. Genes Immun. 2002;3:123–135. doi: 10.1038/sj.gene.6363861. [DOI] [PubMed] [Google Scholar]
- 44.Jeong J., Cho S., Seo M., Lee B.S., Jang Y., Lim S., Park S. Soluble RAGE attenuates Ang II-induced arterial calcification via inhibiting AT1R-HMGB1-RAGE axis. Atherosclerosis. 2022;346:53–62. doi: 10.1016/j.atherosclerosis.2022.02.022. [DOI] [PubMed] [Google Scholar]
- 45.Magna M., Hwang G.H., McIntosh A., Drews-Elger K., Takabatake M., Ikeda A., Mera B.J., Kwak T., Miller P., Lippman M.E., Hudson B.I. RAGE inhibitor TTP488 (Azeliragon) suppresses metastasis in triple-negative breast cancer. NPJ Breast Cancer. 2023;9:59. doi: 10.1038/s41523-023-00564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burstein A.H., Sabbagh M., Andrews R., Valcarce C., Dunn I., Altstiel L. Development of Azeliragon, an Oral Small Molecule Antagonist of the Receptor for Advanced Glycation Endproducts, for the Potential Slowing of Loss of Cognition in Mild Alzheimer's Disease. J Prev Alzheimers Dis. 2018;5:149–154. doi: 10.14283/jpad.2018.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deane R., Singh I., Sagare A.P., Bell R.D., Ross N.T., LaRue B., Love R., Perry S., Paquette N., Deane R.J., et al. A multimodal RAGE-specific inhibitor reduces amyloid β-mediated brain disorder in a mouse model of Alzheimer disease. J. Clin. Investig. 2012;122:1377–1392. doi: 10.1172/jci58642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu M., Wang H., Ding A., Golenbock D.T., Latz E., Czura C.J., Fenton M.J., Tracey K.J., Yang H. HMGB1 signals through toll-like receptor (TLR) 4 and TLR2. Shock. 2006;26:174–179. doi: 10.1097/01.shk.0000225404.51320.82. [DOI] [PubMed] [Google Scholar]
- 50.Pan T., Chen R., He X., Yuan Y., Deng X., Li R., Yan H., Yan S., Liu J., Zhang Y., et al. Infection of wild-type mice by SARS-CoV-2 B.1.351 variant indicates a possible novel cross-species transmission route. Signal Transduct. Target. Ther. 2021;6:420. doi: 10.1038/s41392-021-00848-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yasui F., Matsumoto Y., Yamamoto N., Sanada T., Honda T., Munakata T., Itoh Y., Kohara M. Infection with the SARS-CoV-2 B.1.351 variant is lethal in aged BALB/c mice. Sci. Rep. 2022;12:4150. doi: 10.1038/s41598-022-08104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen R., Huang Y., Quan J., Liu J., Wang H., Billiar T.R., Lotze M.T., Zeh H.J., Kang R., Tang D. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e05672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waraich R.S., Sohail F.A., Khan G., Durr-E-Shahwar S., Khan B., Rafi S., Nasir S. Enhanced Expression of RAGE AXIS Is Associated with Severity of COVID-19 in Patients with Comorbidities. Metab. Syndr. Relat. Disord. 2023;21:141–147. doi: 10.1089/met.2022.0089. [DOI] [PubMed] [Google Scholar]
- 54.Chen L., Long X., Xu Q., Tan J., Wang G., Cao Y., Wei J., Luo H., Zhu H., Huang L., et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:992–994. doi: 10.1038/s41423-020-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan X., Song J.W., Wang S.Y., Cao W.J., Wang X.W., Zhou M.J., Yang T., Zhou C.B., Hou J., Zhang J.Y., et al. Changes of Damage Associated Molecular Patterns in COVID-19 Patients. Infect. Dis. Immun. 2021;1:20–27. doi: 10.1097/01.ID9.0000733572.40970.6c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sivakorn C., Dechsanga J., Jamjumrus L., Boonnak K., Schultz M.J., Dondorp A.M., Phumratanaprapin W., Ratanarat R., Naorungroj T., Wattanawinitchai P., et al. High Mobility Group Box 1 and Interleukin 6 at Intensive Care Unit Admission as Biomarkers in Critically Ill COVID-19 Patients. Am. J. Trop. Med. Hyg. 2021;105:73–80. doi: 10.4269/ajtmh.21-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang J., Zhao Y., Zhang P., Li Y., Yang Y., Yang Y., Zhu J., Song X., Jiang G., Fan J. Hemorrhagic shock primes for lung vascular endothelial cell pyroptosis: role in pulmonary inflammation following LPS. Cell Death Dis. 2016;7:e2363. doi: 10.1038/cddis.2016.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kreutzberger A.J.B., Sanyal A., Saminathan A., Bloyet L.-M., Stumpf S., Liu Z., Ojha R., Patjas M.T., Geneid A., Scanavachi G., et al. SARS-CoV-2 requires acidic pH to infect cells. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2209514119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angioni R., Bonfanti M., Caporale N., Sánchez-Rodríguez R., Munari F., Savino A., Pasqualato S., Buratto D., Pagani I., Bertoldi N., et al. RAGE engagement by SARS-CoV-2 enables monocyte infection and underlies COVID-19 severity. Cell Rep. Med. 2023;4 doi: 10.1016/j.xcrm.2023.101266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Puray-Chavez M., LaPak K.M., Schrank T.P., Elliott J.L., Bhatt D.P., Agajanian M.J., Jasuja R., Lawson D.Q., Davis K., Rothlauf P.W., et al. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gu Y., Cao J., Zhang X., Gao H., Wang Y., Wang J., He J., Jiang X., Zhang J., Shen G., et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 2022;32:24–37. doi: 10.1038/s41422-021-00595-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiappalupi S., Salvadori L., Donato R., Riuzzi F., Sorci G. Hyperactivated RAGE in Comorbidities as a Risk Factor for Severe COVID-19-The Role of RAGE-RAS Crosstalk. Biomolecules. 2021;11:876. doi: 10.3390/biom11060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rubas N.C., Peres R., Kunihiro B.P., Allan N.P., Phankitnirundorn K., Wells R.K., McCracken T., Lee R.H., Umeda L., Conching A., et al. HMGB1 mediates microbiome-immune axis dysregulation underlying reduced neutralization capacity in obesity-related post-acute sequelae of SARS-CoV-2. Sci. Rep. 2024;14:355. doi: 10.1038/s41598-023-50027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oh Y.J., Youn J.H., Ji Y., Lee S.E., Lim K.J., Choi J.E., Shin J.S. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium-dependent mechanism. J. Immunol. 2009;182:5800–5809. doi: 10.4049/jimmunol.0801873. [DOI] [PubMed] [Google Scholar]
- 65.Jain A., Liu R., Ramani B., Arauz E., Ishitsuka Y., Ragunathan K., Park J., Chen J., Xiang Y.K., Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this article will be shared by the lead contact upon request. This article does not report any original code. Additional information required to reanalyze the data reported in this article is available from the lead contact upon request.