Abstract
Rod cyclic nucleotide-gated (CNG) channels are heterotetramers comprised of both CNGA1 and CNGB1 subunits. Calcium/calmodulin (Ca2+/CaM) binds to a site in the N-terminal region of CNGB1 subunits and inhibits the opening conformational change in CNGA1/CNGB1 channels. Here, we show that polypeptides derived from an N-terminal region of CNGB1 form a specific interaction with polypeptides derived from a C-terminal region of CNGA1 that is distal to the cyclic nucleotide-binding domain. Deletion of the Ca2+/CaM-binding site from the N-terminal region of CNGB1 eliminated both Ca2+/CaM modulation of the channel and the intersubunit interaction. Furthermore, the interaction was disrupted by the presence of Ca2+/CaM. These results suggest that Ca2+/CaM-dependent inhibition of rod channels is caused by the direct binding of Ca2+/CaM to a site in the N-terminal region in CNGB1, which disrupts the interaction between this region and a distal C-terminal region of CNGA1. The mechanism underlying Ca2+/CaM modulation of rod channels is distinct from that in olfactory (CNGA2) CNG channels.
Cyclic nucleotide-gated (CNG) channels were first characterized in retinal rods, where they conduct a cation current in response to changes in intracellular levels of cGMP and mediate the electrical response to light (1). CNG channels also are found in olfactory neurons, where they respond to changes in internal cAMP and underlie the electrical response to odorants (2). Evidence exists for a cyclic nucleotide-dependent conductance in taste receptors (3, 4). CNG channels are present in a variety of other tissues, including heart, aorta, and kidney (for a review see ref. 5).
CNG channels were first cloned from retina (6) and olfactory neurons (7). Four channel subunits are arranged to form a tetramer with a central pore (8, 9). Subunits have six proposed membrane-spanning domains, a pore-loop domain, and intracellular N- and C-terminal regions, a topology similar to that of the voltage-activated potassium channels (10). However, CNG channels are only weakly sensitive to membrane voltage. Instead, they contain a large C-terminal cyclic nucleotide-binding domain (CNBD) that exhibits sequence similarity with other cyclic nucleotide-binding proteins (11, 12). CNG channels are activated by the direct binding of cyclic nucleotides to the CNBD (13).
At present, there are six types of mammalian CNG channel genes. The genes are grouped according to sequence similarity into two subtypes, CNGA and CNGB (14). CNGA1, CNGA2, and CNGA3 subunits form functional homomeric channels when expressed alone, whereas CNGB1, CNGB3, and CNGA4 subunits do not appear to form functional homomeric channels when expressed alone. Instead, CNGB1, CNGB3, and CNGA4 subunits form heteromeric channels when coexpressed with CNGA1, CNGA2, or CNGA3 subunits (15).
Native retinal rod channels comprise CNGA1 (formerly CNG1; Rod α) and CNGB1 (formerly CNG4; Rod β) subunits. CNGA1/CNGB1 heterotetramers contain two CNGA1 subunits and two CNGB1 subunits (16, 17). Compared with CNGA1 homomers, CNGA1/CNGB1 heteromers exhibit several new properties, including sensitivity to l-cis diltiazem, slight outward rectification of the current-voltage relationship, a 10-fold increase in the current activated by cAMP, and modulation by calcium/calmodulin (Ca2+/CaM) (16–23).
In addition to inhibiting CNGA1/CNGB1 channels, Ca2+/CaM inhibits CNGA2 (formerly CNG2; olfactory α) channels (20). The mechanism underlying this inhibition is understood in some detail. Ca2+/CaM binds to an N-terminal domain of CNGA2 and reduces the apparent affinity of the channels for cyclic nucleotide by 10-fold. Deletion of the CaM binding site also reduces the apparent affinity by 10-fold (24). The N-terminal region forms an interaction with the C-terminal CNBD of CNGA2 subunits, and the CaM-binding domain is necessary for this interaction. Ca2+/CaM disrupts this interaction, suggesting a mechanism for inhibition whereby Ca2+/CaM prevents a potentiating interaction of the N-terminal region with the C-terminal region (25). Ca2+/CaM inhibition of olfactory CNG channels is thought to underlie olfactory adaptation (26).
Previous work has identified a short domain in the CNGB1 N-terminal region that binds to Ca2+/CaM (27, 28). When this short domain is deleted, Ca2+/CaM does not inhibit these CNG channels. However, Ca2+/CaM inhibition of CNGA1/CNGB1 channels is not as well understood as Ca2+/CaM inhibition of CNGA2 channels. In this study we have investigated the mechanism underlying Ca2+/CaM-dependent inhibition in CNGA1/CNGB1 channels. Unlike the case for CNGA2 channels, we find that the N-terminal region of CNGB1 interacts with a C-terminal region of CNGA1 distal to the CNBD. This CNGA1/CNGB1 interaction was prevented by deletion of the Ca2+/CaM binding site or the presence of Ca2+/CaM. These results suggest a molecular mechanism for rod channel inhibition by Ca2+/CaM where an intersubunit N- and C-terminal region interaction is disrupted by Ca2+/CaM, leading to channel inhibition.
Methods
Molecular Biology and Mutagenesis.
We used a bovine CNGA1 clone as described (29) that was identical to the original isolate (6). We added an 8-aa FLAG epitope tag (DYKDDDYK) in place of the final five amino acids (DSTQD) of CNGA1, but this process did not change any properties measured here (data not shown). The bovine CNGB1 clone (22) was a gift from R. Molday, University of British Columbia. An I2V change was made in CNGB1 for ease of cloning, but it did not affect any characteristics determined here (data not shown). The CNGA2 clone (7) was a gift from R. Reed, The Johns Hopkins University, Baltimore. CNG channel cDNAs were subcloned into the pGEMHE vector (a gift from E. Liman, University of Southern California) for expression in Xenopus oocytes. RNA was made with the Message Machine kit (Ambion, Austin, TX). CNGB1 deletion mutants were made with an oligonucleotide-directed approach and confirmed by fluorescent-based sequencing.
Electrophysiology and Analysis.
In preparation for patch-clamp recording, Xenopus oocytes were microinjected with CNG channel RNA. CNGA1 and CNGB1 RNAs were coinjected at a 1:4 ratio, which produced a pure population of heteromeric channels (16). Injected oocytes were incubated at 16°C and patch-clamped within 5–8 days. We recorded currents by using the excised, inside-out patch-clamp configuration (30) with an Axopatch 200A (Axon Instruments, Foster City, CA) patch-clamp coupled to an ITC-16 AD converter (Instrutech, Great Neck, NY). Data were recorded and analyzed with pulse software (Instrutech) and additionally analyzed with igor software (WaveMetrics, Lake Oswego, OR). Currents in Fig. 1 were elicted by a pulse to 60 mV after a prepulse to −60 mV from a holding potential of 0 mV in subsaturating (50 μM) cGMP. The data in Fig. 2 were determined from peak currents elicited by a pulse to 60 mV from a holding potential of 0 mV in saturating cAMP (16 mM) and saturating (2.5 mM) cGMP.
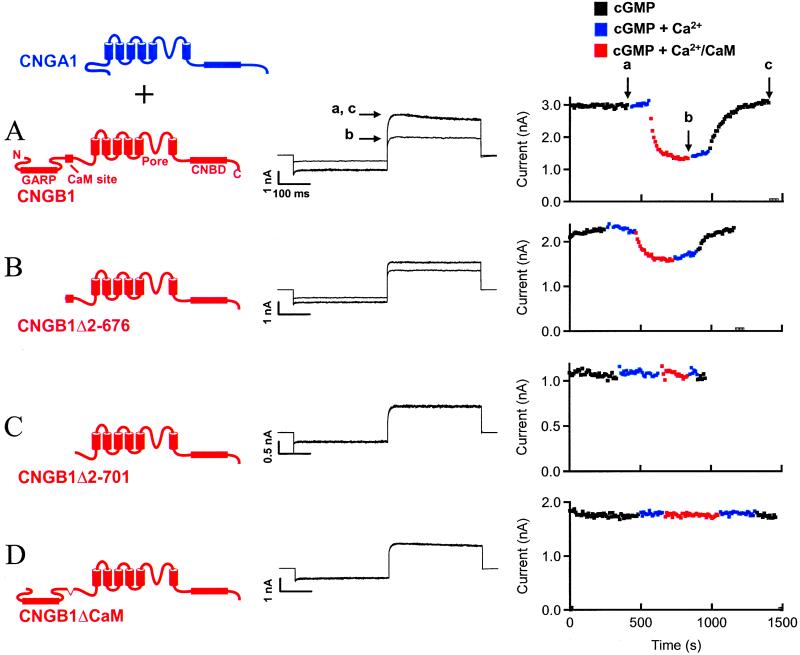
Figure 1.
Localization of a region in rod CNG channels necessary for inhibition by Ca2+/CaM. (A) (Left) Cartoon image depicting coexpression of CNGA1 and CNGB1 subunits. (Center) cGMP-activated currents from wild-type CNGA1/CNGB1 channels in the absence (a and c) and presence (b) of Ca2+/CaM. (Right) Time course of current inhibition by Ca2+/CaM. Arrows labeled a–c indicate points that correspond to the current traces (Center). (B) cGMP-activated currents from CNGA1/CNGB1Δ2–676 channels and time course of inhibition in the presence of Ca2+/CaM. (C) Lack of effect of Ca2+/CaM on CNGA1/CNGB1Δ2–701 channels. (D) Lack of effect of Ca2+/CaM on CNGA1/CNGB1ΔCaM channels. Several CNG channel domains are labeled in A. GARP, glutamic acid-rich protein.
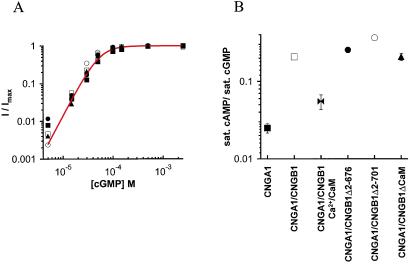
Figure 2.
Functional properties of CNG channels. (A) cGMP dose–response relationships from a representative patch for CNGA1 (■), CNGA1/CNGB1 (□), CNGA1/CNGB1Δ2–676 (●), CNGA1/CNGB1Δ2–701 (○), and CNGA1/CNGB1ΔCaM (▴). Currents were measured at 60 mV, normalized to the maximum value in saturating cGMP (2.5 mM), and superimposed with the Hill equation, I/Imax = 1/(1+ (K1/2/[cGMP])n), with K1/2 = 55 μM and n = 2.5. (B) Fractional activation of CNG channels by cAMP, a partial agonist. Each channel construct is depicted by the same symbol as in A, except for Ca2+/CaM with CNGA1/CNGB1 (bowties). Values were determined by normalizing the current at 60 mV in saturating (16 mM) cAMP (or saturating cAMP with Ca2+/CaM) to that in saturating (2.5 mM) cGMP. Error bars represent the SEM; those not visible are within the symbols. n is ≥ 3 for each point.
For patch-clamp recording, the internal (bath) solution contained 130 mM NaCl, 3 mM Hepes, 0.2 mM EDTA, pH 7.2. The external (pipette) solution was identical but contained 0.5 mM niflumic acid to block endogenous Cl− currents. Niflumic acid had no effect on CNG channels (data not shown). cGMP or cAMP was added to the internal solution at the desired concentration. Internal solutions containing Ca2+ were buffered with 0.2 mM nitrilotriacetic acid such that the free Ca2+ concentration was 1 μM as determined with the winmaxc program (31). The CaM (Calbiochem) concentration was 250 nM in internal solutions. All chemicals were from Sigma unless otherwise noted. Internal solutions were applied with an RSC-100 solution changer (Molecular Kinetics, Pullman, WA).
Biochemical Pull-Down Interaction Assays.
Biochemical pull-down experiments consisted of a “bait” polypeptide immobilized on glutathione beads and a “fish” polypeptide with a FLAG epitope tag that enabled detection. These fusion proteins were prepared by using procedures as described (25, 32). To test for interactions between polyhistidine and glutathione S-transferase (GST)-linked fusion proteins, bait and fish were combined in buffer S [50 mM Tris⋅HCl/150 mM NaCl/25 mM imidazole/1% NDSB-256/0.5% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate)/0.25% Tween-20/1 mM β-mercaptoethanol, pH 7.8] and incubated with shaking for >18 h. The final concentration of bait and fish in solution was 0.5 μM and 1.0 μM, respectively. Protein concentration was determined with a spectrophotometer for a given protein sample in 6 M guanidine-HCl and calculated by using the extinction coefficient. For pull-down assays in which fluorescent CaM (CaM-488; Molecular Probes) was used as fish, the concentration of CaM-488 was 100 μM. For pull-downs where CaM (Calbiochem) was incubated with bait and fish derived from the channel, the free Ca2+ concentration was 1 mM, EGTA was 1 mM, and CaM was 15 μM, in the indicated combinations. Fusion proteins associated with beads were recovered by centrifugation at 2,000 × g and washed for 5 min in 0.5 ml buffer S while shaking at 4°C. The wash and recovery step was repeated five times. Samples were loaded and run on SDS/PAGE gels and subsequently blotted as described (32). Western blots were developed with the Dura Extended chemiluminescence system (Pierce) and visualized with a FluorChem imaging system (Alpha Innotech, San Leandro, CA). For experiments with CaM-488, gels were visualized directly by exciting the CaM-488 fluorophore, and the emission was also detected with the FluorChem system.
Results
Ca2+/CaM Inhibition of Rod CNG Channels.
Heteromeric rod CNG channels were formed by coexpressing CNGA1 subunits and CNGB1 subunits in Xenopus oocytes, as depicted in Fig. 1 Left. Throughout this article, CNGA1 subunits (and polypeptide fragments) will be shown in blue and CNGB1 subunits (and polypeptide fragments) will be shown in red. Currents were recorded from excised, inside-out patches and measured after a voltage step to 60 mV, after a prepulse to −60 mV, in the presence of internal, subsaturating (50 μM) cGMP. Leak currents in the absence of cyclic nucleotide were subtracted from all records.
Ca2+/CaM inhibits rod CNG channels (19–23). The current in the presence of 50 μM cGMP (Fig. 1A Center, trace a) was reduced about 2-fold after internal application of Ca2+/CaM (trace b). Channel inhibition was completely reversed by removal of Ca2+/CaM (trace c). The time course of channel response to Ca2+/CaM also was determined (Fig. 1A Right). Peak current levels at 60 mV in 50 μM cGMP were determined every 10 s (black symbols) and were stable. Addition of 1 μM Ca2+ (blue symbols) did not affect the peak current, indicating that this amount of Ca2+ produced very little channel block. However, addition of 1 μM Ca2+ with 250 nM CaM inhibited channels (red symbols) over a period of a few hundred seconds. Inhibition was maintained after removal of CaM (blue symbols), but reversed slowly upon subsequent removal of Ca2+ (black symbols). The points indicated by a-c in Fig. 1A Right correspond to the currents shown in the center panel.
Ca2+/CaM-dependent inhibition remained intact in heteromeric channels when a large N-terminal region that includes the glutamic acid-rich protein region (amino acids 2–676) was deleted from CNGB1 (Fig. 1B), ruling out the deleted region as a determinant of inhibition. However, successive deletion of the next 24 aa from the N-terminal region of CNGB1 (amino acids 676–701) resulted in channels that were insensitive to Ca2+/CaM (Fig. 1C). As a second test of the importance of this region we made an internal deletion of just 20 aa from CNGB1 (amino acids 682–701), because this segment was sufficient for binding Ca2+/CaM (27, 28). These channels were also insensitive to Ca2+/CaM (Fig. 1D). Consistent with previous work (27, 28), these results suggest that a short region in the N-terminal domain of CNGB1 subunits, including amino acids 682–701, was necessary for Ca2+/CaM-dependent channel inhibition of heteromeric channels.
To determine whether mutations in CNGB1 altered the allosteric transition leading to channel opening, we measured the cGMP dose–response relation for all of the heteromeric channels shown in Fig. 1 and CNGA1 homomers. A fit of the Hill equation to the dose–response relations for cGMP gave similar apparent affinities for each channel type (Fig. 2A, Table 1), indicating that mutations apparently did not affect cyclic nucleotide-binding or channel opening. We also determined the fractional activation of these channels by a saturating concentration of cAMP, a partial agonist that opens the channels poorly compared with cGMP (33, 34). CNGA1 homomers had a characteristic low fractional activation by cAMP compared with cGMP, whereas CNGA1/CNGB1 heteromers had a 10-fold higher fractional activation (Fig. 2B). In the presence of Ca2+/CaM the fractional activation of CNGA1/CNGB1 decreased about 4-fold, indicating inhibition of the allosteric transition. Heteromers containing mutant CNGB1 subunits also had a high fractional activation by cAMP, like that of wild-type heteromers (Fig. 2B). These results indicate that mutant CNGB1 subunits were fully able to form heteromeric channels with CNGA1 subunits and, unlike for CNGA2 channels, deletions in the N-terminal region did not mimic the functional effects of Ca2+/CaM.
Table 1.
Gating parameters of CNG channels
| Channel | K1/2 (μM) for cGMP |
|---|---|
| CNGA1 | 62.4 ± 8.2 (n = 5) |
| CNGA1/CNGB1 | 43.8 ± 2.0 (n = 8) |
| CNGA1/CNGB1Δ2-676 | 50.7 ± 8.3 (n = 4) |
| CNGA1/CNGB1Δ2-701 | 50.0 ± 10.8 (n = 3) |
| CNGA1/CNGB1ΔCaM | 50.8 ± 6.3 (n = 3) |
All data are mean ± SEM.
Molecular Determinants for N-and C-Terminal Interactions in CNGA1/CNGB1 Channels.
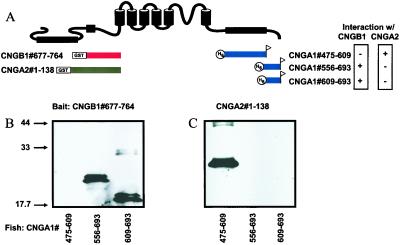
In CNGA2 channels, the mechanism underlying Ca2+/CaM-dependent inhibition is disruption of a physical interaction between N- and C-terminal regions by Ca2+/CaM (25). Does the Ca2+/CaM-dependent inhibition of rod CNGA1/CNGB1 channels, as shown in Fig. 1, involve an N- and C-terminal interaction? To investigate this, we performed biochemical pull-down interaction assays between an N-terminal region of CNGB1 and C-terminal regions of CNGA1 as depicted in Fig. 3A. Bait fusion proteins were constructed by linking N-terminal fragments of CNGB1 or CNGA2 channels in-frame with GST and immobilizing them on glutathione beads. Bait proteins were incubated with fish fusion proteins that were derived from C-terminal regions of CNGA1, linked in-frame with polyhistidine and containing a FLAG epitope tag at their C terminus for detection. Bait and fish proteins are depicted directly beneath regions of a generic CNG subunit to which they correspond, with the exact range of amino acids given within the name of each construct (Fig. 3A). Bait and fish were allowed to interact, washed extensively, recovered by centrifugation, and separated with SDS/PAGE. Fish proteins that interacted with bait were detected by immunoblotting (see Methods). In side-by-side biochemical pull-down experiments using fish proteins corresponding to subregions of the CNGA1 C-terminal region, we found that the CNGB1 N-terminal region did not interact with the CNBD, but did form a strong, specific interaction with proteins containing the distal C-terminal region of CNGA1. Just this region, from amino acids 609 to 693, proved necessary and sufficient for the physical interaction with CNGB1 (Fig. 3B). This interaction has also been recently shown to regulate trafficking of heteromeric channels (32). In contrast, the N-terminal region of CNGA2 channels interacted specifically with the CNBD of CNGA1 subunits, but not the distal C-terminal region (Fig. 3C). Thus, while both CNGA1/CNGB1 channels and CNGA2 channels exhibit a interaction between the N- and C-terminal regions, the interactions are distinguished both because the N-terminal regions of CNGB1 and CNGA2 display no sequence similarity and these N-terminal regions bind to different C-terminal regions in CNGA1 subunits. In intact CNGA1/CNGB1 channels, this N- and C-terminal region interaction must be an intersubunit interaction.
Figure 3.
Difference in molecular determinants for interdomain interactions in CNGA1/CNGB1 channels versus CNGA2 channels. (A) Cartoon showing bait proteins derived from CNGA2 (gray) and CNGB1 (red) and fish proteins derived from CNGA1 (blue) located directly beneath the corresponding region of a generic CNG subunit. The exact amino acid composition of the fusion proteins are given by the numbers within each associated name. (B) Western blot of biochemical pull-down interaction using CNGB1#677–764 as bait and CNGA2#1–138 as fish. (C) Western blot using CNGA2-derived protein as bait and the same fish proteins as in B.
Direct Binding of Ca2+/CaM to CNGB1 Subunits.
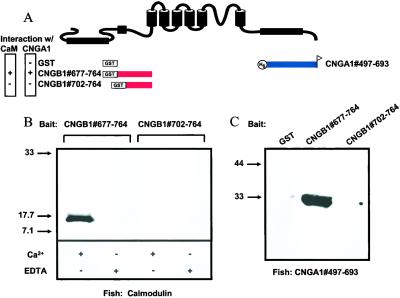
Previously, it was shown that Ca2+/CaM could bind to a short N-terminal region of CNGB1 subunits (27, 28). To investigate binding in our deletion constructs we performed biochemical pull-down interaction assays, in which bait proteins derived from the CNGB1 N-terminal region, as shown in Fig. 4A, were tested for an interaction with a fluorescently labeled CaM. Fluorescent CaMs that interacted with bait proteins were detected by using a fluorescent imaging system after SDS/PAGE. We found that CaM formed a physical interaction with CNGB1#677–764 in the presence of Ca2+ but not in the absence of Ca2+ (Fig. 4B). In contrast, CaM did not form an interaction with CNGB1#702–764, in either the presence or absence of Ca2+ (Fig. 4B). Thus, an ≈20-aa stretch (from amino acids 677 to 701) in the N-terminal region of CNGB1 was necessary for both binding to Ca2+/CaM and inhibition of CNGA1/CNGB1 channels by Ca2+/CaM. In a previous study, no Ca2+ dependence was found for the CaM interaction with CNGB1, but Ca2+ was required for channel inhibition by CaM (27). Our results suggest a mechanism whereby Ca2+/CaM forms a physical interaction with an N-terminal region of CNGB1 subunits, including a necessary region of ≈20 aa, and inhibits CNGA1/CNGB1 channels. This interpretation is generally consistent with previous conclusions (27, 28).
Figure 4.
Requirement of N-terminal Ca2+/CaM binding site in CNGB1 for binding both Ca2+/CaM and a C-terminal domain of CNGA1. (A) Cartoon showing bait and fish fusion proteins and their position relative to a generic CNG channel subunit. (B) Gel showing results of biochemical pull-down interaction assay between bait proteins CNGB1#677–764 and CNGB1#702–764 and fluorescently labeled CaM in the presence of Ca2+ or EDTA. (C) Western blot showing results of pull-down assays with bait proteins CNGB1#677–764 and CNGB1#702–764 and the fish protein CNGA1#497–693.
CNGB1 N-Terminal Ca2+/CaM Binding Site Is Required for Interaction with CNGA1.
To determine whether the interaction between CNGA1 and CNGB1 was involved in Ca2+/CaM modulation, we performed additional interaction assays. In these experiments, we used the same bait proteins used in Fig. 4B (CNGB1#677–764 and CNGB1#702–764) to test for an interaction with a fish protein derived from a C-terminal region of CNGA1 (CNGA1#497–693; Fig. 4A). We detected a strong interaction between CNGB1#677–764 and CNGA1#497–693 (Fig. 4C). In contrast, CNGB1#702–764, which lacked the Ca2+/CaM-binding domain, did not interact with the C-terminal region of CNGA1 (Fig. 4C). A control experiment using GST alone as bait also did not produce an interaction with CNGA1#497–693. These results show that the CNGB1 N-terminal region necessary for Ca2+/CaM binding was also necessary for the interaction with the C-terminal domain of CNGA1.
Ca2+/CaM Disrupts an Intersubunit Interaction.
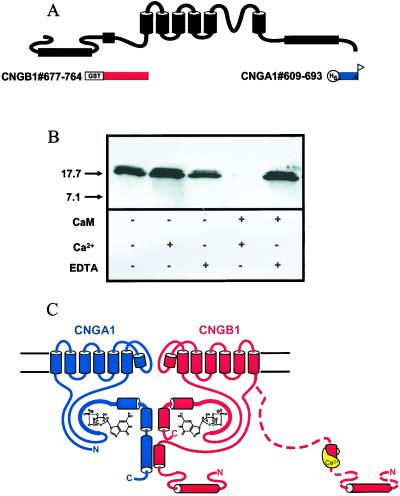
As an N-terminal region of CNGB1 was necessary for an interaction with both Ca2+/CaM and a C-terminal region of CNGA1, we investigated whether Ca2+/CaM could interfere with the CNGA1/CNGB1 intersubunit interaction. We performed the interaction experiment between CNGB1#677–764 and CNGA1#609–693 (Fig. 5A) in the absence or presence of EDTA, Ca2+, and CaM as indicated in Fig. 5B. When both Ca2+ and CaM were present, the interaction between CNGB1#677–764 and CNGA1#609–693 was not formed (Fig. 5B). The interaction remained intact in the presence of EDTA and CaM, indicating that Ca2+-bound CaM was indeed required to perturb the interaction. Thus, Ca2+/CaM disrupted the interaction between the N-terminal region of CNGB1 and the distal C-terminal region of CNGA1.
Figure 5.
Disruption of the interaction between CNGA1 and CNGB1 by Ca2+/CaM. (A) Bait proteins derived from CNGB1 (red) and fish protein derived from CNGA1 (blue). (B) Western blot of pull-down experiments with the bait protein CNGB1#677–764 and the fish protein CNGA1#609–693, in the presence of EDTA, Ca2+, or CaM. (C) An intersubunit interaction between the distal C-terminal domain of CNGA1 and an N-terminal domain of CNGB1 and prevention of the interaction by Ca2+/CaM.
Discussion
We have shown evidence for an interaction between the distal C-terminal region of CNGA1 and an N-terminal region of CNGB1 that includes the Ca2+/CaM-binding site (Fig. 5C). Here, we propose that this intersubunit interaction is critical for Ca2+/CaM modulation of rod CNGA1/CNGB1 heteromers. Ca2+/CaM binds to a site in the CNGB1 N-terminal region that is also necessary for forming the intersubunit interaction with CNGA1. Ca2+/CaM prevents the CNGA1/CNGB1 intersubunit interaction in intact channels, in turn causing channel inhibition (Fig. 5C).
The model for Ca2+/CaM-dependent inhibition in CNGA1/CNGB1 heteromers presented here predicts that deletion of the distal C-terminal region from CNGA1 would result in channels insensitive to Ca2+/CaM. However, when we performed this experiment, heteromeric channels were not expressed at the membrane surface (32). Thus, the N- and C-terminal region interaction between CNGA1 and CNGB1 subunits may be critical not only for modulation of channels by Ca2+/CaM, but also for proper trafficking of heteromeric channels to the cell surface.
Ca2+/CaM modulates a host of ion channels by a variety of mechanisms (for a review see ref. 35). For example, the SK class of Ca2+-activated K+ channels and L-type voltage-activated Ca2+ channels are constitutively bound by CaM in the absence of Ca2+ ion. Gating of the CaM-channel complex is affected only upon introduction of free Ca2+ ion, which activates SK channels, but inhibits Ca2+ channels (36–38). N-methyl-d-aspartate receptors are inhibited when Ca2+/CaM binds to a C-terminal region and displaces a channel domain that normally binds to the cytoskeleton (39).
CNG channels are modulated by Ca2+/CaM via mechanisms distinct from those above. In a general sense, Ca2+/CaM inhibits both retinal rod and olfactory CNG channels by disrupting N- and C-terminal interactions. However, there are several differences between the mechanisms of modulation in CNGA1/CNGB1 channels and CNGA2 channels. (i) In CNGA2 channels, Ca2+/CaM binds an N-terminal sequence (FQRIVRLVGVIRDW) with hallmark features of a “1–8-14” type of Ca2+/CaM binding motif (24). In this type of motif, hydrophobic (often aromatic) residues at the 1 and 14 positions flank internal long-chain hydrophobic residues at the 8 (and also often 5) position (for a review see ref. 40). In CNGA1/CNGB1 channels, the region sufficient to bind Ca2+/CaM (682-LQELVKLFKERTEKVKEKLI-701) bears more similarity to the consensus sequence for IQ motifs (IQxxxRGxxxRxxF/W) but, notably, lacks a final hydrophobic residue. Also, the CNGB1 region is not amphipathic, unlike IQ motifs (27). These differences, plus the finding that IQ motifs usually do not require Ca2+ to bind CaM indicates that the CNGB1 motif is perhaps best characterized as an unconventional type of CaM-binding site, as has been suggested (28). (ii) Ca2+/CaM decreases the apparent affinity for cyclic nucleotide by 10-fold in CNGA2 channels (24, 25) but only about 2-fold for CNGA1/CNGB1 channels (27, 28). (iii) In CNGA2 channels, deletion of the 1–8-14 domain decreases the apparent affinity for cyclic nucleotides about 10-fold, suggesting that Ca2+/CaM disrupts a potentiating effect of this region (24, 25). In contrast, deletion of the unconventional CaM-binding domain from CNGB1 subunits in CNGA1/CNGB1 heteromers does not appear to affect channel gating (i.e., the apparent affinity for cGMP and the fractional activation by cAMP remain similar to that for wild-type channels). It should be noted that a 2-fold shift in the apparent affinity for cGMP upon deletion of the unconventional Ca2+/CaM site (once anticipated based on the 2-fold shift in the presence of Ca2+/CaM) might be difficult to detect, as it approaches the normal range of variability for this value in wild-type channels; however, the fractional activation by cAMP is a more sensitive measure for a potential change in the allosteric opening transition. In CNGA1/CNGB1 heteromers the fractional activation by cAMP was reduced about 4-fold in the presence of Ca2+/CaM, but there was not a significant reduction in the fractional activation by cAMP in heteromeric channels containing CNGB1 subunits with the Ca2+/CaM site deleted. (iv) The N-terminal region of CNGA2 interacts extensively with the C-terminal region of CNGA2, including the C-linker region (the region between the S6 transmembrane domain and the CNBD) and the CNBD, but not the distal C-terminal region (25). In marked contrast, the CNGB1 N-terminal region does not interact with its own C-terminal region (32), or the CNBD of CNGA1, but rather interacts specifically with the distal C-terminal region of the CNGA1 subunit. Surprisingly, CNGA1/CNGB1 and CNGA2 channels have apparently evolved independent mechanisms for inhibition by Ca2+/CaM.
The mechanism of Ca2+/CaM-dependent inhibition in CNGA1/CNGB1 does not appear to involve disruption of an internal autopotentiating interaction. As discussed above, the N-terminal region does not appear to potentiate channel opening, nor does the distal C-terminal region of CNGA1 appear to be a region intimately involved in gating; its deletion does not significantly affect gating of CNGA1 homomers (32). However, we were unable to test this idea more completely in heteromeric CNGA1/CNGB1 channels, because they were not detected at the membrane surface when the CNGA1 subunit had the distal C-terminal region deleted (32). Instead of disrupting a potentiating interaction, Ca2+/CaM disruption of the intersubunit interaction may directly inhibit the opening allosteric transition of CNGA1/CNGB1 channels.
In addition to being a main component of retinal rod channels, a truncated form of CNGB1 (CNGB1b) is expressed in olfactory sensory neurons where it forms channels with both CNGA2 and CNGA4 subunits (41, 42). Although the stoichiometry and arrangement of subunits in CNG channels with three different subunits (CNGA2/CNGA4/CNGB1b) is unknown, such channels have the elements to form N- and C-terminal interactions among CNGA2 subunits and between the CNG1B N-terminal region and distal C-terminal regions of CNGA2 (because of homology with CNGA1). Thus, two different N- and C-terminal interactions involving two distinct Ca2+/CaM binding sites may form in CNGA2/CNGA4/CNGB1b channels, in turn leading to a distinct form of modulation by Ca2+/CaM. This may explain the finding that these channels exhibit faster Ca2+/CaM-dependent inhibition than that in either CNGA2, CNGA2/CNGA4, or CNGA2/CNGB1b channels (43, 44).
A physiological role for Ca2+/CaM inhibition of native retinal rod CNG channels is not completely understood; however, one proposed mechanism is as follows. In the dark, when Ca2+ levels are high (≈500 nM), Ca2+/CaM is associated with the channel, allowing the channel to respond to changes in higher levels of cGMP. In the light, cGMP levels drop, CNG channels close, leading to lower Ca2+ levels (≈50 nM), Ca2+/CaM dissociates from CNG channels, and channels become sensitized to changes in lower levels of cGMP and thus reopen. By this negative feedback mechanism, Ca2+ (acting through CaM) would help reset the circulating photoreceptor current after the light response (19). Such a mechanism contains the components to play a role in light adaptation. However, it has been proposed that because adaptation occurs over a 100-fold range in amphibian rods and Ca2+-dependent channel inhibition is over only a 2.5-fold range the contribution of this mechanism to adaptation may be relatively small (45). Nonetheless, an important physiological role for Ca2+/CaM modulation of CNG channels in adaptation cannot be ruled out (46). Interestingly, an endogenous CaM-like factor is bound to native rod channels in vivo (47, 48). This factor competes with exogenous Ca2+/CaM, indicating that they likely act through a similar mechanism. The endogenous factor may be more physiologically significant than Ca2+/CaM for modulation of CNG channels in native rods.
Acknowledgments
We thank Kimberley Craven, Kim Matulef, Galen Flynn, J. P. Johnson, Jr., Noah Shuart, Jie Zheng, and Mike Varnum for critical reading of the manuscript and Mike Varnum for helpful discussions about interaction assays. This work was supported by a grant to W.N.Z (T32 EY07031) from the National Institutes of Health. M.C.T is an Associate and W.N.Z. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- CNG
cyclic nucleotide-gated
- CaM
calmodulin
- CNBD
cyclic nucleotide-binding domain
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fesenko E E, Kolesnikov S S, Lyubarsky A L. Nature (London) 1985;313:310–313. doi: 10.1038/313310a0. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura T, Gold G H. Nature (London) 1987;325:442–444. doi: 10.1038/325442a0. [DOI] [PubMed] [Google Scholar]
- 3.Kolesnikov S S, Margolskee R F. Nature (London) 1995;376:85–88. doi: 10.1038/376085a0. [DOI] [PubMed] [Google Scholar]
- 4.Misaka T, Kusakabe Y, Emori Y, Gonoi T, Arai S, Abe K. J Biol Chem. 1997;272:22623–22639. doi: 10.1074/jbc.272.36.22623. [DOI] [PubMed] [Google Scholar]
- 5.Richards M J, Gordon S E. Biochemistry. 2000;39:14003–14011. doi: 10.1021/bi001639i. [DOI] [PubMed] [Google Scholar]
- 6.Kaupp U B, Niidome T, Tanabe T, Terada S, Bonigk W, Stuhmer W, Cook N J, Kangawa K, Matsuo H, Hirose T, et al. Nature (London) 1989;342:762–766. doi: 10.1038/342762a0. [DOI] [PubMed] [Google Scholar]
- 7.Dhallan R S, Yau K W, Schrader K A, Reed R R. Nature (London) 1990;347:184–187. doi: 10.1038/347184a0. [DOI] [PubMed] [Google Scholar]
- 8.Liu D T, Tibbs G R, Siegelbaum S A. Neuron. 1996;16:983–990. doi: 10.1016/s0896-6273(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 9.Varnum M D, Zagotta W N. Biophys J. 1996;70:2667–2679. doi: 10.1016/S0006-3495(96)79836-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jan L Y, Jan Y N. Nature (London) 1990;345:672. doi: 10.1038/345672a0. [DOI] [PubMed] [Google Scholar]
- 11.Weber I T, Steitz T A. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 12.Su Y, Dostmann W R, Herberg F W, Durick K, Xuong N H, Ten Eyck L, Taylor S S, Varughese K I. Science. 1995;269:807–813. doi: 10.1126/science.7638597. [DOI] [PubMed] [Google Scholar]
- 13.Zagotta W N, Siegelbaum S A. Annu Rev Neurosci. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
- 14.Bradley J, Frings S, Yau K-W, Reed R R. Science. 2001;294:2095. doi: 10.1126/science.294.5549.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn J T, Krautwurst D, Schroeder J E, Chen T Y, Reed R R, Yau K W. Biophys J. 1998;74:1333–1345. doi: 10.1016/S0006-3495(98)77846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shammat I M, Gordon S E. Neuron. 1999;23:809–819. doi: 10.1016/s0896-6273(01)80038-6. [DOI] [PubMed] [Google Scholar]
- 17.He Y, Ruiz M, Karpen J W. Proc Natl Acad Sci USA. 2000;97:895–900. doi: 10.1073/pnas.97.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen T Y, Peng Y W, Dhallan R S, Ahamed B, Reed R R, Yau K W. Nature (London) 1993;362:764–767. doi: 10.1038/362764a0. [DOI] [PubMed] [Google Scholar]
- 19.Hsu Y T, Molday R S. Nature (London) 1993;361:76–79. doi: 10.1038/361076a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen T Y, Yau K W. Nature (London) 1994;368:545–548. doi: 10.1038/368545a0. [DOI] [PubMed] [Google Scholar]
- 21.Hsu Y T, Molday R S. J Biol Chem. 1994;269:29765–29770. [PubMed] [Google Scholar]
- 22.Korschen H G, Illing M, Seifert R, Sesti F, Williams A, Gotzes S, Colville C, Muller F, Dose A, Godde M, et al. Neuron. 1995;15:627–636. doi: 10.1016/0896-6273(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 23.Bauer P J. J Physiol (London) 1996;494:675–685. doi: 10.1113/jphysiol.1996.sp021523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M, Chen T Y, Ahamed B, Li J, Yau K W. Science. 1994;266:1348–1354. doi: 10.1126/science.266.5189.1348. [DOI] [PubMed] [Google Scholar]
- 25.Varnum M D, Zagotta W N. Science. 1997;278:110–113. doi: 10.1126/science.278.5335.110. [DOI] [PubMed] [Google Scholar]
- 26.Kurahashi T, Menini A. Nature (London) 1997;385:725–729. doi: 10.1038/385725a0. [DOI] [PubMed] [Google Scholar]
- 27.Grunwald M E, Yu W P, Yu H H, Yau K W. J Biol Chem. 1998;273:9148–9157. doi: 10.1074/jbc.273.15.9148. [DOI] [PubMed] [Google Scholar]
- 28.Weitz D, Zoche M, Muller F, Beyermann M, Korschen H G, Kaupp U B, Koch K W. EMBO J. 1998;17:2273–2284. doi: 10.1093/emboj/17.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon S E, Zagotta W N. Neuron. 1995;14:177–183. doi: 10.1016/0896-6273(95)90252-x. [DOI] [PubMed] [Google Scholar]
- 30.Hamill O P, Marty A, Neher E, Sakmann B, Sigworth F J. Pflügers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 31.Bers D, Patton C, Nuccitelli R. Methods Cell Biol. 1994;40:3–29. doi: 10.1016/s0091-679x(08)61108-5. [DOI] [PubMed] [Google Scholar]
- 32.Trudeau M C, Zagotta W N. Neuron. 2002;34:197–207. doi: 10.1016/s0896-6273(02)00647-5. [DOI] [PubMed] [Google Scholar]
- 33.Ildefonse M, Crouzy S, Bennett N. J Membr Biol. 1992;130:91–104. doi: 10.1007/BF00233741. [DOI] [PubMed] [Google Scholar]
- 34.Sunderman E R, Zagotta W N. J Gen Physiol. 1999;113:601–620. doi: 10.1085/jgp.113.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levitan I B. Neuron. 1999;22:645–648. doi: 10.1016/s0896-6273(00)80722-9. [DOI] [PubMed] [Google Scholar]
- 36.Xia X M, Fakler B, Rivard A, Wayman G, Johnson-Pais T, Keen J E, Ishii T, Hirschberg B, Bond C T, Lutsenko S, et al. Nature (London) 1998;395:503–507. doi: 10.1038/26758. [DOI] [PubMed] [Google Scholar]
- 37.Qin N, Olcese R, Bransby M, Lin T, Birnbaumer L. Proc Natl Acad Sci USA. 1999;96:2435–2438. doi: 10.1073/pnas.96.5.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson B Z, DeMaria C D, Adelman J P, Yue D T. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S, Ehlers M D, Bernhardt J P, Su C T, Huganir R L. Neuron. 1998;21:443–453. doi: 10.1016/s0896-6273(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 40.Rhoads A R, Friedberg F. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 41.Sautter A, Zong X, Hofmann F, Biel M. Proc Natl Acad Sci USA. 1998;95:4696–4701. doi: 10.1073/pnas.95.8.4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonigk W, Bradley J, Muller F, Sesti F, Boekhoff I, Ronnett G V, Kaupp U B, Frings S. J Neurosci. 1999;19:5332–5347. doi: 10.1523/JNEUROSCI.19-13-05332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munger S D, Lane A P, Zhong H, Leinders-Zufall T, Yau K W, Zufall F, Reed R R. Science. 2001;294:2172–2175. doi: 10.1126/science.1063224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradley J, Reuter D, Frings S. Science. 2001;294:2176–2178. doi: 10.1126/science.1063415. [DOI] [PubMed] [Google Scholar]
- 45.Nakatani K, Koutalos Y, Yau K W. J Physiol (London) 1995;484:69–76. doi: 10.1113/jphysiol.1995.sp020648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pugh E N, Jr, Nikonov S, Lamb T D. Curr Opin Neurobiol. 1999;9:410–418. doi: 10.1016/S0959-4388(99)80062-2. [DOI] [PubMed] [Google Scholar]
- 47.Gordon S E, Downing-Park J, Zimmerman A L. J Physiol (London) 1995;486:533–546. doi: 10.1113/jphysiol.1995.sp020832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rebrik T I, Korenbrot J I. J Gen Physiol. 1998;112:537–548. doi: 10.1085/jgp.112.5.537. [DOI] [PMC free article] [PubMed] [Google Scholar]