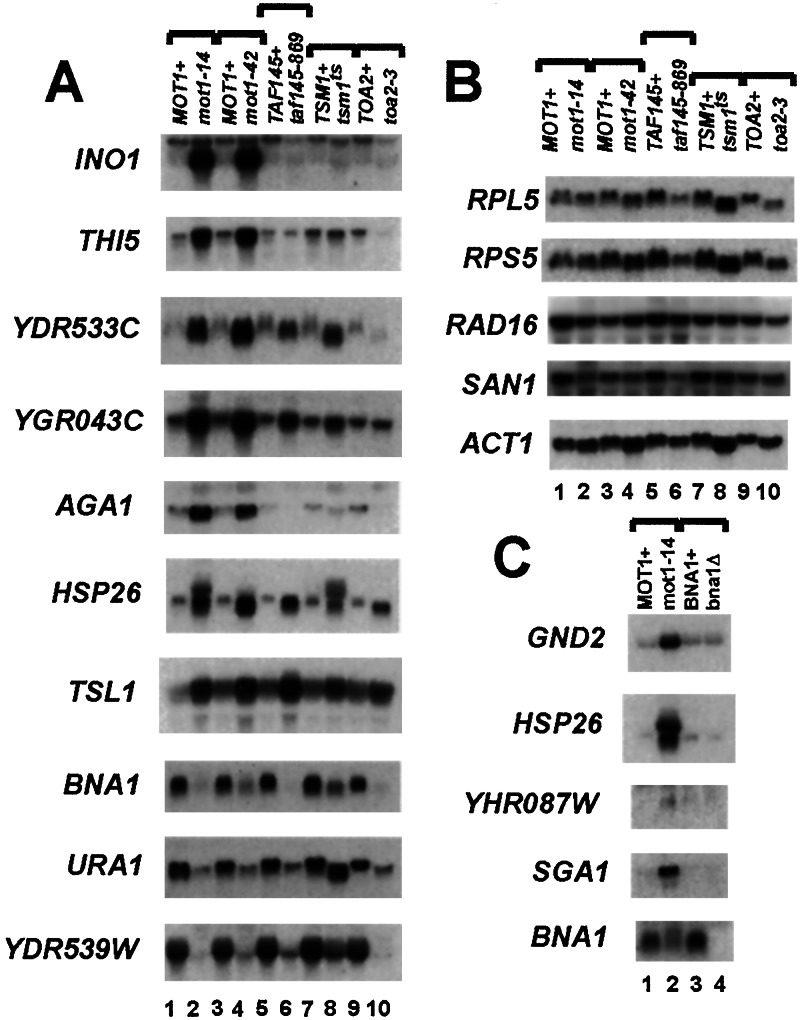
BIOCHEMISTRY. For the article “Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms,” by Arindam Dasgupta, Russell P. Darst, Karla J. Martin, Cynthia A. Afshari, and David T. Auble, which appeared in number 5, March 5, 2002, of Proc. Natl. Acad. Sci. USA (99, 2666–2671), some lane labels were omitted from Fig. 1. The corrected figure and its legend appear below.
Figure 1.
Mot1, TFIIA, and TAF dependence of selected genes. The indicated strains were grown at 30°C to an OD600 of ≈1.0, then heat-shocked for 45 min at 35°C, and total RNA was harvested. Twenty micrograms of RNA from each strain was then resolved by electrophoresis, transferred to nitrocellulose, and probed with radiolabeled DNAs encoding the indicated ORFs. (A) Mot1-repressed and activated genes identified by microarray analysis. (B) Mot1-independent genes. (C) Comparison of message levels in wild-type, mot1–14, and bna1Δ strains. Inactivation of Mot1 led to variable effects on transcript levels for some genes. For instance, BNA1 message levels were reduced 4-fold in A but only ≈2-fold in C. These effects fall roughly within the range defined by microarray analysis (3.7 ± 1.4-fold).