Abstract
Maintaining low levels of sodium ions in the cell cytosol is critical for plant growth and development. Biochemical studies suggest that Na+/H+ exchangers in the plasma membrane of plant cells contribute to cellular sodium homeostasis by transporting sodium ions out of the cell; however, these exchangers have not been identified at the molecular level. Genetic analysis has linked components of the salt overly sensitive pathway (SOS1–3) to salt tolerance in Arabidopsis thaliana. The predicted SOS1 protein sequence and comparisons of sodium ion accumulation in wild-type and sos1 plants suggest that SOS1 is involved directly in the transport of sodium ions across the plasma membrane. To demonstrate the transport capability of SOS1, we studied Na+/H+-exchange activity in wild-type and sos plants using highly purified plasma membrane vesicles. The results showed that plasma membrane Na+/H+-exchange activity was present in wild-type plants treated with 250 mM NaCl, but this transport activity was reduced by 80% in similarly treated sos1 plants. In vitro addition of activated SOS2 protein (a protein kinase) increased Na+/H+-exchange activity in salt-treated wild-type plants 2-fold relative to transport without added protein. However, the addition of activated SOS2 did not have any stimulatory effect on the exchange activity in sos1 plants. Although vesicles of sos2 and sos3 plants had reduced plasma membrane Na+/H+-exchange activity, transport activity in both increased with the addition of activated SOS2 protein. These results demonstrate that SOS1 contributes to plasma membrane Na+/H+ exchange and that SOS2 and SOS3 regulate SOS1 transport activity.
Soil salinity is one of the most significant abiotic stresses facing crop plants in agricultural fields worldwide (1). High levels of sodium ions (Na+) are toxic to plants because of their adverse effects on cellular metabolism and ion homeostasis (2, 3). Therefore, maintaining low levels of Na+ in the cell, specifically in the cell cytoplasm, is essential for plants (3, 4).
Plants are thought to remove Na+ from the cytoplasm by transporting it into the vacuole or out of the cell using Na+/H+ exchangers localized in the vacuolar and plasma membranes, respectively (4–6). Na+/H+ exchangers are membrane proteins that transport protons (H+) across a membrane in exchange for Na+ (7–9). In plants, this exchange activity is driven by the H+ electrochemical gradient generated by the H+ pumps such as the plasma membrane H+-ATPase or the vacuolar membrane H+-ATPase and H+-pyrophosphatase (6).
The transport activity of the plant vacuolar Na+/H+ exchanger has been well characterized (5, 6, 10, 11). In addition, the molecular mechanism underlying vacuolar Na+ compartmentation by the exchanger and its function in plant salt tolerance have been demonstrated recently (10, 12).
Transport studies have provided evidence for Na+/H+ exchangers in the plasma membrane of numerous plants including barley, tobacco, red beet, tomato, and wheat (6). However, plant plasma membrane Na+/H+ exchangers have not been identified at the molecular level. In a genetic screen designed to identify components of the cellular machinery that contribute to salt tolerance in Arabidopsis, three salt overly sensitive genes (SOS1, SOS2, and SOS3) have been found to function in a common pathway (13–16). Based on sequence analysis, the predicted SOS1 gene product is a 127-kDa membrane protein with 12 putative membrane-spanning domains and a long hydrophilic tail at the C-terminal end of the protein (17). The predicted membrane-spanning domains in the SOS1 protein display significant similarity to domains of the plasma membrane-localized Na+/H+ exchangers from animal, bacterial, and fungal cells (17). Sequence analysis also indicates that SOS1 is distinct from the Arabidopsis family of vacuolar membrane Na+/H+ exchangers and represents a class of exchanger that may function in the plasma membrane. Support for the plasma membrane localization of SOS1 comes from recent studies in which an SOS1-green fluorescent protein (GFP) fusion protein was found to localize to the plasma membrane (18). Steady-state SOS1 transcript levels increase significantly in roots and to a much lesser extent in shoots when seedlings are exposed to high levels of NaCl. This regulation by salt is mediated, at least in part, by the other identified components of the SOS pathway, SOS3 (a calcineurin B-like calcium-binding protein) and SOS2 (a serine/threonine protein kinase) (17).
To demonstrate the transport activity of SOS1 and characterize its regulation, we measured Na+/H+-exchange activity by using highly purified plasma membrane vesicles isolated from wild-type and sos plants. Relative to activity in wild-type plants, plasma membrane Na+/H+ exchange was reduced in sos1 plants, and this activity could not be restored by in vitro addition of activated SOS2 protein. Mutations in the SOS2 and SOS3 genes led to reductions in plasma membrane Na+/H+-exchange activity; however, transport in these mutants increased with the addition of activated SOS2 protein in vitro. These results (i) provide a molecular identification of a plasma membrane Na+/H+ exchanger in plants, (ii) functionally identify one of the targets of the SOS regulatory pathway and link the activity of this plasma membrane Na+/H+ exchanger to a role in plant salt tolerance, (iii) demonstrate that the activity of the plasma membrane Na+/H+ exchanger is controlled by the SOS2/SOS3 regulatory pathway, and (iv) provide evidence for ion transport in plasma membrane vesicles isolated from Arabidopsis, thereby laying the foundation for studies addressing the role of plasma membrane transporters in plant growth and development.
Materials and Methods
Plant Material.
Arabidopsis thaliana ecotype Columbia was used in all experiments. sos1-1, sos2-2, and sos3-1 plants were described previously by Zhu et al. (15). Plants were grown in potting soil (Metro-Mix, Grace Sierra Horticultural Products, Milpitas, CA) in a growth room with a cycle of 16 h light (≈100 microeinstein⋅m−2⋅s−1) at 22°C and 8 h dark at 20°C. Plants were watered with tap water three times a week. After 4 weeks, plants were treated with 250 mM NaCl for 3 days by placing the pots in a tray containing the NaCl solution. Rosette leaves were harvested and used for membrane isolation.
Plasma Membrane Isolation and Characterization.
Plasma membrane vesicles were isolated using two-phase partitioning (19) with the following modifications. All steps were carried out at 4°C or on ice. Leaves were homogenized in isolation medium containing 0.33 M sucrose, 10% (wt/vol) glycerol, 0.2% (wt/vol) BSA, 5 mM EDTA, 5 mM DTT, 5 mM ascorbate, 0.2% (wt/vol) casein, 0.6% polyvinylpyrrolidone, 1 mM PMSF, 3 μg/ml leupeptin, 1 μg/ml pepstatin A, and 50 mM Hepes-KOH (pH 7.5). Two to four milliliters of homogenization buffer were used per gram of tissue. The homogenate was filtered through one layer of miracloth and centrifuged at 13,000 × g for 10 min. The supernatant then was centrifuged for 50 min at 80,000 × g to obtain a microsomal pellet that was resuspended in a buffer containing 0.33 M sucrose, 3 mM KCl, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, and 5 mM potassium phosphate (pH 7.8). The suspension was added to a phase mixture to obtain a phase system consisting of 6.2% (wt/wt) Dextran T-500 and 6.2% (wt/wt) polyethylene glycol 3350 in 5 mM potassium phosphate (pH 7.8)/0.33 M sucrose/3 mM KCl. The final upper phases were collected, diluted with resuspension buffer [0.33 M sucrose/10% (wt/vol) glycerol/0.1% (wt/vol) BSA/0.1 mM EDTA/2 mM DTT/1 μg/ml leupeptin/1 μg/ml pepstatin A/20 mM Hepes-KOH (pH 7.5)], and centrifuged for 50 min at 100,000 × g. The resulting pellet was collected and resuspended with the above-described resuspension buffer containing 1 mM EDTA.
The membrane identity and transport competence of the vesicles were assessed with measurements of the H+-transport activity of the plasma membrane H+-ATPase (19). An inside-acid pH gradient (ΔpH) was formed in the vesicles by the activity of the H+ pump and measured as a decrease (quench) in the fluorescence of quinacrine (a pH-sensitive fluorescent probe; refs. 20–22). Assays (1 ml) contained 5 μM quinacrine, 3 mM ATP, 100 mM KCl, 25 mM 1,3-bis[Tris(hydroxylmethyl)methylamino]propane-Hepes (pH 6.5), 250 mM mannitol, and 50 μg of plasma membrane protein. Reactions were mixed by inversion several times and then placed in a dark chamber in a luminescence spectrophotometer (Perkin–Elmer model LS-5B). Reactions were equilibrated in the dark with stirring for 5 min before beginning fluorescence readings, and all additions to reactions were made in a darkened room. Assays were initiated with the addition of MgSO4 (to a final concentration of 4 mM), and formation of ΔpH was measured at excitation and emission wavelengths of 430 and 500 nm, respectively.
Control experiments were conducted to monitor potential effects of organic solvents used with inhibitors and ionophores in H+-transport assays; no solvent effects were observed (data not shown).
Na+/H+-Exchange Assays.
Na+/H+-exchange activity was measured as a Na+-induced dissipation of ΔpH (i.e., a Na+-induced increase in quinacrine fluorescence; ref. 23). When the maximum ΔpH was formed (reached steady state), NaCl was added to initiate Na+ transport. To determine initial rates of Na+/H+ exchange (change in fluorescence per minute, Δ%F/min), changes in relative fluorescence were measured 15 sec after the addition of Na+. Specific activity was calculated by dividing the initial rate by the mass of plasma membrane protein in the reaction (Δ%F/min per mg of protein). Unless indicated, all data represent means ± standard errors of at least three replicate experiments.
Preparation of Constitutively Active SOS2 Recombinant Protein.
Three glutathione S-transferase (GST) fusions proteins with varying levels of constitutive SOS2 serine/threonine kinase activity (GST-SOS2DF, GST-T/DSOS2, and GST-T/DSOS2DF) were used in phosphorylation assays. To produce GST-T/DSOS2DF, a plasmid construct containing the GST-T/DSOS2 sequence (24) was used as a template for PCR amplification. The primer pair forward primer 5′-GAGAGAAATGATGAAGGGCCCAGGCGACAGGATTTTGTTAAAAG-3′ and reverse primer 5′-CCTGGGCCCTTCATCATTTCTCTC-3′ was used; the 5′ half of the forward primer is a complement to the reverse primer. The resulting PCR product was digested with the methylation-sensitive restriction enzyme DpnI to linearize the template plasmid. The circularized PCR product was transformed into Escherichia coli strain DH5α cells. The GST-SOS2DF fusion protein was produced by using the same primer pairs and a plasmid containing the SOS2 sequence fused to GST (25) as a PCR template. The constructs were sequenced fully to verify that there were no PCR or cloning errors.
The induction and purification of the GST-SOS2 fusion proteins were carried out according to Guo et al. (24). Kinase assays, both autophosphorylation and those using the artificial substrate peptide p3, were performed by using these fusion proteins as described previously (25).
The constitutively active form of the SOS2 protein, GST-T/DSOS2DF, was used in transport assays in which 0.2 μg of the protein was preincubated with membrane vesicles for 7 min at room temperature before formation of the ΔpH.
Protein Determination.
The protein content of isolated membrane vesicles and the GST-fusion proteins was determined by the method of Bradford (26) using BSA as a standard.
Results
Arabidopsis Two-Phase Membrane Vesicles Are Transport-Competent and Enriched in Plasma Membrane.
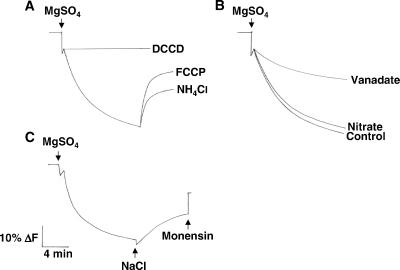
Measurements of H+ transport in membrane vesicles isolated from leaves of wild-type Arabidopsis using aqueous two-phase partitioning demonstrated that the vesicles were transport-competent and enriched in plasma membrane. Quenching of quinacrine fluorescence was observed when ATP/Mg2+ was added to the vesicles (Fig. 1A). The fluorescence quench was inhibited if the H+-channel inhibitor N,N′-dicyclohexylcarbodiimide was added before the initiation of ΔpH formation, and quinacrine fluorescence recovered if either the protonophore carbonylcyanide p-trifluoro-methoxyphenylhydrazone or uncoupler NH4Cl were added after ΔpH had formed (Fig. 1A). These results indicate that the ATP/Mg2+-induced fluorescence quench was the result of the transport of H+ and reflects the formation of a ΔpH.
Figure 1.
Vesicles isolated from wild-type plants are transport-competent and enriched for plasma membrane. Plasma membrane vesicles were isolated by two-phase partitioning from the leaves of wild-type plants treated with 250 mM NaCl for 3 days. Transport assays were performed as described in Materials and Methods. (A) When added at the start of the reaction, the proton channel inhibitor N,N′-dicyclohexylcarbodiimide (DCCD, 20 μM) prevented the formation of ΔpH. When added after the ΔpH formation reached steady state, the protonophore carbonylcyanide p-trifluoro-methoxyphenylhydrazone (FCCP, 5 μM) and the uncoupler NH4Cl (1 mM) dissipated the existing ΔpH. (B) When added at the start of the reaction, 100 μM vanadate (a plasma membrane H+-ATPase inhibitor) reduced ΔpH formation 48%, while control levels of ΔpH formation were measured in the presence of 50 mM nitrate (a vacuolar H+-ATPase inhibitor). (C) After the formation of ΔpH, NaCl (50 mM) was added to initiate Na+/H+-exchange activity (dissipation of ΔpH). The electroneutral Na+/H+ exchanger, monensin (150 μM), was added at the end of the assay to eliminate any remaining ΔpH. The data shown in A–C are representative at least three experiments.
H+ transport was sensitive to the plasma membrane H+-ATPase inhibitor vanadate (≈50% inhibition in the presence of 100 μM vanadate) and insensitive to the vacuolar membrane H+-ATPase inhibitor nitrate (Fig. 1B), indicating that this transport activity was generated by the plasma membrane H+-ATPase. These assays provide evidence for ion transport in plasma membrane vesicles of Arabidopsis, providing an important approach for studies of the role of plasma membrane processes in plant biology.
Wild-Type Arabidopsis Plants Have Plasma Membrane Na+/H+-Exchange Activity.
Na+/H+-exchange activity was examined by using the highly purified plasma membrane vesicles. As shown in Fig. 1C, a dissipation of the ΔpH was induced by the addition of Na+ to vesicles that had been isolated from wild-type plants treated with 250 mM NaCl for 3 days. No activity could be measured when Na+ was added to vesicles that had been isolated from wild-type plants grown in the absence of NaCl (data not shown). Dissipation of the ΔpH in vesicles isolated from salt-treated wild-type plants depends on the Na+ concentration in the assay (Fig. 2) and is specific for Na+, because other monovalent cations were unable to dissipate the ΔpH (data not shown).
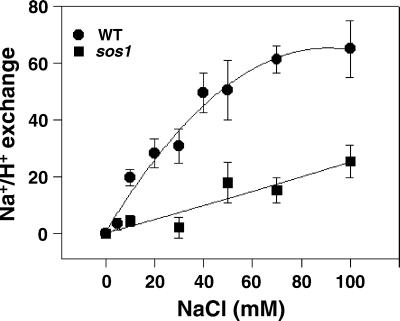
Figure 2.
Vesicles isolated from sos1 plants have reduced plasma membrane Na+/H+-exchange activity relative to that of vesicles isolated from wild-type (WT) plants. Plasma membrane vesicles were isolated from the leaves of wild-type and sos1 plants treated with 250 mM NaCl for 3 days. Transport assays were performed as described in Materials and Methods. When ΔpH reached steady state, NaCl was added over a range of final concentrations (0–100 mM), and initial rates of dissipation were measured. ● and ■, vesicles isolated from wild-type and sos1 plants, respectively. The units of Na+/H+-exchange activity are Δ%F/min per mg of protein.
sos Plants Have Reduced Plasma Membrane Na+/H+-Exchange Activity.
To determine whether SOS1 encodes a Na+/H+ exchanger, Na+-induced dissipation of ΔpH was examined in plasma membrane vesicles isolated from the leaves of wild-type and sos1 plants treated with 250 mM NaCl for 3 days. As shown in Fig. 2, the initial rate of dissipation of ΔpH by 100 mM NaCl was 65 units/min per mg of protein in vesicles isolated from leaves of salt-treated wild-type plants. However, in vesicles isolated from the salt-treated sos1 plants, no dissipation was observed at low salt concentrations (10–30 mM), and only a slight dissipation was observed when the Na+ concentration was greater than 30 mM (25.3 units/min per mg of protein by 100 mM NaCl). The significantly reduced plasma membrane Na+/H+-exchange activity (≈80% reduction) in vesicles isolated from sos1 plants provides strong evidence that SOS1 is a plasma membrane Na+/H+ exchanger.
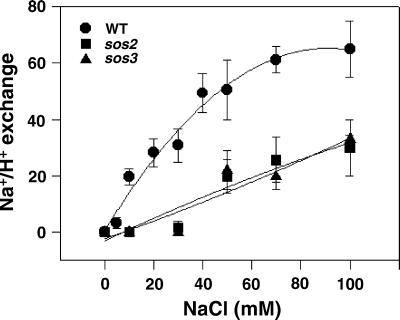
SOS1 is the only identified target of the SOS pathway to date. Molecular studies have shown that SOS1 gene expression is regulated by SOS2 and SOS3 (17); however, regulation at the level of activity (ion transport) has not been shown. For this reason, Na+/H+-exchange activity was measured in vesicles isolated from salt-treated sos2 and sos3 plants. No exchange activity was observed when Na+ was added to the reaction to a low final concentration (10–30 mM; Fig. 3). A low level of activity was observed when the Na+ concentration was increased above 30 mM. The reduced Na+/H+-exchange activity in the sos2 and sos3 plants demonstrates that SOS2 and SOS3 are involved in the regulation of SOS1 activity.
Figure 3.
Vesicles isolated from sos2 and sos3 plants have reduced plasma membrane Na+/H+-exchange activity relative to that of vesicles isolated from wild-type (WT) plants. Plasma membrane vesicles were isolated from the leaves of wild-type, sos2, and sos3 plants treated with 250 mM NaCl for 3 days. Transport assays were performed as described in Materials and Methods. When ΔpH reached steady state, NaCl was added over a range of final concentrations (0–100 mM), and the initial rates of dissipation were measured. ●, ■, and ▴, vesicles isolated from wild-type, sos2, and sos3 plants, respectively. The units of Na+/H+-exchange activity are Δ%F/min per mg of protein.
Constitutively Active SOS2 Protein Stimulates Plasma Membrane Na+/H+-Exchange Activity of Wild-Type Plants in Vitro.
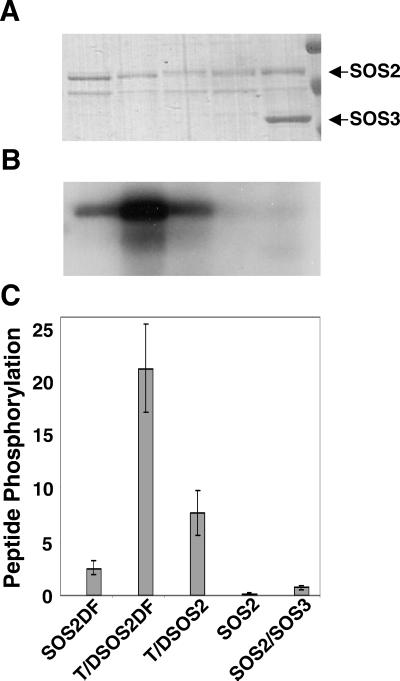
To provide additional evidence that SOS1 encodes a plasma membrane-localized Na+/H+ exchanger regulated by the SOS pathway, constitutively active SOS2 protein was added to Na+/H+-exchange assays. Previous work has shown that SOS2 activation requires SOS3 and Ca2+ (25). However, a Thr-168-to-Asp amino acid change in the kinase activation loop and deletion of the C-terminal end of the SOS2 protein produced an altered form of SOS2 (T/DSOS308) with constitutive kinase activity and, therefore, no requirement for either SOS3 or Ca2+ (24). In this study, we used a form that had been altered further, T/DSOS2DF. Unlike the T/DSOS308 fusion protein in which the entire C-terminal portion of SOS2 has been deleted, only the short FISL motif conferring autoinhibition and SOS3 binding has been removed to produce SOS2DF and T/DSOS2DF. Therefore, these proteins retain most of the C-terminal sequence and any important functional domains therein. Whereas autophosphorylation of GST-SOS2 (Fig. 4 A and B) and its ability to phosphorylate the p3 peptide substrate in vitro (Fig. 4C) depended on SOS3, with GST-SOS2DF and GST-T/DSOS2 these activities were seen in the absence of SOS3. GST-T/DSOS2DF shows even greater activity in the absence of SOS3 for both autophosphorylation (Fig. 4 A and B) and in vitro peptide phosphorylation (Fig. 4C) and was used in subsequent transport assays.
Figure 4.
T/DSOS2DF is a constitutive, highly active form of the SOS2 serine/threonine protein kinase. The kinase activities (autophosphorylation and phosphorylation of an in vitro substrate) of altered forms of the serine/threonine kinase SOS2 (GST-fusion proteins of SOS2DF, T/DSOS2DF, and T/DSOS2) were evaluated. After the autophosphorylation assays, protein (100 ng per lane) was separated by SDS/PAGE, and the gel was stained with Coomassie blue (A) and exposed to x-ray film (B). The arrows indicate SOS2 and SOS3 proteins. (C) The ability of the same GST-SOS2 fusion proteins to phosphorylate the peptide substrate p3 (400 pmol per assay) was determined. The units of protein phosphorylation are nmol/min per mg of protein.
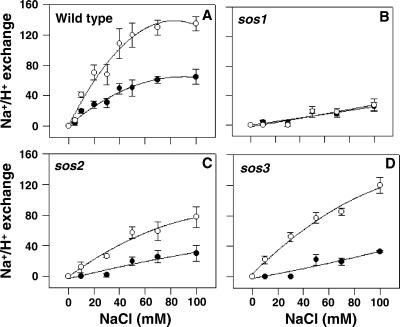
In the presence of added GST-T/DSOS2DF protein, plasma membrane exchange activity in vesicles isolated from wild-type plants increased with increasing NaCl concentration and reached a 2-fold stimulation relative to activity without the added protein (Fig. 5A). Stimulation of exchange activity increased as a function of the amount of added GST-T/DSOS2DF protein up to 0.2 μg; higher amounts of the protein dissipated the ΔpH in the absence of added Na+ (data not shown). SOS2 protein in which kinase activity had not been activated (either unmodified wild-type GST-SOS2 recombinant protein or boiled GST-T/DSOS2DF protein) did not have any effect on exchange activity (data not shown).
Figure 5.
Constitutively active recombinant SOS2 protein stimulates plasma membrane Na+/H+-exchange activity in vesicles isolated from wild-type, sos2, and sos3 plants but not in those isolated from sos1 plants. Transport assays were performed as described in Materials and Methods. ΔpH was formed in the absence (●) or presence (○) of active recombinant SOS2 protein (GST-T/DSOS2DF). When ΔpH reached steady state, NaCl was added over a range of final concentrations (0–100 mM), and the initial rates of dissipation were measured. (A) Wild-type. (B) sos1. (C) sos2. (D) sos3. The units of Na+/H+-exchange activity are Δ%F/min per mg of protein.
Constitutively Active SOS2 Does Not Stimulate Plasma Membrane Na+/H+-Exchange Activity of sos1 Plants in Vitro.
The regulation of SOS1 exchange activity by SOS2 was examined further by monitoring the effect of the constitutively active SOS2 on the activity of the Na+/H+ exchanger of sos1 plants. Exchange activity did not increase in the presence of added GST-T/DSOS2DF at any of the GST-T/DSOS2DF (data not shown) or NaCl concentrations tested (Fig. 5B). These data provide additional evidence that a major plasma membrane Na+/H+ exchanger is absent in sos1 plants and that this exchanger is the target of SOS2 regulation.
Constitutively Active SOS2 Protein Stimulates Plasma Membrane Na+/H+-Exchange Activity of sos2 and sos3 Plants in Vitro.
Unlike transport in sos1 plants, when GST-T/DSOS2DF was added to vesicles isolated from either the sos2 or sos3 plants, Na+/H+-exchange activity was stimulated (Figs. 5 C and D). As was observed when GST-T/DSOS2DF was added to vesicles isolated from wild-type plants, stimulation of activity was evident over the entire range of NaCl concentrations used in the assay (Figs. 5 C and D), and addition of inactive SOS2 did not stimulate plasma membrane-exchange activity (data not shown). After addition of GST-T/DSOS2DF, Na+/H+-exchange activity in sos2 or sos3 vesicles was higher than that measured in wild-type vesicles without the added protein; however, the level and properties of the stimulation differed from that measured with wild-type vesicles. The overall stimulation of activity in sos2 vesicles was reduced and slower at each concentration of NaCl tested, whereas the overall stimulation of activity in sos3 vesicles approached the stimulated activity in wild-type vesicles, but the response was slower at all concentrations measured. Stimulation of Na+/H+-exchange activities in the sos2 and sos3 vesicles by the constitutively active protein provides additional evidence that SOS1 activity is regulated by SOS2 and SOS3 and that SOS1 activation is an output of the SOS signaling pathway.
Discussion
SOS1 Is a Plasma Membrane Na+/H+ Exchanger That Is Regulated by the SOS Pathway.
In our previous studies, we provided evidence that pointed strongly to a role for SOS1 as a plasma membrane Na+/H+ exchanger. We found that (i) mutations in SOS1 render Arabidopsis extremely sensitive to growth in high levels of NaCl (13, 15), (ii) under severe salt stress, sos1 plants accumulate higher levels of Na+ than do wild-type plants (18), and (iii) the predicted protein sequence of SOS1 shares significant sequence and domain similarity with plasma membrane Na+/H+ exchangers from animal, bacterial, and fungal cells (17).
In the present study, we have provided direct evidence that SOS1 is a plasma membrane Na+/H+ exchanger and that it is regulated by the SOS pathway. (i) Exchange activity was reduced significantly in the sos1 plants relative to that in wild-type plants (Fig. 2). (ii) The absence of SOS1 in sos1 plants resulted in a lack of stimulation of transport activity when activated SOS2 protein was added in vitro (Fig. 5B). (iii) The absence of regulatory components in the sos2 and sos3 plants led to a reduction in exchanger activity (Fig. 3). This reduction could be overcome by the addition of activated SOS2 protein, thus adding back the component absent in sos2 plants (Fig. 5C) and bypassing the defect in sos3 plants (Fig. 5D).
Our findings are supported further by the work of Shi et al. (18). Cells of Saccharomyces cerevisiae with mutations in the endogenous Na+/H+ exchangers NHA1 and NHX1 are unable to grow in media containing NaCl levels tolerated by wild-type cells. However, when these cells expressed the Arabidopsis SOS1 gene, growth in NaCl was restored to wild-type levels. A subsequent study showed that S. cerevisiae cells expressing all three identified SOS pathway members (SOS1, SOS2, and SOS3) were much more salt-tolerant than cells expressing only SOS1 (data not shown). Positive regulation of SOS1 activity by SOS2 and SOS3 is consistent with results from our previous genetic analyses (14, 15, 25) showing that the three identified SOS genes function in the same salt-tolerance pathway. The stimulation of Na+/H+-exchange activity in sos3 plants by the activated SOS2 protein suggests that SOS3 does not regulate SOS1 activity directly but that it operates through SOS2, which is in agreement with our previous findings that SOS3 physically interacts with SOS2 and activates the substrate phosphorylation activity of SOS2 (25).
Multiple Na+/H+ Exchangers Exist in the Plasma Membrane of Arabidopsis.
The low level of transport activity seen in sos1 vesicles when higher levels of NaCl were added to assays (Figs. 2 and 5B) suggests the presence of additional Na+/H+ exchangers in the plasma membrane of Arabidopsis. To determine whether this activity was caused by additional ΔpH-dependent transport activities or to the electrical potential (ΔΨ) of the membrane, we measured transport activity in the presence and absence of the K+ ionophore, valinomycin. K+ in the presence of valinomycin eliminates all ΔΨ, and thus any difference in Na+-induced dissipation of ΔpH with and without valinomycin indicates the degree of contribution of ΔΨ. There was no difference in the Na+/H+-exchange activity measured (data not shown), indicating that Na+ transport likely represents Na+/H+ exchange originating from another as-yet-unidentified exchanger(s). If present, this transporter(s) contributes only a small portion of the Na+/H+-exchange activity measured in plasma membrane vesicles of Arabidopsis (Fig. 2), and it is not a target of the SOS regulatory pathway (Figs. 3 and 5).
Common Signaling Components Regulate Plasma Membrane Na+/H+-Exchange Activity in Plants and Animals.
The current model of the signal transduction events in the SOS pathway suggests that SOS3 (a calcineurin B-like Ca2+-binding protein) perceives changes in cytoplasmic Ca2+ levels elicited by salt stress. SOS3 subsequently transmits the signal to SOS2, a serine/threonine protein kinase (16). Our current studies showing that Na+/H+-exchange activity is reduced in sos3 plants and that addition of active SOS2 protein to vesicles isolated from these plants restores Na+/H+-exchange activity suggest that the exchanger is an indirect target of a calcineurin B-like protein. Na+/H+-exchange regulation in animal cells also has been shown to involve calcineurin B-like proteins; however, this regulation takes place through direct interaction with the exchanger (27, 28).
Modulation of Na+/H+-exchange activity by changes in phosphorylation may represent another common point of regulation in plant and animal cells. Our observation that in vitro addition of the constitutively active SOS2 kinase-stimulated plasma membrane-exchange activity in vesicles from salt-treated sos2 and sos3 plants (Fig. 5) suggests that SOS2 and SOS3 activate SOS1 activity via phosphorylation of the exchanger. Phosphorylation by protein kinases also has been shown to be an important mechanism underlying regulation of the animal Na+/H+ exchangers (29, 30).
Complex Interactions Define the SOS2/SOS3 Regulatory Pathway and Influence the Activity of SOS1.
Measurements of Na+/H+-exchange activity in the presence of constitutively active SOS2 protein added in vitro suggest that complex interactions regulate SOS1 activity. In vesicles isolated from wild-type plants, the addition of active SOS2 protein in vitro stimulated exchange activity more than 2-fold, suggesting that, in vivo, either SOS1 activity is regulated tightly or levels of active SOS2 may be limiting. As would be expected for a mutant in which SOS1 is absent, no stimulation of activity was observed after the addition of constitutively active SOS2 protein to vesicles isolated from sos1 plants. In contrast, the addition of active SOS2 protein to vesicles isolated from the sos2 and sos3 plants led to stimulation of Na+/H+-exchange activity; however, the response of the exchanger at each concentration of substrate was lower than was measured with wild-type vesicles and differed for the two mutants. Measurements of SOS1 transcript levels in shoots of wild-type and sos2 and sos3 plants (17) suggest that the effects of addition of active SOS2 protein to vesicles isolated from these plants cannot be explained merely by changes in levels of Na+/H+-exchanger protein. Instead, our present results suggest that the SOS1 protein is present but that the biochemical activity of an element or elements essential for formation of fully responsive SOS1 is missing or altered (and that the biochemical activities of different components may be altered or lacking in the two mutants). Identification of these components and the underlying mechanisms await further experimentation.
Acknowledgments
We thank Dr. Edgar Spalding for helpful discussions and suggestions. Work in our laboratories has been supported by National Institutes of Health Grant R01GM59138 (to J.-K.Z.) and the Southwest Consortium on Plant Genetics and Water Resources (J.-K.Z. and K.S.S.). Q.-S.Q. was supported in part by Major State Basic Research and Development Plan of the People's Republic of China Grant G1999011705.
Abbreviations
- SOS
salt overly sensitive
- ΔpH
pH gradient
- GST
glutathione S-transferase
References
- 1.Rhoades J D, Loveday J. In: Irrigation of Agricultural Crops. Steward B A, Nielsen D R, editors. Vol. 30. Madison, WI: Am. Soc. Agronomists; 1990. pp. 1089–1142. [Google Scholar]
- 2.Niu X, Bressan R A, Hasegawa P M, Pardo J M. Plant Physiol. 1995;109:735–742. doi: 10.1104/pp.109.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacoby B. In: Handbook of Plant and Crop Stress. Pessarakli M, editor. New York: Dekker; 1999. pp. 97–123. [Google Scholar]
- 4.Hasegawa P M, Bressan R A, Zhu J-K, Bohnert H J. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- 5.Blumwald E. Curr Opin Cell Biol. 2000;12:431–434. doi: 10.1016/s0955-0674(00)00112-5. [DOI] [PubMed] [Google Scholar]
- 6.Blumwald E, Aharon G S, Apse M P. Biochim Biophys Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 7.Counillon L, Pouyssegur J. J Biol Chem. 2000;275:1–4. doi: 10.1074/jbc.275.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Padan E, Venturi M, Gerchman Y, Dover N. Biochim Biophys Acta. 2001;1505:144–157. doi: 10.1016/s0005-2728(00)00284-x. [DOI] [PubMed] [Google Scholar]
- 9.Wiebe C A, Dibattista E R, Fliegel L. Biochem J. 2001;357:1–10. doi: 10.1042/0264-6021:3570001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaxiola R A, Rao R, Sherman A, Grisafi P, Alper S L, Fink G R. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darley C P, van Wuytswinkel O C M, van de Woude K, Mager W H. Biochem J. 2000;351:241–249. doi: 10.1042/0264-6021:3510241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apse M P, Aharon G S, Snedden W A, Blumwald E. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 13.Wu S-J, Lei D, Zhu J-K. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Zhu J-K. Proc Natl Acad Sci USA. 1997;94:14960–14964. doi: 10.1073/pnas.94.26.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J-K, Liu J, Xiong L. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu J-K. Plant Physiol. 2000;124:941–948. doi: 10.1104/pp.124.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H, Ishitani M, Kim C, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Qintero F J, Pardo J M, Zhu J-K. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu Q S, Su X F. Aust J Plant Physiol. 1998;25:923–928. [Google Scholar]
- 20.Bennett A B, Spanswick R M. J Membr Biol. 1983;71:95–107. [Google Scholar]
- 21.Qiu QS. J Plant Physiol. 1999;154:628–633. [Google Scholar]
- 22.Parks G E, Dietrich M A, Schumaker K S. J Exp Bot. 2002;53:1055–1065. doi: 10.1093/jexbot/53.371.1055. [DOI] [PubMed] [Google Scholar]
- 23.Blumwald E, Pool R. Plant Physiol. 1985;78:163–167. doi: 10.1104/pp.78.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y, Halfter U, Ishitani M, Zhu J-K. Plant Cell. 2001;13:1383–1399. doi: 10.1105/tpc.13.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halfter U, Ishitani M, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Lin X, Barber D L. Proc Natl Acad Sci USA. 1996;93:12631–12636. doi: 10.1073/pnas.93.22.12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang T, Su X, Wakabayashi S, Shigekawa M. J Biol Chem. 2001;276:17367–17372. doi: 10.1074/jbc.M100296200. [DOI] [PubMed] [Google Scholar]
- 29.Fliegel L, Walsh M P, Singh D, Wong C, Barr A. Biochem J. 1992;282:139–145. doi: 10.1042/bj2820139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi E, Jun-ichi A, Gallis B, Aebersold R, Spring D J, Krebs E G, Berk B C. J Biol Chem. 1999;274:20206–20214. doi: 10.1074/jbc.274.29.20206. [DOI] [PubMed] [Google Scholar]