Abstract
The opportunistic pathogen Pseudomonas aeruginosa secretes a capsule-like polysaccharide called alginate that is important for evasion of host defenses, especially during chronic pulmonary disease of patients with cystic fibrosis (CF). Most proteins for alginate biosynthesis are encoded by the 12-gene algD operon. Interestingly, this operon also encodes AlgL, a lyase that degrades alginate. Mutants lacking AlgG, AlgK, or AlgX, also encoded by the operon, synthesize alginate polymers that are digested by the coregulated protein AlgL. We examined the phenotype of an ΔalgL mutation in the highly mucoid CF isolate FRD1. Generating a true ΔalgL mutant was possible only when the algD operon was under the control of a LacIq-repressed trc promoter. Upon induction of alginate production with isopropyl-β-d-thiogalactopyranoside, the ΔalgL mutant cells were lysed within a few hours. Electron micrographs of the ΔalgL mutant showed that alginate polymers accumulated in the periplasm, which ultimately burst the bacterial cell wall. The requirement of AlgL in an alginate-overproducing strain led to a new model for alginate secretion in which a multiprotein secretion complex (or scaffold, that includes AlgG, AlgK, AlgX, and AlgL) guides new polymers through the periplasm for secretion across the outer membrane. In this model, AlgL is bifunctional with a structural role in the scaffold and a role in degrading free alginate polymers in the periplasm.
Pseudomonas aeruginosa is an opportunistic pathogen par excellence that causes severe and life-threatening infections in immunocompromised hosts such as patients with respiratory diseases, burns, cancers undergoing chemotherapy, and cystic fibrosis (CF). Virulence factors produced by P. aeruginosa include numerous extracellular toxins, proteases, hemolysins, and exopolysaccharides. The most striking feature of P. aeruginosa strains infecting the CF pulmonary tract is their highly mucoid phenotype, which is due to alginate overproduction (Alg+) (10, 24). Mutations in mucA, encoding a transmembrane anti-sigma factor, are typically responsible for the mucoid conversion observed in clinical isolates of CF patients (25, 26).
The Alg+ phenotype correlates with the ability of P. aeruginosa to persist in the lungs of CF patients and cause chronic bronchopulmonary infections (18). About 80% of the P. aeruginosa isolates from CF patients undergo mucoid conversion in vivo, whereas only about 1% of clinical P. aeruginosa isolates from other types of infections are Alg+ (7, 8, 46). Thus, alginate appears to have an important role in the unique host-parasite relationship between the CF patient and P. aeruginosa. Alg+ may confer several selective advantages on the bacterial invader, which have been reviewed (17) and include increased resistance to phagocytosis (a property typically associated with bacterial capsules) and reduced susceptibility to antibody-dependent bactericidal mechanisms (1, 36, 37, 43). Alginate also provides a polyanionic barrier that may exclude cationic peptide antibiotics (34). The biofilms of mucoid organisms in the CF lung show an unusual microcolony morphology (23), which is unique. Although alginate is not required for biofilm formation, an algD mutant (i.e., defective in alginate biosynthesis) shows a lag in initial biofilm development, suggesting its role in early attachment (30).
Structurally, alginate is a simple, unbranched polysaccharide of very high molecular weight that is composed of two uronic acids: β-d-mannuronic acid and its C5 epimer α-l-guluronic acid. In addition to Pseudomonas sp., members of the bacterial genus Azotobacter also synthesize alginate, where it forms part of a protective coat during cyst formation. Alginate is commonly found in brown algal seaweed as part of its gelatinous cell wall (19). The viscous nature of the algal alginate in aqueous solution makes it a commercially important product, especially in the food industry. In P. aeruginosa, alginate is produced as a capsule-like exopolysaccharide that loosely adheres to P. aeruginosa cells, and so most of it is found in the culture supernatant.
In P. aeruginosa, all but one of the known genes encoding the alginate biosynthetic machinery are clustered in an operon encoding 12 gene products: AlgD-8-44-K-E-G-X-L-I-J-F-A (5). The early pathway of biosynthesis utilizes AlgA, AlgD, and AlgC (unlinked to the operon) to form GDP-mannuronic acid, the primary precursor of alginate (28). The polymerase for alginate has not been positively identified, but the most likely candidate is Alg8, which shows structural homology to β-glycosyltransferases (41). The remainder of the proteins encoded by the operon bear little resemblance to known enzymes for capsule biosynthesis in other bacteria. AlgG was shown to be a periplasmic d-mannuronate C5 epimerase in P. aeruginosa, which is responsible for converting d-mannuronates to l-guluronates at the polymer level (4, 12). Other periplasmic proteins include AlgK (21) and AlgX (29), and mutants lacking these proteins are Alg−. Secretion of polymer appears to occur through the outer membrane protein AlgE (38). AlgI, AlgJ, and AlgF are required to modify alginate with O-acetyl groups, but these proteins are not required for polymer formation (13-15). In addition, it is curious that the operon for alginate biosynthesis also includes a gene, algL, that encodes an alginate lyase, which can efficiently degrade alginate (42).
We have shown that, in addition to its C-5 polymer level epimerase activity, AlgG is bifunctional, having another role in protecting alginate from degradation by AlgL during transport across the periplasm (16, 20). Similarly, a nonpolar ΔalgK mutation, like an ΔalgG mutation, results in the secretion of small alginate fragments due to AlgL-mediated polymer degradation (21), and so AlgK appears to share this second role. It was recently shown that an algX mutation also results in the secretion of AlgL-degraded alginate (40). Thus, AlgG, AlgK, and AlgX may be part of a periplasmic protein complex (scaffold) that guides alginate polymers to AlgE in the outer membrane. Furthermore, it appears that breakdown of this scaffold due to any one missing component protein leads to degradation of the freed polymer by AlgL depolymerase activity. In this study, we sought to evaluate the phenotype of a ΔalgL mutation. We show evidence that despite the alginate-degrading activity of AlgL, it too is important for alginate secretion. This became evident through the discovery that alginate polymers are trapped in the periplasm in strains lacking AlgL, which ultimately leads to cell lysis. Our genetic analysis has led to a new model for alginate secretion whereby a protein scaffold that includes the alginate-degrading protein AlgL transports the growing alginate polymer chain through the periplasm.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
P. aeruginosa FRD1 is an Alg+ CF isolate (31). Bacterial strains were routinely grown in L broth (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter). MAP defined medium (14) was used to reduce the background reaction when measuring the production of extracellular uronic acids. Isopropyl-β-d-thiogalactopyranoside (IPTG) was generally used with P. aeruginosa at a concentration of 5 mM to induce the Ptrc promoter. Antibiotics, when used, were at the following concentrations (per milliliter): ampicillin at 100 μg, carbenicillin at 300 μg, gentamicin at 15 μg for Escherichia coli and 300 μg for P. aeruginosa, and kanamycin at 40 μg. Sucrose (7.5%) was added to L agar to counterselect against the sucrose sensitivity caused by sacB. Plasmids were conjugated from E. coli to P. aeruginosa by triparental matings using the conjugative helper plasmid pRK2013 [ColE1-Tra(RK2) Kmr] (11) with selection on a 1:1 mixture of Pseudomonas Isolation Agar (Difco) and L Agar (Sigma).
Construction of Ptrc-algD mutant strain FRD1050.
To control alginate biosynthesis, the algD promoter was replaced with an inducible Ptrc promoter as follows. A 600-bp fragment at the 5′ end of algD was obtained by PCR amplification using primers that placed the algD start ATG codon within an NcoI site, and the 3′ end contained an XbaI site. The fragment was cloned into the NcoI-XbaI sites and immediately downstream of the Ptrc promoter in expression vector pMF53 [ori(pBR322) bla oriT(RK2) lacIq Ptrc], which was expression vector pMF54 (12) but without a stabilization fragment for replication in Pseudomonas. This plasmid, pJLS3, was conjugated into FRD1, and its bla marker for carbenicillin resistance permitted selection for exconjugants with single-crossover events by homologous recombination within algD on the chromosome. The carbenicillin-resistant merodiploids had the algD-A operon under Ptrc control, while the 0.6-kb algD fragment was under the control of the native PalgD promoter. One such merodiploid, called FRD1050, was used in this study. The integration of pJLS3 at algD in the chromosome was confirmed by PCR amplification of genomic DNA with primers specific for vector sequences upstream of Ptrc and downstream to the terminator of the algD gene. DNA amplified by these primers with FRD1050 produced the expected 2,265-bp fragment, whereas no product was amplified with wild-type FRD1 or E. coli(pJLS3) (data not shown).
Construction of ΔalgL Gmr mutant FRD1300.
pCC27 was pCP19 (IncP1, broad host range, Tcr) with 23 kb of P. aeruginosa FRD1 DNA containing algD-algF (4). A 3.4-kb EcoRI fragment containing ′algX-algL-algI from FRD1 was cloned into the SmaI site of the gene replacement vector pEX100T (bla sacB oriT) (44). Then, the 1,208-bp KpnI-XbaI fragment containing algL was replaced with a nonpolar gentamicin resistance cassette on a 785-bp KpnI-XbaI fragment from pSJ12 (21). The resulting plasmid, pSJ243, contained the ΔalgL::Gmr allele and was conjugated into FRD1050 with selection for gentamicin resistance. Colonies were screened for loss of sucrose sensitivity, which typically indicates a double-crossover event and thus gene replacement. In one isolate, called FRD1300, the ΔalgL::Gmr mutation was confirmed by PCR amplification of genomic DNA with primers specific for sequences upstream and downstream of the algL gene. DNA amplified by these primers with wild-type FRD1 produced the expected 1,485-bp fragment, and FRD1300 produced the predicted ∼900-bp fragment (data not shown).
Alginate assay.
To assay the amount of alginate secreted, cultures were grown for 18 h at 37°C in 10 ml of MAP medium or L broth. Cells were removed by centrifugation, and culture supernatants of L broth cultures were exhaustively dialyzed against saline. Samples were assayed for alginate concentration using a colorimetric test for uronic acids (22), with alginic acid from Macrocystis pyrifera (Sigma) used as the standard.
Electron microscopy.
Samples of log-phase bacterial cultures were withdrawn periodically, and cells were collected by centrifugation in a Microfuge at a low speed (3,000 rpm). The pelleted cells were washed with saline and resuspended in glutaraldehyde. The samples were thin sectioned, stained, and examined using a Zeiss EM10CA transmission electron microscope in the Virginia Commonwealth University Core Electron Microscopy Facility.
RESULTS
Attempted construction of a ΔalgL mutant of P. aeruginosa FRD1.
Strain FRD1 was used because it is a CF clinical isolate of P. aeruginosa that displays the typical Alg+ phenotype, and it is genetically manipulatable (20, 21). Previous studies have shown that mutants lacking AlgG, AlgK, or AlgX synthesize alginate polymers which are rapidly degraded by AlgL, an alginate lyase in the same operon as and thus coregulated with AlgG and AlgK (16, 20, 21, 40). Here we sought to evaluate the phenotype of a nonpolar ΔalgL mutation in FRD1. The strategy used to construct an algL mutant was similar to one we have used previously (21). A clone was modified by replacing algL sequences with a nonpolar gentamicin resistance cassette, followed by allelic exchange using a suicide vector (pEX100T) that contains sacB, which confers sucrose sensitivity and thus provides counterselection on sucrose. However, when the resulting plasmid (pSJ243) was transferred to FRD1 with selection for Gmr, an unusually low number of gene replacement candidates (i.e., sucrose-resistant colonies) was obtained. Among several algL mutants of FRD1 tested, all of which were nonmucoid, none could be complemented to the Alg+ phenotype by a plasmid expressing algL in trans. This suggested that secondary suppressor mutations to block alginate biosynthesis are required for viability in a ΔalgL mutant. Thus, this inability to construct a true ΔalgL mutant suggested that AlgL might be required for cell viability in a highly mucoid strain like FRD1.
Placement of the alginate operon under IPTG control in P. aeruginosa.
To test the hypothesis that a ΔalgL mutation is lethal in a highly mucoid P. aeruginosa strain, a derivative of FRD1 was constructed for controlled expression of the Alg+ phenotype. The 12-gene algD operon for alginate biosynthesis is under the control of a complex hierarchy of gene regulators (6, 32). In order to turn alginate on or off at will, a genetic technique was employed that placed the chromosomal algD operon under Ptrc control, which could then be repressed by the LacIq repressor but derepressed in the presence of IPTG. To accomplish this, a 5′-terminal fragment of algD′ was placed under the control of a Ptrc promoter in a suicide plasmid (pJLS3) and integrated into the FRD1 chromosome by homologous recombination at algD (Fig. 1). The merodiploids thus formed had the algD operon under Ptrc control, and the 0.6-kb algD′ fragment was then under the control of the native PalgD promoter. The merodiploids obtained were nonmucoid on L agar but produced the Alg+ phenotype when plated on L agar plus IPTG (Fig. 2). One of these FRD1 derivatives, showing alginate production only in the presence of IPTG, was called FRD1050 and used as described below. Under typical conditions in L broth, cultures of FRD1 accumulated about 1,000 ± 50 μg of alginate per ml. Similar cultures of FRD1050 accumulated 700 ± 20 μg of alginate per ml with IPTG and 80 ± 10 μg of alginate per ml without IPTG. Thus, Ptac was nearly as strong a promoter as PalgD in P. aeruginosa.
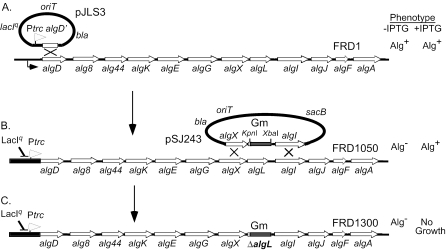
FIG. 1.
Genetic manipulations of the algD operon encoding proteins for alginate biosynthesis in P. aeruginosa FRD1. (A). Suicide plasmid pJLS3 contained a Ptrc-algD′ fragment and, when integrated into the chromosome of FRD1 at algD, formed strain FRD1050, which put the operon under LacIq repression. (B) Suicide plasmid pSJ243, containing a fragment of the algD operon with a nonpolar ΔalgL::Gmr allele, was used to generate a ΔalgL mutant of FRD1050 by allelic exchange. (C) Among the Gmr transconjugants, FRD1300 was a ΔalgL mutant that had lost the vector-encoded (Sucs) phenotype. Induction of the algD operon in FRD1300 with IPTG prevented cell growth.
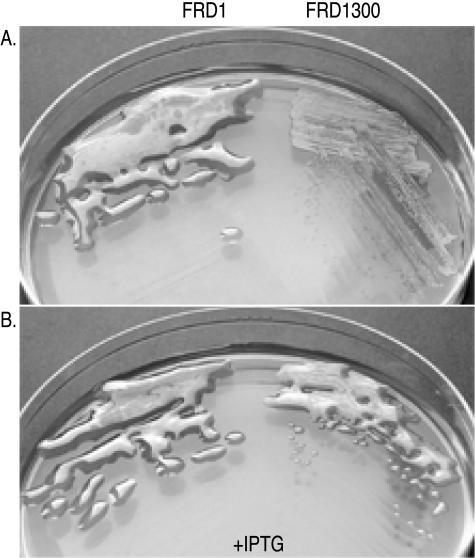
FIG. 2.
Alginate phenotypes of FRD1 derivatives. (A) The mucoid (Alg+) phenotype on L agar was observed with parent strain FRD1 but not with FRD1050 (Ptrc-algD-A), which has the algD operon repressed by LacIq. (B) The mucoid phenotype on L agar plus IPTG (1 mM) was observed with both FRD1 and FRD1050 (Ptrc-algD-A), which has the algD operon induced.
Construction of a conditionally lethal ΔalgL mutant of P. aeruginosa.
FRD1050 was grown under conditions in which alginate was not produced (L broth without IPTG), and the ΔalgL::Gmr gene replacement plasmid described above, pSJ243, was introduced to replace the chromosomal algL gene (Fig. 1B). Unlike the experience with Alg+ FRD1, using FRD1050 as a host produced numerous ΔalgL::Gmr mutants. One potential ΔalgL::Gmr mutant, called FRD1300, was selected, and its mutation was confirmed at the DNA level by PCR analysis (see Materials and Methods). When this mutant was grown on agar medium lacking IPTG, it displayed a nonmucoid phenotype like its parent FRD1050. However, when incubated on L agar containing IPTG, it was unable to grow. When a self-replicating plasmid expressing algL in trans was introduced into FRD1300, it restored both viability on L agar with IPTG and the Alg+ phenotype. Thus, AlgL (i.e., an alginate lyase) was required for viability in Alg+ strain FRD1.
Kinetics of cell death due to ΔalgL mutation in Alg+ P. aeruginosa.
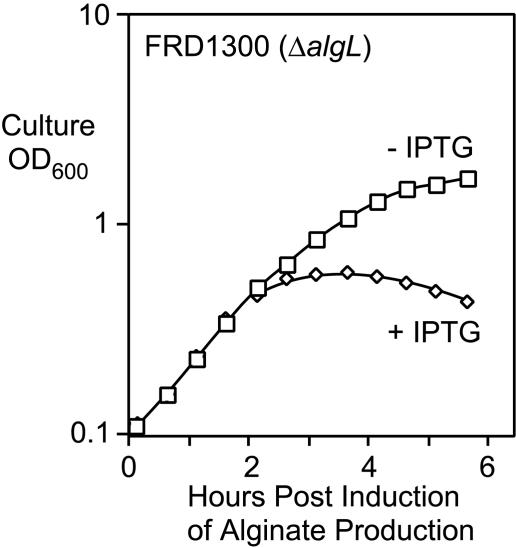
To better understand the nature of the growth defect due to the ΔalgL mutation, cultures were examined to determine the effect of alginate induction on growth over time. FRD1300 (Ptrc-algD-A ΔalgL) was grown for 1 h in L broth with aeration, and then IPTG was added to half of the divided culture. At 3 h postinduction, the growth of the alginate-induced culture was clearly slower than that of the uninduced culture (Fig. 3). By 4 h postinduction, the density of the induced culture actually began to decline, suggesting rapidly dying cells (Fig. 3). Thus, a ΔalgL mutation resulted in a lethal phenotype in mucoid P. aeruginosa FRD1. In that FRD1300 (an AlgL− mutant) was viable as long as it was nonmucoid (i.e., IPTG was not included), then apparently AlgL is not required for viability as long as alginate is not being overproduced.
FIG. 3.
Effect of the induction of alginate production on the growth of FRD1300 (Ptrc-algD-A ΔalgL::Gmr). A 0.3-ml sample of an FRD1300 overnight culture was inoculated into 25 ml L broth, which was incubated with aeration at 37°C. After 1 h, the culture was split and 5 mM IPTG was added to one flask. A plot of culture density (OD600) versus minutes postinduction is shown.
Loss of AlgL results in accumulation of periplasmic alginate.
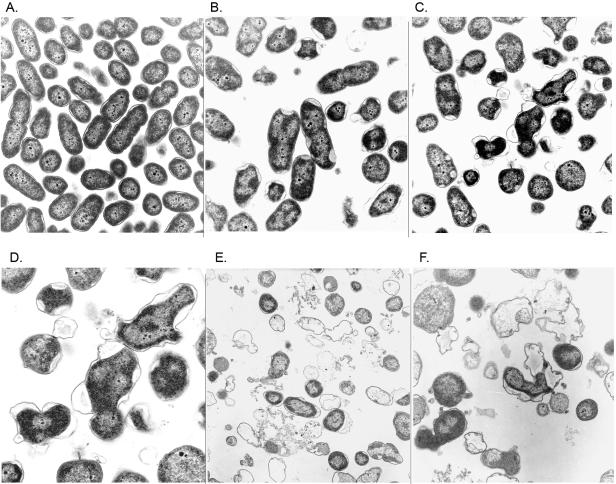
Transmission electron microscopy was used to investigate the cause of cell lysis upon induction of alginate biosynthesis in the ΔalgL mutant. Samples were withdrawn over time from a culture of FRD1300 (Ptrc-algD-A ΔalgL) following IPTG induction, and thin sections were prepared for transmission electron microscopy. Prior to IPTG induction, these gram-negative bacterial cells showed a normal cell structure and morphology (Fig. 4A). However, by 2 h of IPTG induction to induce alginate production, most cells of the ΔalgL mutant showed obvious zones of separation between the inner and outer membranes (Fig. 4B). By 4 h of IPTG induction, the periplasm of the ΔalgL mutant was dramatically swollen (Fig. 4C). Increasing the magnification (Fig. 4D) showed more clearly that the periplasm had accumulated a material (i.e., alginate) that caused large separations between the inner and outer membranes. By 6 h of induction, nearly all of the ΔalgL mutant cells had been lysed and few showed remnants of their original cell morphology (Fig. 4E and F). These observations suggested that the loss of AlgL in Alg+ FRD1 prevented newly formed alginate polymers in the periplasm from being transported through the outer membrane, which ultimately led to periplasmic accumulation of alginate and caused the cell walls to burst from the pressure.
FIG. 4.
Electron micrographs of thin-sectioned FRD1300 (Ptrc-algD-A ΔalgL::Gmr) following incubation in L broth and comparing cells at 0, 2, 4, and 6 h post IPTG induction of the algD operon. (A) Uninduced, normal-appearing cells (×40,000). P. aeruginosa rods are approximately 1 by 2 μm in size. (B) Induction for 2 h shows zones of separation between membranes. (C) Induction for 4 h shows larger zones of separation. (D) Induction for 4 h; this enlargement of panel C shows periplasmic polymer accumulation. (E) Induction for 6 h showed general lysis of the cells. (F) Enlarged image of FRD1300 cells after 6 h of induction with IPTG.
Reduced Ptac.
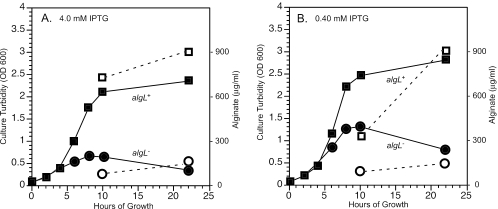
algD expression in the ΔalgL mutant causes lysis. We took advantage of the Ptrc-algD inducible system and reduced the amount of IPTG to reduce alginate production to see if this would allow the ΔalgL mutant to survive. Nearly identical cultures of FRD1050 (algL+) and FRD1300 (ΔalgL) were grown in the presence of 4.0 mM (high expression) or 0.4 mM (lower expression) IPTG and then compared for cell growth (optical density at 600 nm [OD600]) and amounts of alginate produced over time. This 10-fold difference in IPTG resulted in approximately a twofold difference in the amount of alginate made by the Alg+ strain during the log phase of growth: 4.0 mM and 0.4 mM IPTG induction for 10 h in the algL+ strain resulted in 725 ± 75 μg/ml and 410 ± 35 μg/ml alginate, respectively (Fig. 5A and B). However, alginate accumulation was approximately the same with either IPTG concentration, ∼900 ± 75 μg/ml, after 22 h. Lower concentrations of IPTG were also tested, but they did not induce detectable alginate production in the algL+ strain (data not shown). With FRD1300 (ΔalgL), 4.0 mM and 0.40 mM IPTG both induced approximately 95 ± 25 μg/ml alginate at 10 h postinduction and 150 ± 20 μg/ml alginate at 22 h postinduction (Fig. 5). The log-phase cells of FRD1300 were able to grow to a higher OD with the lower IPTG level but were still lysed at approximately the same time. Thus, reducing the level of alginate production in FRD1300 did not appear to abrogate the lethal phenotype, suggesting that AlgL still plays a role in strains producing lower levels of alginate.
FIG. 5.
Effect of IPTG concentrations on growth and alginate production in FRD1050 and FRD1300. Overnight cultures were used to inoculate MAP medium and incubated with aeration at 37°C to an OD600 of 0.20. Twenty-five-milliliter volumes of log-phase cultures were incubated with 4.0 mM IPTG (A) or 0.4 mM IPTG (B) to induce alginate production. Samples were taken periodically for measurements of growth as OD600 (closed symbols) and alginate accumulation in culture supernatants (open symbols). FRD1050 algL+, squares; FRD1300 ΔalgL, circles. A repeat of this experiment produced similar results.
DISCUSSION
A wide range of organisms is known to produce alginate lyases (alginases), which are enzymes that catalyze the degradation of alginate polymers into unsaturated oligosaccharides. Many marine bacteria produce alginate lyases in order to use the alginate of brown algae (seaweed) as a carbon source (49). Alginate-producing bacteria like Azotobacter and Pseudomonas spp. have AlgL, a periplasmic alginate lyase, but they do not use alginate as a carbon source (39, 42). The AlgL protein (40 kDa) has a signal peptide, which is processed during periplasmic localization (42). Pseudomonas AlgL has been shown to preferentially cleave bonds between pairs containing at least one d-mannuronate residue, which thus classifies it as a d-mannuronate-specific lyase (9).
The role of an alginate-degrading enzyme within the alginate biosynthesis-secretion pathway has not been intuitive, and so the function of AlgL has been somewhat controversial. AlgL may control the length of the polymer produced (27). In support of this, an algL mutant of Azotobacter vinelandii produced an alginate with a higher mean molecular weight than the parent strain, although neither encystment nor cyst germination was affected (47). AlgL may also be important in facilitating dissemination of bacteria; overexpression of algL in mucoid P. aeruginosa leads to a decrease in alginate polymer length and an increase in bacterial detachment from an adherent surface (2). With algL in the operon for alginate biosynthesis, and thus coexpressed with alginate biosynthetic enzymes, it seemed reasonable that a periplasmic AlgL protein might be part of a polymerization or transport complex in the periplasm. In Pseudomonas syringae pv. syringae, the absence of lyase activity reduces alginate production by about 50% (35). In P. aeruginosa, one study reports that AlgL is not required for alginate production by P. aeruginosa (3), yet in another a lyase-negative P. aeruginosa isolate appeared nonmucoid and produced only small amounts of alginate (29).
In this study, we examined the effect of a nonpolar ΔalgL mutation in strain FRD1, a highly mucoid (Alg+) CF clinical isolate of P. aeruginosa. This construction was problematic in that we found such mutants to be very difficult to construct using a well-established gene replacement technology for this organism. This was circumvented by artificially replacing the promoter of the algD operon with a LacIq-repressed tac promoter, which then permitted IPTG induction of the operon. In this background, ΔalgL mutants could readily be constructed (e.g., FRD1300), as long as expression of the operon was low. However, when the algD operon was reactivated with IPTG in the ΔalgL mutant, the cells were lysed within a few hours. Electron microscopy showed over time a dramatic separation of the inner and outer membranes, with accumulation of alginate in the periplasm that ultimately led to cell rupture. Alginate was detected in the supernatant of lysed FRD1300 cultures, indicating that AlgL was not required for alginate production. These microscopic observations are similar to those seen with mutations that block ABC transporter-dependent secretion of K1 polysaccharide out of the cell in E. coli, which leads to cytoplasmic accumulation of polymer (33, 45). In contrast, our observation of periplasmic accumulation of polymer in P. aeruginosa suggests that the AlgL− block in alginate transport is by a distinctly different mechanism. We have isolated IPTG-resistant mutants of FRD1300 which were Alg−, and they presumably have secondary mutations that prevent alginate production, which is currently under study.
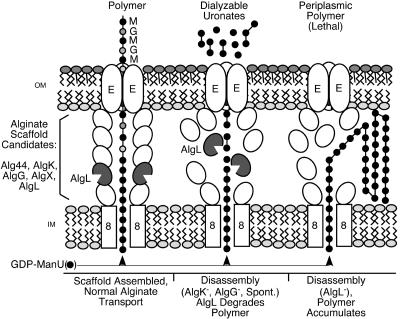
To explain these results, we propose a model for alginate biosynthesis in which AlgL has both an enzymatic function as an alginate lyase and a structural role as a component of a periplasmic transport apparatus, or scaffold, in mucoid P. aeruginosa (Fig. 6, left). The term scaffold has also been applied to the biosynthesis-transport complex for K5 capsule production in E. coli (48). Our previous studies indicate that the periplasmic AlgG protein is also bifunctional, having an enzyme activity (i.e., polymer level C-5 mannuronan epimerase) (12) and a structural role in protecting nascent alginate polymers from the enzymatic attack of the AlgL lyase (20). The AlgK protein is a putative lipoprotein in the periplasm, which likewise is required to protect the polymer from the enzymatic attack of the AlgL lyase (21). AlgX mutants have a similar phenotype (40). Thus, mutants lacking AlgG, AlgK, or AlgX produce polymers that fail to be secreted, but they are rapidly degraded by AlgL. High levels of low-molecular-weight uronic acids, the products of alginate degradation, are found in the culture supernatant of these mutants. Thus, a model for the alginate scaffold includes at least AlgG, AlgK, AlgX, and probably AlgL. It is conceivable that AlgL plays its major role in degrading alginate that accumulates in the periplasm under highly mucoid conditions, and that it has a minor role in transporting polymers across the outer membrane. However, previous studies did not reveal the release of AlgL-degraded alginate in the highly mucoid CF strain FRD1 (21). The attempt here to reduce alginate production levels by FRD1300 by lowering the IPTG induction still resulted in the lysis of cells. Most striking are the electron micrographs here that show periplasmic blebs of alginate after just 2 h of induction of the alginate biosynthetic operon.
FIG. 6.
Model for alginate secretion. (Left) Polymerization of mannuronates (•) from GDP-mannuronate probably occurs via Alg8, which appears to be an inner membrane (IM) glycosyltransferase. A multicomponent protein scaffold is proposed to transport polymer across the periplasm to AlgE in the outer membrane (OM). AlgG, a periplasmic C-5 mannuronan epimerase, converts some d-mannuronate residues to l-guluronate (grey circles). Likely candidates as components of the scaffold complex include the periplasmic proteins Alg44, AlgK, AlgG, AlgX, and AlgL. (Middle) The scaffold fails to assemble correctly when AlgK or AlgG protein is absent. AlgL's alginate lyase activity is then free to digest any newly formed polymer in the periplasm, thus releasing dialyzable uronates into the extracellular environment. Occasionally, the scaffold may disassemble spontaneously in the wild type, allowing AlgL to digest periplasmic polymer. (Right) When AlgL is absent, the scaffold again does not assemble correctly but the absence of periplasmic lyase activity causes polymer to accumulate in the periplasmic space, ultimately bursting the cells.
In concurrence with the mutant analysis, our model for alginate biosynthesis in P. aeruginosa suggests that the periplasmic scaffold does not form correctly when any single component is missing and results in polymer being trapped in the periplasm. We also propose that the depolymerase activity of AlgL is at least partially masked while it is part of the transport scaffold. If the missing component is AlgK, AlgG, or AlgX, the trapped alginate is subject to attack by active AlgL, and this results in the release of small uronic acids (Fig. 6, middle). However, when AlgL is not present due to mutation, then polymer rapidly builds up in the cell's periplasmic space (Fig. 6, right), with lethal effects. Other periplasmic components of the scaffold may include Alg44, which is periplasmic and also required for polymer formation. A putative role for AlgL lyase activity, under normal conditions, is to digest any alginate that remains in the periplasm after spontaneous disassembly of the scaffold. This would indirectly affect polymer size as well. Overall, these data are consistent with the idea that AlgL is a vital part of the alginate transport scaffold, as well as having a role in degrading alginate as a lyase. Future studies will examine the enzymatic role of AlgL in polymer formation and identify the interacting components of the putative alginate transport scaffold.
Acknowledgments
We are grateful to Michael Franklin, Joy Snyder, and Lynn Wood for the construction of plasmids and strains that led to the development of a system for controlled expression of the algD operon in P. aeruginosa. We also acknowledge the Electron Microscopy Core Facility of Virginia Commonwealth University for excellent service and technical assistance.
This work was supported by Veterans Administrations medical research funds (D.E.O) and by Public Health Service grant AI-19146 from the National Institute of Allergy and Infectious Diseases (D.E.O.).
Editor: V. J. DiRita
REFERENCES
- 1.Baltimore, R. S., and M. Mitchell. 1982. Immunologic investigations of mucoid strains of Pseudomonas aeruginosa: comparison of susceptibility to opsonic antibody in mucoid and nonmucoid strains. J. Infect. Dis. 141:238-247. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, A., M. Ghosh, T. B. May, D. Shinabarger, R. Keogh, and A. M. Chakrabarty. 1993. Sequence of the algL gene of Pseudomonas aeruginosa and purification of its alginate lyase product. Gene 131:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Chitnis, C. E., and D. E. Ohman. 1990. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J. Bacteriol. 172:2894-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chitnis, C. E., and D. E. Ohman. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583-590. [DOI] [PubMed] [Google Scholar]
- 6.Deretic, V., M. J. Schurr, J. C. Boucher, V. Deretic, M. J. Schurr, J. C. Boucher, and D. W. Martin. 1994. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J. Bacteriol. 176:2773-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.diSant'Agnese, P. A., and P. B. Davis. 1976. Research in cystic fibrosis. N. Engl. J. Med. 295:597-602. [DOI] [PubMed] [Google Scholar]
- 8.Doggett, R. G., G. M. Harrison, R. N. Stillwell, and E. S. Wallis. 1966. An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J. Pediatr. 68:215-221. [Google Scholar]
- 9.Dunne, W. M., Jr., and F. L. Buckmire. 1985. Partial purification and characterization of a polymannuronic acid depolymerase produced by a mucoid strain of Pseudomonas aeruginosa isolated from a patient with cystic fibrosis. Appl. Environ. Microbiol. 50:562-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, L. R., and A. Linker. 1973. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116:915-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin, M. J., C. E. Chitnis, P. Gacesa, A. Sonesson, D. C. White, and D. E. Ohman. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franklin, M. J., and D. E. Ohman. 1993. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J. Bacteriol. 175:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin, M. J., and D. E. Ohman. 1996. Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O acetylation. J. Bacteriol. 178:2186-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gimmestad, M., H. Sletta, H. Ertesvag, K. Bakkevig, S. Jain, S. J. Suh, G. Skjak-Braek, T. E. Ellingsen, D. E. Ohman, and S. Valla. 2003. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J. Bacteriol. 185:3515-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govan, J. R. W., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govan, J. R. W., and G. S. Harris. 1986. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol. Sci. 3:302-308. [PubMed] [Google Scholar]
- 19.Haug, A., B. Larsen, and O. Smidrod. 1967. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem. Scand. 21:691-704. [Google Scholar]
- 20.Jain, S., M. J. Franklin, H. Ertesvag, S. Valla, and D. E. Ohman. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47:1123-1133. [DOI] [PubMed] [Google Scholar]
- 21.Jain, S., and D. E. Ohman. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutson, C. A., and A. Jeanes. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal. Biochem. 24:470-481. [DOI] [PubMed] [Google Scholar]
- 23.Lam, J., R. Chan, K. Lam, and J. R. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linker, A., and R. S. Jones. 1966. A new polysaccharide resembling alginic acid isolated from pseudomonads. J. Biol. Chem. 241:3845-3851. [PubMed] [Google Scholar]
- 25.Martin, D. W., M. J. Schurr, M. H. Mudd, J. R. W. Govan, B. W. Holloway, and V. Deretic. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 90:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathee, K., C. J. McPherson, and D. E. Ohman. 1997. Posttranslational control of the algT (algU)-encoded σ22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN). J. Bacteriol. 179:3711-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.May, T. B., and A. M. Chakrabarty. 1994. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 2:151-157. [DOI] [PubMed] [Google Scholar]
- 28.May, T. B., D. Shinabarger, R. Maharaj, J. Kato, L. Chu, J. D. DeVault, S. Roychoudhury, N. A. Zielinski, A. Berry, R. K. Rothmel, T. K. Misra, and A. M. Chakrabarty. 1991. Alginate synthesis by Pseudomonas aeruginosa: a key pathogenic factor in chronic pulmonary infections of cystic fibrosis patients. Clin. Microbiol. Rev. 4:191-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monday, S. R., and N. L. Schiller. 1996. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J. Bacteriol. 178:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohman, D. E., and A. M. Chakrabarty. 1981. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect. Immun. 33:142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohman, D. E., K. Mathee, C. J. McPherson, C. A. DeVries, S. Ma, D. J. Wozniak, and M. J. Franklin. 1996. Regulation of the alginate (algD) operon in Pseudomonas aeruginosa, p. 472-483. In T. Nakazawa, K. Furukawa, D. Haas, and S. Silver (ed.), Molecular biology of pseudomonads. ASM Press, Washington, D.C.
- 33.Pavelka, M. S., Jr., S. F. Hayes, and R. P. Silver. 1994. Characterization of KpsT, the ATP-binding component of the ABC-transporter involved with the export of capsular polysialic acid in Escherichia coli K1. J. Biol. Chem. 269:20149-20158. [PubMed] [Google Scholar]
- 34.Pedersen, S. S. 1992. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl. 28:1-79. [PubMed] [Google Scholar]
- 35.Penaloza-Vazquez, A., S. P. Kidambi, A. M. Chakrabarty, and C. L. Bender. 1997. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J. Bacteriol. 179:4464-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pier, G. B., F. Coleman, M. Grout, M. Franklin, and D. E. Ohman. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69:1895-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pier, G. B., G. J. Small, and H. B. Warren. 1990. Protection against mucoid Pseudomonas aeruginosa in rodent models of endobronchial infections. Science 249:537-540. [DOI] [PubMed] [Google Scholar]
- 38.Rehm, B. H., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehm, B. H., H. Ertesvåg, and S. Valla. 1996. A new Azotobacter vinelandii mannuronan C-5-epimerase gene (algG) is part of an alg gene cluster physically organized in a manner similar to that in Pseudomonas aeruginosa. J. Bacteriol. 178:5884-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robles-Price, A., T. Y. Wong, H. Sletta, S. Valla, and N. L. Schiller. 2004. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7369-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxena, I. M., R. M. Brown, M. Fevre, R. A. Geremia, and B. Henrissat. 1995. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schiller, N. L., S. R. Monday, C. M. Boyd, N. T. Keen, and D. E. Ohman. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarzmann, S., and J. R. Boring III. 1971. Antiphagocytic effect of slime from a mucoid strain of Pseudomonas aeruginosa. Infect. Immun. 3:762-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 45.Silver, R. P., K. Prior, C. Nsahlai, and L. F. Wright. 2001. ABC transporters and the export of capsular polysaccharides from gram-negative bacteria. Res. Microbiol. 152:357-364. [DOI] [PubMed] [Google Scholar]
- 46.Thomassen, M. J., C. A. Demko, B. Boserbaum, R. C. Stern, and P. T. Kuchenbrod. 1979. Multiple isolates of Pseudomonas aeruginosa with differing antimicrobial susceptibility patterns from patients with cystic fibrosis. J. Infect. Dis. 140:873-880. [DOI] [PubMed] [Google Scholar]
- 47.Trujillo-Roldan, M. A., S. Moreno, D. Segura, E. Galindo, and G. Espin. 2003. Alginate production by an Azotobacter vinelandii mutant unable to produce alginate lyase. Appl. Microbiol. Biotechnol. 60:733-737. [DOI] [PubMed] [Google Scholar]
- 48.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 49.Wong, T. Y., L. A. Preston, and N. L. Schiller. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54:289-340. [DOI] [PubMed] [Google Scholar]