Abstract
As the primary site of lipogenesis in birds, the liver orchestrates avian lipid metabolism and is pivotal for fat accumulation in chickens. Lipid metabolism during the broiler embryo stage may significantly affect post-hatch growth performance, yet research on this subject remains limited. While long non-coding RNAs (lncRNAs) have been found to regulate liver lipid metabolism in post-hatch chickens, their functions during the embryonic stage remains unclear. This study revealed that, compared to lean line broiler embryos, fat line broiler embryos showed upregulated gene expression related to de novo fatty acid synthesis, glycerol-3-phosphate synthesis, triglyceride synthesis, and the degradation of both fatty acids and cholesterol. Through transcriptome analysis and functional validation, lncRNA1926 and lncRNA3223 were identified as key regulators of lipid metabolism in broiler embryo livers. Knocking down either of lncRNA1926 or lncRNA3223 significantly reduced lipid droplet accumulation, triglyceride levels, and total cholesterol levels in primary hepatocytes of broiler embryos. Our findings demonstrate distinct lipid metabolic gene expression profiles between fat and lean line broiler embryo livers, and highlight lncRNA1926 and lncRNA3223 are key regulators of lipid metabolism during the embryonic stage. This study enhances the scientific understanding of lipid metabolism regulation in chicken livers and provides a theoretical foundation for genetically improving abdominal fat traits in broilers.
Keywords: Broiler, Embryonic stage, Liver, Lipid metabolism, LncRNAs
Introduction
After long term selective breeding, the growth rate and meat yield of broilers have been significantly improved. However, the rapid growth has led to excessive accumulation of body fat in broilers, especially abdominal fat, which reduces feed conversion efficiency and meat quality in broilers (Chen et al., 2021; Shen, Bai et al., 2024), and reproductive performance of broiler breeder (Ma, Cheng et al., 2024). Therefore, controlling excessive fat deposition is a critical issue in broiler breeding and production.
Liver lipid metabolism is crucial in broiler fat deposition, differing from mammals (Emami, Jung et al., 2021). With limited capacity for fatty acid synthesis in adipose tissue, over 70 % of fatty acids are synthesized in liver tissue (O'Hea and Leveille, 1968). With the shortened time to market, the embryonic period accounts for a larger proportion of their productive life (De Oliveira, Uni et al., 2008). The significance of the embryonic phase in broiler genetic potential is increasingly attracting breeder attention. Lipid metabolism in the liver is highly active during chicken embryo growth and development (Romanoff, 1967). In the middle to later stages of hatching, the embryo begins synthesizing cholesterol and triglycerides (Rogers and Bell, 1994). Specifically, in the final week before hatching, liver lipid content increases significantly (Hicks, Porter et al., 2019). Our previous research showed that by embryonic day 12, lipid droplets appear in the livers of both fat and lean lines, increasing in number and size with embryonic age (Na Wei, 2012). Guo et al. demonstrated that fat deposition in abdominal adipose tissue of both fat and lean lines of broilers is observable just 3 days post-hatching. By day 7, there is a clear difference in abdominal fat percentage (AFP) between the two lines. By week 7, the AFP in fat line broilers is 5 times higher than in lean line broilers (Guo, Sun et al., 2011). Thus, we hypothesize that variations in liver lipid metabolism capacity during the embryonic phase in fat and lean line broilers may correlate with postnatal differences in abdominal fat deposition.
Liver lipid metabolism in broilers is regulated by various factors, including long non-coding RNAs (lncRNAs). Although lncRNAs like LncLTR (Li, Gu et al., 2018), lncRNA-FNIP2 (Guo, Chao et al., 2021) and lnc_DHCR24 (Muret, Klopp et al., 2017) have been found to regulate liver lipid metabolism in post-hatch chicken, research in this area remains limited. In particular, the role of lncRNAs during the embryonic stage of chickens is poorly understood.
Northeast Agricultural University broiler lines (NEAUHLF) have been selectively bred since 1996 based on AFP and plasma very low-density lipoprotein (VLDL) levels, successfully established two divergent phenotypic lines (Guo, Sun et al., 2011). These lines serve as a well-established genetic model for investigating the mechanisms of abdominal fat deposition in broilers. Using this model, our study compared lipid metabolism differences in embryonic liver tissues between fat and lean line broilers. Through transcriptomic and functional genomic analyses, we identified the key lncRNAs that regulate embryonic hepatic lipid metabolism and elucidated their basic biological functions. Our findings provide important molecular targets and a theoretical foundation for the genetic improvement of abdominal fat traits in broilers.
Materials and methods
Ethics approval and consent to participate
All animal studies were conducted in strict accordance with the Guidelines for the Care and Use of Experimental Animals established by the Ministry of Science and Technology of the People’s Republic of China (Approval Number: 2006-398). The experimental protocols were reviewed and approved by the Laboratory Animal Management Committee and the Institutional Biosafety Committee of Northeast Agricultural University (Harbin, China).
Experimental birds and liver tissue collection
This study used hens from the 24th generation of the NEAUHLF as experimental materials. Fertilized eggs were collected and incubated. On embryonic days 12, 14, 17, and 21 (E12, E14, E17, and E21), eggs were removed from the incubator, and liver tissues from embryos of both fat and lean lines were immediately collected and stored in liquid nitrogen.
Isolation and culture of primary hepatocytes from broiler embryos
Eggs from the fat line broilers were incubated until embryonic day 17 (E17). Liver tissues were then isolated, and primary embryonic hepatocytes were obtained using a collagenase digestion method (Charni-Natan and Goldstein, 2020). The resulting cell sediment was resuspended and further separated using a Percoll gradient medium (Solarbio, China). The isolated cells were then resuspended in Opti-MEM® I culture medium (Invitrogen, USA) and cultured in an incubator at 37°C with 5 % CO₂.
Cell transfection
Primary hepatocytes isolated from embryos were seeded into a 12-well cell culture plate. When the cells reached 50 % confluency, they were transfected with SmartSilencer or the negative control (SmartSilencer-NC) using Lipofectamine™ 3000 Transfection Reagent (Invitrogen, USA). The interference fragments targeting lncRNA3223 and lncRNA1926 were designed and synthesized by RiboBio Co., Ltd (Guangzhou, China). These fragments included three small interfering RNAs (siRNAs) and three antisense oligonucleotides (ASOs). The specific sequences of the interference fragments are provided in Additional file 1 (Additional file 1: Table S1).
Oil Red O staining (ORO), triglyceride (TG), and total cholesterol (TCHO) assay
Intracellular lipid droplets accumulation was assessed via Oil Red O (ORO) staining. Briefly, cells were fixed with 4 % formaldehyde solution for 30 min, stained with ORO solution (Sigma, USA) for 15 min, and then washed with distilled water to remove unbound stain. The absorbance was measured at 510 nm using a microplate reader (BioTec, USA). Lipid accumulation was normalized to protein content, measured using a BCA Protein Quantitative Analysis Kit (Beyotime, China).
Triglyceride (TG) and total cholesterol (TCHO) contents were measured using commercial assay kits (Jiancheng, China) following the manufacturer's instructions. The TG absorbance was measured at 546 nm, and TCHO absorbance was measured at 510 nm using a microplate reader (BioTec, USA). TG and TCHO contents were calculated based on standard curves generated from known concentrations of triglyceride and cholesterol standards, respectively.
Library construction and sequencing analysis
Total RNA was extracted from embryonic liver tissues using the Trizol Reagent (Invitrogen, USA). High-throughput transcriptome sequencing was carried out by BioNew Technology (BioNew, China). The quality of raw sequencing data was assessed using FastQC software. Clean reads that passed quality control were aligned to the chicken reference genome (bGalGal1.mat.broiler.GRCg7b) using the HISAT2 program (Kim, Langmead et al., 2015).
LncRNAs identification
Known lncRNAs were identified by aligning transcripts with lncRNA databases (lncRNAdb and NONCODE). Novel lncRNAs were identified based on the following criteria: (1) lncRNA transcripta length > 200 nt, > 2 exons, and read coverage > 5; (2) transcript classes: "i," "x," "u"; (3) no alignment with known protein domains in the Pfam database; (4) non-coding potential confirmed by CNCI (score < 0), CPC (c score < 0), and CPAT (score < 0.363) (Kong, Zhang et al., 2007; Sun, Luo et al., 2013; Wang, Park et al., 2013; Luo, Bu et al., 2014; Mistry, Chuguransky et al., 2021). Only transcripts meeting all criteria were recognized as novel lncRNAs. These were merged with known lncRNAs to create a comprehensive collection for embryonic liver tissue in broilers from fat and lean lines at different developmental stages.
Differential expression and functional analysis
DESeq software was used to identify differentially expressed lncRNAs (DElncRNAs) based on Fragments Per Kilobase of exon per Million mapped fragments (FPKM), with thresholds of |log2Fold Change| ≥ 1 and q < 0.05. Potential target genes of lncRNAs were predicted based on positional relationships with and expression correlations. Genes within 100 kb upstream or downstream of a lncRNA were considered cis-target genes, while those with an absolute co-expression correlation coefficient > 0.8 and P < 0.05 were considered trans-target genes. KOBAS 3.0 online tool was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis on these target genes, with P < 0.05 was the threshold for significant enrichment.
Key lncRNAs affecting lipid metabolism in chicken embryo livers
This study identified key lncRNAs affecting lipid metabolism in chicken embryo livers through a four-step pipeline: (1) Identification of DElncRNAs between fat and lean line chicken embryo livers. (2) Chose DElncRNAs expressed differentially in more than two developmental stages (differential expression count >2). (3) Selected the top 10 % of lncRNAs with the highest fold changes in expression. (4) Performed cis and trans target gene prediction and functional enrichment analysis, focusing on those related to lipid metabolism. LncRNAs meeting all four criteria were considered key regulators of lipid metabolism in chicken embryo livers and were selected for further functional studies.
RT-qPCR
Total RNA was extracted using Trizol Reagent (Invitrogen, USA) following the manufacturer’s protocol. cDNA was synthesized with the PrimeScript RT reagent Kit (Takara, China). RT-qPCR was performed with 2 × SYBR Green master mix (Takara, China) on the Mx3000P Real-Time PCR System (Agilent, USA). The relative expression levels of target genes were determined by RT-qPCR using the 2−ΔCt method for liver tissues and 2−ΔΔCt method for cultured cells. TATA-box binding protein (TBP) was used as the internal reference gene (Na, Wang et al., 2021). Primer sequences are listed in Additional file 2 (Additional file 2: Table S2).
Statistical analysis
All experiments were conducted in triplicate, with three biological replicates per group. Data are presented as mean ± standard deviation (SD). Differences between the two groups were analyzed using Student’s t-test. P < 0.05 was considered significant, while P < 0.01 and P < 0.001 were considered highly significant.
Results
Comparison of TG and TCHO content in fat and lean line broiler embryo livers
To compare differences in liver lipid metabolism capacity between fat and lean lines during the embryonic stages (E12, E14, E17, E21), TG and TCHO content were measured in embryo liver tissues. The results showed that TG and TCHO content gradually increased in both lines (Fig. 1). TG content in the fat line was generally higher than in the lean line at all four stages (Fig. 1A). At E12, E14, and E17, TCHO content in the lean line was significantly higher than in the fat line (P < 0.05, Fig. 1B).
Fig. 1.
TG and TCHO content in embryonic liver of fat and lean line broilers. (A) TG content (B) TCHO content. Note: Different capital letters indicate significant differences between embryo stages in the lean line. Different lowercase letters indicate significant differences between embryo stages in the fat line. *P < 0.05, ***P < 0.001; n = 3.
Comparison of gene expression levels related to lipid metabolism in chicken embryo livers from fat and lean lines
To compare the difference in lipid metabolism between fat and lean line embryo livers, RT-qPCR was used to measure the mRNA expression of 10 key genes related to TG and TCHO synthesis, transport, and degradation at four embryonic development stages (E12, E14, E17, E21). These genes included fatty acid synthesis genes ACC (Acetyl-CoA Carboxylase) and FASN (Fatty Acid Synthase), glycerol-3-phosphate synthesis gene PHGDH (Phosphoglycerate Dehydrogenase), TG synthesis gene GPAT (Glycerol-3-Phosphate Acyltransferase), TG transport gene APOB100 (Apolipoprotein B100), triglyceride hydrolysis gene ATGL (Adipose Triglyceride Lipase), fatty acid oxidation gene CPT1 (Carnitine Palmitoyltransferase 1), cholesterol synthesis gene HMGR (HMG-CoA Reductase), cholesterol transport gene APOA1 (Apolipoprotein A1), and cholesterol hydrolysis gene CYP27A1 (cytochrome P450 family 27 subfamily A member 1). Results showed that ACC expression was significantly higher in fat line broilers than in lean line broilers at E17 and E21 (P < 0.05, Fig. 2A). FASN expression was significantly higher in fat line broilers than in lean line broilers at E12 and E17 (P < 0.05, Fig. 2B). PHGDH expression was significantly higher in fat line broilers than in lean line broilers at E17 and E21 (P < 0.05, Fig. 2C). GPAT expression was significantly higher in fat line broilers than in lean line broilers at all four stages (P < 0.05, Fig. 2D). APOB100 expression was significantly higher in fat line broilers than in lean line broilers at E17 (P < 0.01, Fig. 2E). CPT1 expression was significantly higher in fat line broilers than in lean line broilers at E14 and E21 (P < 0.05, Fig. 2G). CYP27A1 expression was significantly higher in fat line broilers than in lean line broilers at all four stages (P < 0.05, Fig. 2J). No significant differences were observed in ATGL, HMGR or APOA1 expression levels (Fig. 2F, 2 H, and 2 I). These findings suggest that fat line broilers have higher expression of genes involved in fatty acid and TG synthesis, stronger TG transport and fatty acid breakdown capacities, and higher cholesterol breakdown gene expression during the embryonic period compared to lean line broilers.
Fig. 2.
Expression of genes related to the synthesis, transport, and degradation of TG in embryonic liver of fat and lean line broilers. Note: *P < 0.05, ***P < 0.001; n = 3.
The screening of important lncRNAs related to lipid metabolism in liver tissue during the chicken embryonic stage
A total of 12,975 lncRNAs were identified in chicken embryonic liver tissue through the lncRNA identification process, of which 5,502 were previously annotated in the chicken reference genome (Additional file 3: Table S3) and 7,473 were identified as novel lncRNAs in this study. To verify the accuracy of the RNA-Seq result, five lncRNAs, LOC107049214, LOC107051926, LOC121109001, LOC112532434, and LOC112533223, were randomly selected for RT-qPCR validation. The RT-qPCR results were consistent with the RNA-Seq data (Fig. 3), confirming the reliability of sequencing results for further analysis.
Fig. 3.
RT-qPCR validation of lncRNAs. (A, C, E, G, I) RT-qPCR Validation Results (B, D, F, H, J) Sequencing Results. Note: *P < 0.05, **P < 0.01; n = 3.
Screening of DElncRNAs in liver tissue at different stages of chicken embryo development
To further understand lncRNA expression differences in broiler embryo liver tissues from fat and lean lines during embryonic development, DESeq software was used to analyze differential expression patterns at critical stages: E12, E14, E17, and E21. At E12, 133 DElncRNAs were identified, with 67 highly expressed in the fat line and 66 in the lean line. At E14, 99 DElncRNAs were detected, with 51 highly expressed in the fat line and 48 in the lean line. At E17, DElncRNAs increased to 182, with 100 highly expressed in the fat line and 82 in the lean line. Finally, at E21, 91 DElncRNAs were identified, with 38 highly expressed in the fat line and 53 in the lean line. In total, 350 DElncRNAs were identified across all four stages (Additional file 3: Table S3).
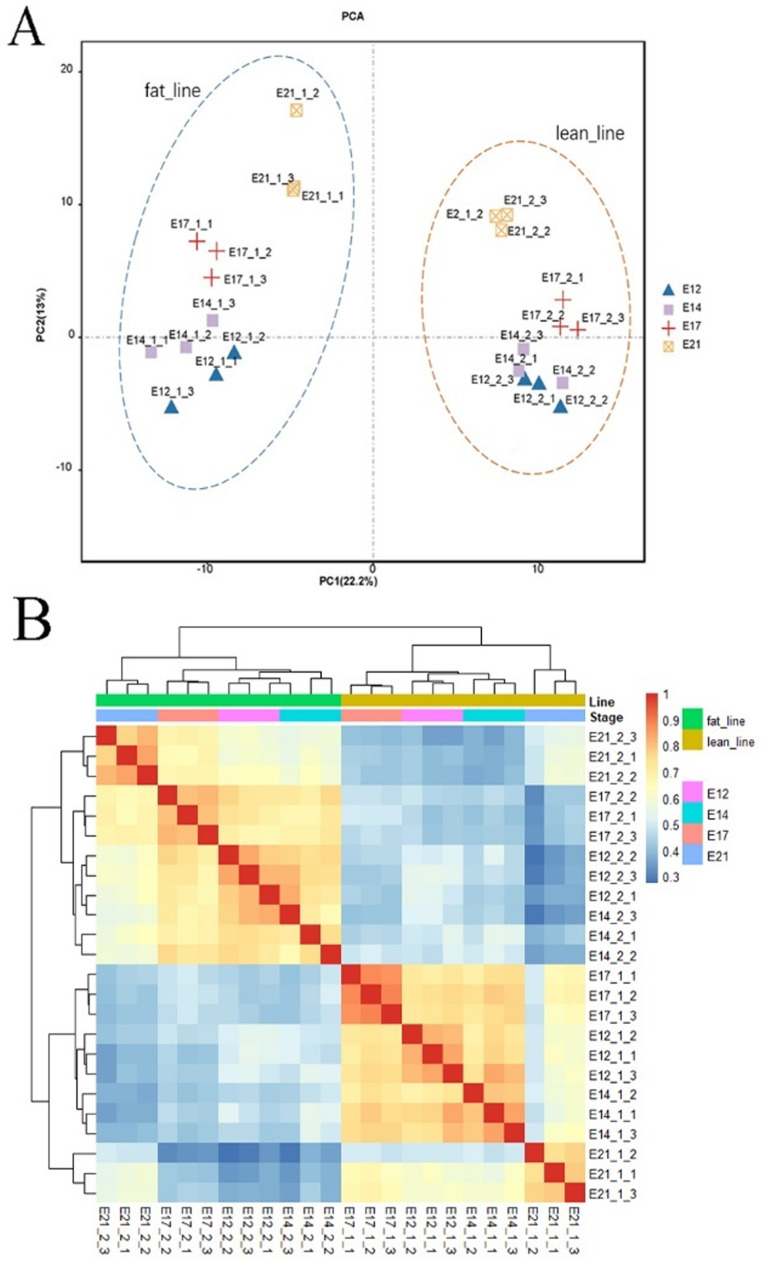
To ensure sample selection reliability, PCA and hierarchical clustering were performed based on DElncRNA expression. PCA showed that the 24 samples were segregated distinctly into fat and lean line groups, with clear clustering of liver samples from different embryonic stages (Fig. 4A). Hierarchical clustering also showed distinct separation into fat and lean groups, with clear discrimination between samples from various developmental stages (Fig. 4B).
Fig. 4.
Principal component and clustering analysis of 24 samples. (A) PCA analysis of all samples (B) Cluster analysis of all samples.
Key lncRNAs affecting lipid metabolism in chicken embryo livers
To identify key lncRNAs affecting lipid metabolism in embryonic broiler livers, a four-step screening process was used. Out of 350 DElncRNAs across four developmental stage, 96 were differentially expressed at multiple embryonic stages. The top 10 lncRNAs were selected based on fold change. Functional enrichment analysis of target genes revealed that two lncRNAs had target genes enriched in pathways associated with metabolic processes, lipid homeostasis, sphingolipid metabolism, triglyceride catabolism, lipid droplet formation, and other lipid metabolism-related pathways (Table S3). Through these four steps, these two annotated lncRNAs were identified: LOC112533223 (named lncRNA3223) and LOC107051926 (named lncRNA1926), which may play key roles in lipid metabolism. According to the NCBI database, lncRNA1926 is an intergenic lncRNA located on the Z chromosome, present in both the cytoplasm and nucleus, while lncRNA3223 is an antisense lncRNA located on chromosome 10, also present in both the cytoplasm and nucleus (Additional file 4: Table S4).
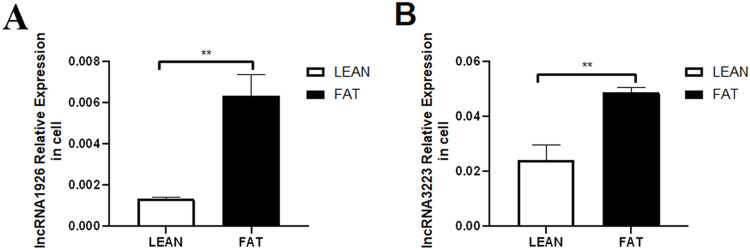
Expression of lncRNA1926 and lncRNA3223 in primary embryonic hepatocytes of fat and lean line broilers
Expression levels of lncRNA1926 and lncRNA3233 were assessed in primary hepatocytes from fat and lean line broiler embryos to select a suitable cell model. RT-qPCR results showed that both lncRNAs had lower expression in lean line hepatocytes than in fat line hepatocytes (Fig. 5). Therefore, primary hepatocytes from fat line broiler embryos were selected as the cellular model to explore the functions of these lncRNAs.
Fig. 5.
Expression of lncRNA1926 and lncRNA3223 in two types of hepatocytes (A) Expression of lncRNA1926 in two types of hepatocytes (B) Expression of lncRNA3223 in two types of hepatocytes. Note: *P < 0.05, **P < 0.01; n = 3.
Effects of lncRNA1926 knockdown on lipid droplet accumulation, TG content, TCHO content, and lipid metabolism-related gene expression in embryonic primary hepatocytes
To investigate the function of lncRNA1926 in liver lipid metabolism during the embryonic stage of broilers, we used a loss-of-function approach. Cells were transfected with lncRNA1926 Smart Silence (si-lncRNA1926) and an unrelated interference fragment (si-NC). The effects of lncRNA1926 knockdown were assessed at 24 and 48 hours (h) post-transfection on lipid droplet accumulation, TG and TCHO content, and expression levels of lipid metabolism-related genes, including ACC, FASN, PHGDH, GPAT, APOB100, CPT1, and CYP27A1.
RT-qPCR analysis showed that lncRNA1926 expression was significantly reduced by 79.7 % at 24 h and 71.1 % at 48 h post-transfection in si-lncRNA1926-treated cells compared to si-NC group (P < 0.05, Fig. 6A). This confirmed that the Smart Silence construct effectively reduced lncRNA1926 expression in hepatocytes from fat line broilers.
Fig. 6.
Verification of interference effects of lncRNA1926 and lncRNA3223 Smart Silence (A) Verification of interference effect of lncRNA1926 interference fragments (B) Verification of interference effect of lncRNA3223 interference fragments. Note: * P < 0.05, **P < 0.01; n = 3.
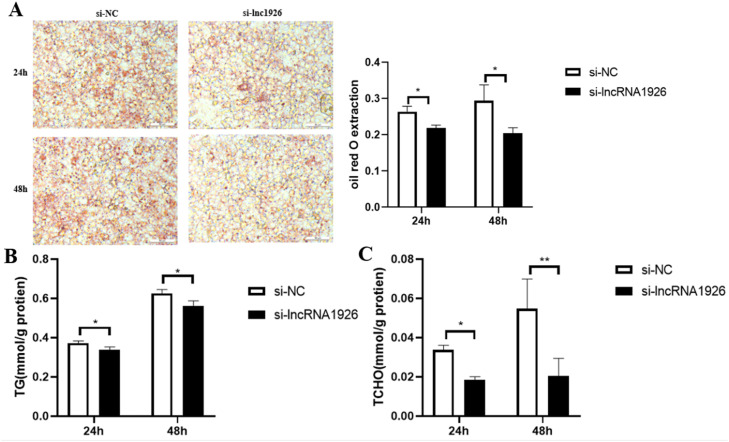
ORO staining and extraction colorimetry results indicated that lipid droplet accumulation was significantly lower in the si-lncRNA1926 group than in the si-NC group at 24 and 48 h post-transfection (P < 0.05, Fig. 7A). Similarly, TG and TCHO content were significantly reduced in the si-lncRNA1926 group compared to the si-NC group at these time points (P < 0.05, Fig. 7B and 7 C). These findings indicate that lncRNA1926 knockdown effectively inhibits lipid droplet formation and reduces TG and TCHO content in primary hepatocytes from broiler embryos.
Fig. 7.
Effects of lncRNA1926 knockdown on lipid droplet deposition, TG and TCHO content in primary hepatocytes of fat line broiler embryos (A) Primary hepatocytes of fat line broiler embryos were stained and extracted colorimetrically; Ruler: 50 μm (B) Effect of knockdown lncRNA1926 on TG content in primary hepatocytes of fat line broiler embryos (C) Effect of knockdown lncRNA1926 on TCHO content in primary hepatocytes of fat line broiler embryos. Note: *P < 0.05, **P < 0.01; n = 3.
RT-qPCR results indicated that, at 24 h post-transfection, the expression levels of fatty acid synthesis related genes ACC and FASN were significantly lower in the si-lncRNA1926 group than in the si-NC group (P < 0.01, Fig. 8A and 8 B). Similarly, the expression levels of TG synthesis related gene GPAT (P < 0.001, Fig. 8D), and fatty acid breakdown related gene CPT1 (P < 0.05, Fig. 8F) were significantly reduced. In contrast, the expression level of cholesterol degradation related gene CYP27A1 was significantly increased compared to the si-NC group (P < 0.05, Fig. 8G). At 48 h post-transfection, the expression level of FASN (P < 0.05, Fig. 8B) and GPAT (P < 0.05, Fig. 8D) were significantly lower in the si-lncRNA1926 group than in the si-NC group, while CYP27A1 expression level was further increased (P < 0.01, Fig. 8G). These results indicate that lncRNA1926 knockdown inhibited the expression of genes involved in fatty acid synthesis, TG synthesis, and fatty acid breakdown, while enhancing the expression of cholesterol breakdown genes. But it did not significantly affect the expression of genes involved in glycerol-3-phosphate synthesis and TG transport.
Fig. 8.
Effect of lncRNA1926 knockdown on the expression of lipid metabolism-related genes in primary hepatocytes of fat line broiler embryos. Note: *P < 0.05, **P < 0.01; n = 3.
Effects of lncRNA3223 knockdown on lipid droplet accumulation, TG content, TCHO content, and lipid metabolism-related gene expression in embryonic primary hepatocytes
RT-qPCR analysis confirmed that lncRNA3223 expression was significantly reduced by 73 % at 24 h and 82.8 % at 48 h post-transfection in si-lncRNA3223 group compared to si-NC group (P < 0.01, Fig. 6B).
ORO staining and extraction colorimetry results showed that lipid droplet accumulation was significantly lower in the si-lncRNA3223 group than in the si-NC group at 24 h and 48 h post-transfection (P < 0.05, Fig. 9A). Similarly, TG and TCHO content were significantly reduced in the si-lncRNA3223 group compared to the si-NC group at these time points (P < 0.05, Fig. 9B and 9C). These results suggest that knocking down lncRNA3223 inhibited lipid droplet accumulation and reduced TG and TCHO content in broiler embryonic primary hepatocytes.
Fig. 9.
Effects of lncRNA3223 knockdown on lipid droplet deposition, TG and TCHO content in primary hepatocytes of fat line broiler embryo (A) Primary hepatocytes of fat line broiler embryos were stained and extracted colorimetrically; Ruler: 50 μm (B) Effect of knockdown lncRNA3223 on triglyceride content in primary hepatocytes of fat line broiler embryos (C) Effect of knockdown lncRNA3223 on total cholesterol content in primary hepatocytes of fat line broiler embryos. Note: *P < 0.05, **P < 0.01; n = 3.
RT-qPCR results showed that lncRNA3223 knockdown inhibited the expression of fatty acid synthesis -related genes ACC and FASN (P < 0.05, Fig. 10A and 10 B), TG synthesis related gene GPAT (P < 0.001, Fig. 10D), fatty acid degradation related gene CPT1 (P < 0.01, Fig. 10F), and cholesterol degradation related gene CYP27A1 (P < 0.01, Fig. 10G) in embryonic primary hepatocytes at both 24 and 48 h post-transfection. However, it did not significantly affect the expression of genes related to glycerol-3-phosphate synthesis or TG transport.
Fig. 10.
Effect of lncRNA3223 knockdown on the expression of lipid metabolism-related genes in primary hepatocytes of fat line broiler embryos. Note: *P < 0.05, **P < 0.01; n = 3.
Discussion
The liver is the primary site for TG and cholesterol synthesis in poultry, playing a crucial role in lipid synthesis, transport, and degradation(Nguyen, Leray et al., 2008), and the accumulation of TG in adipose tissue is directly related to hepatic lipogenesis. Research on the role of lncRNA in the regulation of lipid metabolism in chicken livers is relatively limited, with only a few reports in the post-hatch stage (Wu, Liu et al., 2018; Xu, Zhang et al., 2019; Tan, Liu et al., 2020). However, studies focusing on the embryonic period are virtually non-existent. Therefore, this study used NEAUHLF embryonic liver tissues as the research subject, compared the differences in TG and TCHO content between the fat and lean line broilers, and identified lncRNA1926 and lncRNA3223 as key lncRNAs regulating lipid metabolism in the embryonic liver of broilers.
Research has shown that, as broiler embryos develop, the levels of TG and TCHO in the embryonic liver gradually increase (Sato, Tachibana et al., 2006; Liu, Zhou et al., 2020). Consistent with previous findings, our findings indicate that TG and TCHO content in liver tissues of both fat and lean line broiler embryos increase progressively with embryonic development. Notably, fat line broiler embryos generally have higher TG levels than the lean line broiler embryos across four stages. However, at E12, E14, and E17, TCHO levels in the livers of lean line broiler embryos significantly exceed those of fat line broilers (P < 0.05). These findings suggest that differences in lipid metabolism may exist between the fat and lean broiler lines during embryonic development.
To further compare lipid metabolism differences between the fat and lean lines, we used RT-qPCR to detect the mRNA expression levels of several lipid metabolism-related genes. In lipid synthesis, ACC and FASN are key enzymes for fatty acid synthesis(Wang, Kim et al., 2017) . Bourneuf et al. reported higher ACC and FASN expression in high abdominal fat weight broilers than in low abdominal fat weight broilers (Bourneuf, Hérault et al., 2006). Shi et al. found that ACC and FASN expression in the livers of fat line broilers was significantly higher than in lean line broilers during weeks 1-12 (Mingxin., Hongyan. et al., 2013). Consistent with these findings, our study found that ACC and FASN gene expression of fat line broiler embryos was significantly higher than in lean line broilers. In glycerol triphosphate synthesis, glycerol-3-phosphate is a key precursor, generated from the glucose metabolite dihydroxyacetone phosphate catalyzed by PHGDH. Reducing PHGDH activity in rat preadipocytes decreases TG accumulation in cells (Xu, Mao et al., 2011). Earlier research showed PHGDH expression was upregulated in abdominal fat tissue of fat line broilers at 7 weeks, indicating higher glycerol-3-phosphate synthesis in fat line broilers compared to lean line broilers (Wang, Li et al., 2007; Wang, Leng et al., 2021). Our study found that PHGDH expression in fat line broiler embryonic livers was significantly higher than in lean line broilers. In TG synthesis, fatty acids and glycerol-3-phosphate undergo esterification to form TG, with genes GPAT playing key roles (Weiss, Kennedy et al., 1960; Coleman, Lewin et al., 2000; Kavadia, Yadav et al., 2018). Studies have shown that GPAT levels in fat line broiler livers were generally higher than in lean line broilers (Mingxin., Hongyan. et al., 2013). In line with previous findings, our study found that GPAT expression in the embryonic livers of fat line broilers is significantly higher than in lean line broilers. Combining findings from this and earlier studies, fat line broilers exhibit stronger fatty acid synthesis, glycerol-3-phosphate synthesis, and TG synthesis capabilities in both the embryonic and postnatal periods, which may be one of the reasons for the differences in abdominal fat rates between broiler lines.
APOB100 is an essential component of very low-density lipoprotein, and changes in its expression levels may affect TG transport (Chen, Dettipponpong et al., 2025). Studies show that fat line broiler embryos have stronger fat transport capacity in the liver than lean line broilers (Na, Wu et al., 2018; Wu, Chen et al., 2021) . Consistent with these findings, our study also found significantly higher APOB100 expression in the embryonic livers of fat line broilers. This suggests that fat line broilers have enhanced fat transport in both embryonic and post-hatch tissues compared to lean line broilers.
ATGL is the rate-limiting enzyme for TG hydrolysis (Obrowsky, Chandak et al., 2013). Studies have shown that at 7 weeks of age, there is no significant difference in ATGL gene expression levels in the adipose tissue of high-fat and low-fat broilers (Mingxin., Hongyan. et al., 2013). Our study found that there was also no significant difference in ATGL gene expression in the embryonic livers of the two broiler lines. These findings are consistent with previous research, indicating that there is no significant difference in TG hydrolysis capacity between fat and lean line broilers in both embryonic liver tissue and postnatal adipose tissue. CPT1 is the key rate-limiting enzyme that catalyzes fatty acid breakdown (Karagianni and Talianidis, 2015). Our study showed that CPT1 gene expression was significantly higher in the livers of fat line broiler embryos than in lean line broilers, indicating stronger fatty acid breakdown capacity in fat line broilers. Notably, despite enhanced lipid metabolism in the embryonic livers of fat line broilers, their cholesterol content is lower than that of lean line broilers. This may be due to significantly higher expression of the CYP27A1 gene in the embryonic livers of fat line broilers, indicating stronger cholesterol breakdown capacity. In summary, fat line broilers show higher gene expression related to fatty acid synthesis, TG synthesis, fatty acid breakdown, and TG transport in embryonic livers than lean line broilers. This may explain why there's no significant difference in TG content between the two lines during the embryonic period. Fat line broilers also have higher gene expression for cholesterol breakdown, which might explain the lower cholesterol levels in their embryonic livers compared to lean line broilers.
LncRNAs are much less abundant than mRNAs in cells and can be found in both the nucleus and cytoplasm (Cabili, Trapnell et al., 2011; Mattick, Amaral et al., 2023). Because of this, siRNAs used for mRNA knockdown may not work well on lncRNAs. A Smart Silencer, which consists of a mixture of siRNAs and ASOs, offers higher sensitivity and inhibition efficiency than siRNAs alone and can achieve simultaneous knockdown of lncRNAs in both the nucleus and cytoplasm (Deng, Han et al., 2019). In this study, since lncRNA1926 and lncRNA3223 localize in both the nucleus and cytoplasm, we selected Smart Silencers to achieve a more effective knockdown of these targets. TG and cholesterol are essential precursors for lipid droplet formation (Farese and Walther, 2009). Our findings showed that knocking down lncRNA1926 and lncRNA3223 reduced lipid droplet deposition, as well as the TG and TCHO content in primary embryonic broiler hepatocytes. Our findings are consistent with previous research on the embryonic liver of Arbor Acres (AA) broilers (Li, Zhang et al., 2023). Thus, our findings suggest that lncRNA1926 and lncRNA3223 promote lipid droplet deposition in primary broiler embryo hepatocytes by increasing intracellular TG and TCHO content. Our results demonstrated that knocking down lncRNA1926 in primary hepatocytes of fat line broilers significantly decreased ACC, FAS, GPAT and CPT1 expressions, but did not affect PHGDH or APOB100 expression levels. Our findings are consistent with previous research on the embryonic liver of Arbor Acres (AA) broilers (Li, Zhang et al., 2023). In summary, lncRNA1926 knockdown reduced de novo fatty acid synthesis and TG synthesis capacity in primary hepatocytes of fat lean line broilers, without impacting glycerol-3-phosphate synthesis or TG transport capacity. Similarly, knocking down lncRNA3223 in embryonic primary hepatocytes of fat line broilers significantly decreased ACC, FAS, GPAT and CPT1 expressions, while PHGDH and APOB100 expressions remained unaffected. In summary, lncRNA3223 knockdown reduced de novo fatty acid synthesis and TG synthesis capacity in embryonic primary hepatocytes of broilers, without affecting glycerol-3-phosphate synthesis or TG transport. These results indicate that lncRNA1926 and lncRNA3223 positively regulate genes involved in de novo fatty acid synthesis and TG synthesis in embryonic primary hepatocytes of fat line broilers.
Approximately 50 % of cholesterol in animals is broken down and oxidized into bile acids in the liver. About 40 % of the bile is transported from the liver to the small intestine, where it is ultimately excreted in the feces along with bile acids and phospholipids. The remaining roughly 10 % of cholesterol is used for the synthesis of steroid hormones(Ma, 2019 (in Chinese)). The CYP27A1 gene is crucial for cholesterol conversion into bile acids (Dubrac, Lear et al., 2005). Our study found that knocking down lncRNA1926 significantly upregulated CYP27A1 in primary hepatocytes derived from fat line broiler embryos and decreased TCHO content. This suggests that lncRNA1926 may act as a negative regulator of cholesterol metabolism. Conversely, knocking down lncRNA3223 reduced CYP27A1 expression and unexpectedly decreased TCHO content, implying that lncRNA3223 may be a positive regulator of cholesterol metabolism. Although knocking down lncRNA3223 might inhibit cholesterol metabolism, further research is needed to clarify the underlying mechanisms. In summary, lncRNA1926 negatively regulates genes related to cholesterol catabolism in primary hepatocytes of fat line broiler embryos, while lncRNA3223 positively regulates these genes.
Although this study systematically analyzed the differences in hepatic lipid metabolism between fat and lean line broiler during the embryonic stage, identified lncRNAs that may play key roles, and conducted preliminary functional analyses, many questions remain. For example, whether the differential expression of proteins related to lipid metabolism in the livers of high- and low-fat broiler embryos correlates with mRNA levels, and how lncRNA1926 and lncRNA3223 regulate lipid metabolism in primary hepatocytes of chicken embryos, are areas for further investigation.
Conclusion
The study concludes that fat line broiler embryo livers have superior capabilities for de novo fatty acid synthesis, glycerol-3-phosphate synthesis, triglyceride synthesis, and the degradation of fatty acids and cholesterol, compared to lean line broilers. LncRNA1926 and lncRNA3223 are key regulators of lipid metabolism in broiler embryo livers. Knocking down either lncRNA1926 or lncRNA3223 significantly reduces lipid droplet accumulation, TG, and TCHO levels in primary broiler embryo hepatocytes. LncRNA1926 acts as a positive regulator of genes involved in de novo fatty acid synthesis, triglyceride synthesis, and fatty acid degradation, while negatively regulating genes related to cholesterol degradation in primary chicken embryo hepatocytes. Similarly, lncRNA3223 acts as a positive regulator of genes involved in de novo fatty acid synthesis, triglyceride synthesis, and cholesterol degradation in these cells.
Funding
This work was supported by the National Key R&D Program Young Scientist Project (No. 2022YFF1000800), Heilongjiang Province Postdoctoral General Program (LBH-Z20111), the Joint Guidance Project of Heilongjiang Natural Science Foundation (No. LH2021C037), the Earmarked Fund for CARS-41 (No. CARS-41).
Declaration of competing interest
All authors have approved the final version of the manuscript and declare no conflicts of interest.
Acknowledgements
The authors extend their sincere gratitude to the members of the Poultry Breeding group at Northeast Agricultural University for their invaluable assistance in bird management and sample collection. The authors also wish to express their appreciation to BioNew Technology (Inner Mongolia, China) for providing sequencing services.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2025.105261.
Appendix. Supplementary materials
References
- Bourneuf E., Hérault F., Chicault C., Carré W., Assaf S., Monnier A., Mottier S., Lagarrigue S., Douaire M., Mosser J., Diot C. Microarray analysis of differential gene expression in the liver of lean and fat chickens. Gene. 2006;372:162–170. doi: 10.1016/j.gene.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Cabili M.N., Trapnell C.., Goff L., Koziol M., Tazon-Vega B., Regev A., Rinn J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charni-Natan M., Goldstein I. Protocol for primary mouse hepatocyte isolation. STAR. Protoc. 2020;1(2) doi: 10.1016/j.xpro.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Su Z., Li Y., Luan P., Wang S., Zhang H., Xiao F., Guo H., Cao Z., Li H., Leng L. Estimation of the genetic parameters of traits relevant to feed efficiency: result from broiler lines divergent for high or low abdominal fat content. Poult. Sci. 2021;100(2):461–466. doi: 10.1016/j.psj.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-H., Dettipponpong P., Sin M.-Y., Chang L.-C., Cheng C.-Y., Huang S.-Y., Walzem R.L., Cheng H.-C., Chen S.-E. Ovarian expression of functional MTTP and apoB for VLDL assembly and secretion in chickens. Poult. Sci. 2025;104(5) doi: 10.1016/j.psj.2025.104993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R.A., Lewin T..M., Muoio D.M. Physiological and nutritional regulation of enzymes of triacylglycerol synthesis. Annu. Rev. Nutr. 2000;20:77–103. doi: 10.1146/annurev.nutr.20.1.77. [DOI] [PubMed] [Google Scholar]
- De Oliveira J.E., Uni Z.., Ferket P.R. Important metabolic pathways in poultry embryos prior to hatch. Poult. Sci. J. 2008;64(4):488–499. World's. [Google Scholar]
- Deng R., Han C., Zhao L., Zhang Q., Yan B., Cheng R., Wei B., Meng P., Mao T., Zhang Y., Liu J. Identification and characterization of ERV transcripts in goat embryos. Reproduction. 2019;157(1):115–126. doi: 10.1530/REP-18-0336. [DOI] [PubMed] [Google Scholar]
- Dubrac S., Lear S.R., Ananthanarayanan M., Balasubramaniyan N., Bollineni J., Shefer S., Hyogo H., Cohen D.E., Blanche P.J., Krauss R.M., Batta A.K., Salen G., Suchy F.J., Maeda N., Erickson S.K. Role of CYP27A in cholesterol and bile acid metabolism. J. Lipid Res. 2005;46(1):76–85. doi: 10.1194/jlr.M400219-JLR200. [DOI] [PubMed] [Google Scholar]
- Emami N.K., Jung U.., Voy B., Dridi S. Radical response: effects of heat stress-induced oxidative stress on lipid metabolism in the avian liver. Antioxidants. 2021;10(1):35. doi: 10.3390/antiox10010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese R.V., Jr, Walther T.C. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139(5):855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Chao X., Huang W., Li Z., Luan K., Ye M., Zhang S., Liu M., Li H., Luo W., Nie Q., Zhang X., Luo Q. Whole transcriptome analysis reveals a potential regulatory mechanism of LncRNA-FNIP2/miR-24-3p/FNIP2 axis in chicken adipogenesis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.653798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun B., Shang Z., Leng L., Wang Y., Wang N., Li H. Comparison of adipose tissue cellularity in chicken lines divergently selected for fatness. Poult. Sci. 2011;90(9):2024–2034. doi: 10.3382/ps.2010-00863. [DOI] [PubMed] [Google Scholar]
- Hicks J.A., Porter T..E., Sunny N.E., Liu H.C. Delayed feeding alters transcriptional and post-transcriptional regulation of hepatic metabolic pathways in Peri-hatch broiler chicks. Genes. (Basel) 2019;10(4):272. doi: 10.3390/genes10040272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagianni P., Talianidis I. Transcription factor networks regulating hepatic fatty acid metabolism. Biochim. Biophys. Acta (BBA) - Mol. Cell Biol. Lipids. 2015;1851(1):2–8. doi: 10.1016/j.bbalip.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Kavadia M.R., Yadav M..G., Odaneth A.A., Lali A.M. Synthesis of designer triglycerides by enzymatic acidolysis. Biotechnol. Rep. (Amst) 2018;18 doi: 10.1016/j.btre.2018.e00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L., Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic. Acids. Res. 2007;35(suppl_2):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Gu Z., Yang L., Tian Y., Kang X., Liu X. Transcriptome profile analysis reveals an estrogen induced LncRNA associated with lipid metabolism and carcass traits in chickens (Gallus Gallus) Cell Physiol. Biochem. 2018;50(5):1638–1658. doi: 10.1159/000494785. [DOI] [PubMed] [Google Scholar]
- Li H., Zhang H., Wang Y., Cheng B., Leng L., Luan P., Li Y., Bai X. Functional study on lncRNA1926 and lncRNA3223 in lipid metabolism of broiler embryonic primary hepatocytes. J. Northeast Agric. Univ. 2023;54(08):17–26. Chinese. [Google Scholar]
- Liu Y., Zhou J., Musa B.B., Khawar H., Yang X., Cao Y., Yang X. Developmental changes in hepatic lipid metabolism of chicks during the embryonic periods and the first week of posthatch. Poult. Sci. 2020;99(3):1655–1662. doi: 10.1016/j.psj.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Bu D., Sun L., Chen R., Zhao Y. De novo approach to classify protein-coding and noncoding transcripts based on sequence composition. Methods Mol. Biol. 2014;1182:203–207. doi: 10.1007/978-1-4939-1062-5_18. [DOI] [PubMed] [Google Scholar]
- Ma Y., Cheng B., Zhou S., Wang Y., Jing Y., Leng L., Wang S., Li Y., Luan P., Cao Z., Li H. Comparative analyses of laying performance and follicular development characteristics between fat and lean broiler lines. Poult. Sci. 2024;103(1) doi: 10.1016/j.psj.2023.103250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z. Henan Agricultural University; 2019. STUDY on EXPRESSION CHARACTERISTICS ANDREGULATION MECHANISM of CHOLESTEROL SYNTHESISPATHWAY RELATED GENES in CHICKEN LIVER. Doctor. Chinese. [Google Scholar]
- Mattick J.S., Amaral P..P., Carninci P., Carpenter S., Chang H.Y., Chen L.-L., Chen R., Dean C., Dinger M.E., Fitzgerald K.A., Gingeras T.R., Guttman M., Hirose T., Huarte M., Johnson R., Kanduri C., Kapranov P., Lawrence J.B., Lee J.T., Mendell J.T., Mercer T.R., Moore K.J., Nakagawa S., Rinn J.L., Spector D.L., Ulitsky I., Wan Y., Wilusz J.E., Wu M. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023;24(6):430–447. doi: 10.1038/s41580-022-00566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingxin S., Hongyan S., Hui L., Ning W. Expression analysis of lipid synthesis-related genes in the livers of fat and lean broilers. J. Agric. Biotechnol. 2013;21(03):306–312. Chinese. [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., Finn R.D., Bateman A. Pfam: the protein families database in 2021. Nucleic. Acids. Res. 2021;49(D1):D412–d419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muret K., Klopp C., Wucher V., Esquerré D., Legeai F., Lecerf F., Désert C., Boutin M., Jehl F., Acloque H., Giuffra E., Djebali S., Foissac S., Derrien T., Lagarrigue S. Long noncoding RNA repertoire in chicken liver and adipose tissue. Genet. Sel. Evol. 2017;49(1):6. doi: 10.1186/s12711-016-0275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na W., Wang Y., Gong P., Zhang X., Zhang K., Zhang H., Wang N., Li H. Screening of reference genes for RT-qPCR in chicken adipose tissue and adipocytes. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.676864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na W., Wu Y.Y., Gong P.F., Wu C.Y., Cheng B.H., Wang Y.X., Wang N., Du Z.Q., Li H. Embryonic transcriptome and proteome analyses on hepatic lipid metabolism in chickens divergently selected for abdominal fat content. BMC Genom. 2018;19(1):384. doi: 10.1186/s12864-018-4776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Wei W.Y., Yuxiang W., Hui L.i. Astrological observations on the broiler liver tissue during embryonic period. China Anim. Husb. Vet. Med. 2012;39(11):111–115. Chinese. [Google Scholar]
- Nguyen P., Leray V., Diez M., Serisier S., Bloc’h J.L., Siliart B., Dumon H. Liver lipid metabolism. J. Anim. Physiol. Anim. Nutr. 2008;92(3):272–283. doi: 10.1111/j.1439-0396.2007.00752.x. [DOI] [PubMed] [Google Scholar]
- O'Hea E.K., Leveille G.A. Lipogenesis in isolated adipose tissue of the domestic chick (Gallus domesticus) Comp. Biochem. Physiol. 1968;26(1):111–120. doi: 10.1016/0010-406x(68)90317-4. [DOI] [PubMed] [Google Scholar]
- Obrowsky S., Chandak P.G., Patankar J.V., Povoden S., Schlager S., Kershaw E.E., Bogner-Strauss J.G., Hoefler G., Levak-Frank S., Kratky D. Adipose triglyceride lipase is a TG hydrolase of the small intestine and regulates intestinal pparα signaling. J. Lipid Res. 2013;54(2):425–435. doi: 10.1194/jlr.M031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers L.J., Bell G.A. Changes in metabolic activity in the hyperstriatum of the chick before and after hatching. Int. J. Dev. Neurosci. 1994;12(6):557–566. doi: 10.1016/0736-5748(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Romanoff, A. L. (.1967). "Biochemistry of the avian embryo: a quantitative analysis of prenatal development".
- Sato M., Tachibana T., Furuse M. Total lipid and triacylglycerol contents in the liver of broiler and layer chickens at embryonic stages and hatching. Anim. Sci. J. 2006;77(5):526–531. [Google Scholar]
- Shen L., Bai X., Zhao L., Zhou J., Chang C., Li X., Cao Z., Li Y., Luan P., Li H., Zhang H. Integrative 3D genomics with multi-omics analysis and functional validation of genetic regulatory mechanisms of abdominal fat deposition in chickens. Nat. Commun. 2024;15(1):9274. doi: 10.1038/s41467-024-53692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic. Acids. Res. 2013;41(17):e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Liu R., Xing S., Zhang Y., Li Q., Zheng M., Zhao G., Wen J. Genome-wide detection of key genes and epigenetic markers for chicken fatty liver. Int. J. Mol. Sci. 2020;21(5):1800. doi: 10.3390/ijms21051800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Kim W.K., Cline M.A., Gilbert E.R. Factors affecting adipose tissue development in chickens: a review. Poult. Sci. 2017;96(10):3687–3699. doi: 10.3382/ps/pex184. [DOI] [PubMed] [Google Scholar]
- Wang H.B., Li H.., Wang Q.G., Zhang X.Y., Wang S.Z., Wang Y.X., Wang X.P. Profiling of chicken adipose tissue gene expression by genome array. BMC Genom. 2007;8:193. doi: 10.1186/1471-2164-8-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Leng L., Ding R., Gong P., Liu C., Wang N., Li H., Du Z.Q., Cheng B. Integrated transcriptome and proteome analysis reveals potential mechanisms for differential abdominal fat deposition between divergently selected chicken lines. J. Proteom. 2021;241 doi: 10.1016/j.jprot.2021.104242. [DOI] [PubMed] [Google Scholar]
- Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic. Acids. Res. 2013;41(6):e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S.B., Kennedy E..P., Kiyasu J.Y. The enzymatic synthesis of triglycerides. J. Biol. Chem. 1960;235:40–44. [PubMed] [Google Scholar]
- Wu C., Chen C., Liu Y., Li H., Cheng B. Proteomic analysis of liver tissue between fat and lean broiler lines. Br. Poult. Sci. 2021;62(2):211–218. doi: 10.1080/00071668.2020.1847253. [DOI] [PubMed] [Google Scholar]
- Wu S., Liu Y., Guo W., Cheng X., Ren X., Chen S., Li X., Duan Y., Sun Q., Yang X. Identification and characterization of long noncoding RNAs and mRNAs expression profiles related to postnatal liver maturation of breeder roosters using Ribo-zero RNA sequencing. BMC Genom. 2018;19(1):498. doi: 10.1186/s12864-018-4891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu E., Zhang L., Yang H., Shen L., Feng Y., Ren M., Xiao Y. Transcriptome profiling of the liver among the prenatal and postnatal stages in chickens. Poult. Sci. 2019;98(12):7030–7040. doi: 10.3382/ps/pez434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.P., Mao X..Y., Ren F.Z., Che H.L. Attenuating effect of casein glycomacropeptide on proliferation, differentiation, and lipid accumulation of in vitro Sprague-Dawley rat preadipocytes. J. Dairy. Sci. 2011;94(2):676–683. doi: 10.3168/jds.2010-3827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.