Abstract
Listeria monocytogenes expresses surface proteins covalently anchored to the peptidoglycan by sortase enzymes. Inactivation of srtA attenuates Listeria virulence in mice (H. Bierne, S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. Garcia-del Portillo, O. Schneewind, and P. Cossart, Mol. Microbiol. 43:869-881, 2002). We show here that an srtA mutant is more attenuated than an internalin mutant in orally infected guinea pigs and transgenic mice expressing human E-cadherin (hEcad mice), indicating the involvement of other SrtA substrates, LPXTG proteins, in food-borne listeriosis. Data recently generated with a listerial DNA macroarray identified two LPXTG protein-encoding genes present in the genomes of L. monocytogenes strains and absent from all other Listeria species, inlI (lmo0333) and inlJ (lmo2821). They also revealed two other LPXTG protein-encoding genes, ORF29 and ORF2568, present only in a subclass of L. monocytogenes serovars, including the epidemic serovar 4b. We report here that an inlJ deletion mutant, in contrast to inlI and ORF29 mutants, is significantly attenuated in virulence after intravenous infection of mice or oral inoculation of hEcad mice. Interestingly, a ΔORF2568 strain showed a slight increase in virulence. inlJ encodes a leucine-rich repeat (LRR) protein that is structurally related to the listerial invasion factor internalin. However, the consensus sequence of the InlJ LRR defines a novel subfamily of cysteine-containing LRRs in bacteria. In conclusion, this postgenomic approach identified InlJ as a new virulence factor among the proteins belonging to the internalin family in L. monocytogenes.
Listeria monocytogenes is a bacterial pathogen that causes listeriosis, a severe food-borne disease in humans (21). It is a facultative intracellular pathogen that is able to enter and multiply in both professional phagocytes (45) and nonphagocytic cells (24, 35). Following ingestion of contaminated food, some bacteria cross the intestinal barrier and gain access to the liver and spleen via the bloodstream. In these two organs the majority of bacteria are eliminated by the immune system. In immunocompromised individuals and pregnant women bacteria can cross the tight blood-brain barrier and the maternofetal barrier, respectively, and reach the central nervous system and the placenta.
Several listerial surface proteins play key roles in listerial interactions with mammalian host cells (8). One of these proteins, internalin (InlA), promotes Listeria internalization in epithelial cells by interacting with the host adhesion protein E-cadherin (51) and is required for invasion of intestinal and placental villi in vivo (38, 40). InlA possesses an N-terminal leucine-rich repeat (LRR) domain (59) and a C-terminal sorting signal, including an LPXTG motif. Anchoring of InlA to the bacterial surface is achieved by sortase A (SrtA) (6), a transpeptidase that cleaves the LPXTG motif between the threonine and glycine residues and covalently links the threonine to the peptidoglycan (12, 16, 48, 61). Inactivation of srtA in L. monocytogenes inhibits anchoring of LPXTG proteins to the cell wall and attenuates virulence following intravenous or oral infection in mice (6, 27). The Listeria genome encodes another sortase, SrtB, which seems to have only two substrates carrying an NXZTN motif instead of LPXTG and which is not required for the infectious process in mice (5).
A better understanding of listeriosis pathophysiology relies on identification of new bacterial virulence factors and their characterization in adapted animal models. Intravenous inoculation of mice has proven to be a good route to study the virulence factors involved in the intracellular life of L. monocytogenes (18, 35). However, bacterial translocation across the intestinal barrier is inefficient in the mouse (40). In contrast, in guinea pigs and in transgenic mice expressing human E-cadherin, the internalin receptor (40), in enterocytes (hEcad mice), bacteria can efficiently cross the intestinal epithelial barrier and gain access to deeper tissues. These two models thus appear to be more appropriate than a normal mouse model for studying listeriosis acquired via the oral route (37), which is the natural route for infection in humans (57).
In the past few years, major listerial virulence factors have been identified by using classical genetic approaches, such as generation of transposition mutants and characterization of these mutants in infection experiments in cultured cells (18, 35). Determination and comparison of the genome sequences of L. monocytogenes and the nonpathogenic closely related species Listeria innocua (28) have now paved the way for identifying new virulence factors (8, 14, 28, 53). Such a comparative approach has, for instance, led to the identification of a bile salt hydrolase (17) and an autolysin (9) as new factors involved in Listeria infections. Besides whole-genome comparisons, a Listeria DNA array has recently been generated to study inter- and intraspecies diversity (14). This macroarray, which harbors specific genes from three sequenced strains, two pathogenic L. monocytogenes strains (serovars 1/2a and 4b) and a nonpathogenic L. innocua strain, was used to analyze the DNA content and genomic biodiversity of 113 Listeria strains of different species and serovars. Hybridization results identified L. monocytogenes-specific marker genes, including inlA and other previously identified virulence factors, such as hly, actA, or inlB, as well as serovar-specific marker genes. It is now well established that L. monocytogenes strains do not appear to be equally able to generate disease in humans (30). Only 4 of the 13 serovars identified in this species, serovars 1/2a, 1/2c, 1/2b, and 4b, are responsible for the reported human listeriosis cases (31). Notably, serotype 4b strains are overrepresented compared with the strains of other serotypes among the organisms responsible for outbreaks and sporadic cases of listeriosis (63).
In this work, we first confirmed that srtA inactivation strongly attenuates Listeria virulence in orally acquired listeriosis, supporting the role of LPXTG surface proteins in the infectious process. Then, by exploiting the genome and biodiversity array data, we addressed the role of four previously uncharacterized LPXTG protein-encoding genes, two L. monocytogenes-specific genes and two genes present in a subset of serovars, including serovar 4b. We generated deletion mutants and analyzed them in vitro and in vivo. This work revealed the role of the lmo2821 gene product, InlJ, an LRR cysteine-containing protein, in orally acquired listeriosis. In addition, inactivation of the ORF2568 gene in L. monocytogenes serovar 4b slightly increased bacterial virulence, suggesting that the gene product has a role in virulence modulation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cell lines.
The strains and plasmids used in this study are listed in Table 1. Bacteria were grown at 37°C in brain heart infusion (BHI) medium (Difco) supplemented with erythromycin (5 μg ml−1) when they carried pMAD derivatives. The cell lines used were as follows: human choriocarcinoma JEG-3 cells (ATCC HTB-36), colon carcinoma Caco-2 cells (ATCC HTB-37) and LoVo cells (ATCC CCL-229), hepatocarcinoma HepG2 cells (ATCC HB-8065), cervix carcinoma HeLa cells (ATCC CCL-2), human embryonic kidney HEK 293 epithelial cells (ATCC CRL-1573), umbilical vein HUVEC endothelial cells (ATCC CRL-1730), and RAW 264.7 (ATCC TIB-71) and J774 (ATCC TIB-67) murine macrophages. Cell lines were cultured in modified Eagle medium (Gibco), Dulbecco modified Eagle medium (DMEM), or Ham's F12K medium supplemented with fetal bovine serum (10% or 20%), 1 mM pyruvate, 2 mM glutamine, and 1% nonessential amino acids (Gibco) according to American Type Culture Collection recommendations. HUVEC cells were cultured with 0.1 mg · ml−1 of heparin (Sigma) and 0.05 mg · ml−1 of endothelial growth factor. Macrophages were cultured with decomplemented fetal bovine serum. Cells were incubated at 37°C in the presence of 10% CO2.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Property | Reference |
|---|---|---|
| Strains | ||
| BUG 1600 | L. monocytogenes EGDe wild-type strain | 28 |
| BUG 1777 | Isogenic ΔsrtA strain | 6 |
| BUG 1877 | Isogenic ΔsrtB strain | 5 |
| BUG 1454 | Isogenic ΔinlA strain | 42 |
| BUG 2158 | Isogenic ΔinlI strain | This study |
| BUG 2159 | Isogenic ΔinlJ strain | This study |
| BUG 2099 | L. monocytogenes CLIP80459 seravor 4b wild-type strain | 14 |
| BUG 2194 | Isogenic ΔORF29 strain | This study |
| BUG 2195 | Isogenic ΔORF2568 strain | This study |
| Plasmids | ||
| pMAD | 1 | |
| pMAD-ΔinlI | Thermosensitive plasmid pMAD carrying coligated 650-bp fragments flanking inlI | This study |
| pMAD-ΔinlJ | Thermosensitive plasmid pMAD carrying coligated 650-bp fragments flanking inlJ | This study |
| pMAD-ΔORF29 | Thermosensitive plasmid pMAD carrying coligated 650-bp fragments flanking ORF29 | This study |
| pMAD-ΔORF2568 | Thermosensitive plasmid pMAD carrying coligated 650-bp fragments flanking ORF2568 | This study |
Chromosomal inactivation of the inlI, inlJ, ORF29, and ORF2568 genes. (i) Construction of the in-frame ΔinlI and ΔinlJ L. monocytogenes EGDe deletion mutants.
Two ∼650-bp fragments flanking the target gene (inlI or inlJ) were amplified by PCR from EGDe chromosomal DNA with primers inside and outside the inlI and inlJ loci. The primers used for the inlI 5′ flanking fragment were i1 (5′-CGCGGATCCTGCGGTATCAAGAAGTTTATC-3′) and i2 (5′-AAAAGGCCTCTTTTTCAAGACTTTCCCTCT-3′), and the primers used for the 3′ fragment were i3 (5′-AAAAGGCCTACGAACCAATAAATGGATAAAC-3′) and i4 (5′-TCCCCCGGGCGGAGTTGCTGTGTATCTAGT-3′). The primers used for the inlJ 5′ flanking fragment were j1 (5′-CGCGGATCCTAGCAGAAGATGGAACCC-3′) and j2 (5′-AAAAGGCCTAGTTTTCAATTTGGCGCACTC-3′), and the primers used for the 3′ fragment were j3 (5′-AAAAGGCCTAAATAGTAAAAAAGCCGGACA-3′) and j4 (5′-CCGGAATTCCAAATGATGTTATCGGTCAGC-3′). After restriction of the amplified 5′ and 3′ fragments with BamHI and StuI or StuI and SmaI (for ΔinlI construction) and with BamHI and StuI or StuI and EcoRI (for ΔinlJ construction), 5′ and 3′ fragments were coligated in the thermosensitive plasmid pMAD digested by BamHI and SmaI or by BamHI and EcoRI, yielding the pMAD-ΔinlI and pMAD-ΔinlJ plasmids. These plasmids were electroporated into L. monocytogenes strain EGDe, and gene replacement was performed as described previously (1) but at a nonpermissive temperature growth, 43.5°C, resulting in in-frame deletions of the inlI or inlJ genes.
(ii) Construction of the in-frame ΔORF29 and ΔORF2568 deletion mutants in L. monocytogenes serovar 4b strain.
ΔORF29 and ΔORF2568 were constructed as described above for the ΔinlI and ΔinlJ strains, with the following modifications. The primers used for ΔORF29 construction were k1 (5′-CGCGGATCCGTGAGCCTATGTACGGTC-3′), k2 (5′-AAAAGGCCTCACAGAAAAGCCCCCTTA-3′), k3 (5′-AAAAGGCCTAGAAGTTAAACCACTCCAT-3′) for the 5′ and 3′ flanking fragments, respectively, and k4 (5′-CCGGAATTCCCATTAAAGAAGCAGAACA-3′), and the primers used for ΔORF2568 construction were l1 (5′-CGCGGATCCCGTCATATATCAATCTCCAT-3′), l2 (5′-AAAAGGCCTCATTTTTTTTGCCCCTTCAA-3′), l3 (5′-AAAAGGCCTTAAAAAAACGCCCAGTATTTG-3′), and l4 (5′-TCCCCCGGGTCTCCAAGCCACAATCAC-3′) for the 5′ and 3′ flanking fragments, respectively. The enzymes used for the construction of pMAD-ΔORF29 and pMAD-ΔORF2568 were BamHI, StuI, and EcoRI for pMAD-ΔORF29 and BamHI, StuI, and SmaI for pMAD-ΔORF2568.
Mutant verification by PCR and sequencing.
Mutants were verified by PCR analysis of chromosomal DNA using pairs of oligonucleotide primers inside genes, including i7 (5′-CGGAGCGCCTGTCATTTC-3′) and i8 (5′-TACGCCGTCTTCATTCGT-3′) for ΔinlI, j7 (5′-GAGGCTGAAGGGCAAACAATC-3′) and j8 (5′-CATCCGCCAATTTTTCTCCTT-3′) for ΔinlJ, k7 (5′-GGGAGGAACTCTTTTAGTGTC-3′) and k8 (5′-GCTTGGTACAATGAGTGTACT-3′) for ΔORF29, and l7 (5′-CATCTTCAAGATCAAGCATTA-3′) and l8 (5′-GCTAACTTATTTCCATCACCA-3′) for ΔORF2568, and pairs of oligonucleotide primers in regions flanking the genes, including i5 (5′-GCGTACTCATTTAAGACGAAT-3′) and i6 (5′-AAGCCTCTCTTTAATGGACAG-3′) for ΔinlI, j5 (5′-ATAAATACGCCTCGCTTA-3′) and j6 (5′-GACTAGTTTTGATGTGGA-3′) for ΔinlJ, k5 (5′-TCTAGCGGGAATACTTGGGTT-3′) and k6 (5′-GCAGGAGCAACATTCGGTTGG-3′) for ΔORF29, and l5 (5′-AGGCGAGCTAGAACATAATAC-3′) and l6 (5′-CTGCTTCCAAAAGCAACAATC-3′) for ΔORF2568. Amplified fragments from ΔinlI, ΔinlJ, ΔORF29, and ΔORF2568 strain chromosomal DNA, obtained by using primers i5 and i6, primers j5 and j6, primers k5 and k6, and primers l5 and l6, respectively, were verified by sequencing.
RNA techniques.
RNA from Listeria was isolated and purified with an RNAeasy kit (QIAGEN). Briefly, cultures of Listeria were centrifuged at 4,000 × g for 6 min. The pellets were resuspended in 100 μl of Tris-EDTA and 250 μl of RNAeasy lysis buffer and transferred to a Bead Beater tube containing 0.4 g of mini glass beads (Sigma). Bacteria were broken mechanically using a Fast Prep apparatus (Bio 101) and centrifuged (13,000 × g, 1 min), and the supernatants were transferred and treated according to the manufacturer's procedure. DNA contaminants were eliminated with a DNase kit (Ambion). The presence of the inlI, inlJ, ORF29, and ORF2568 genes in wild-type and mutants strains was assed by amplification of internal fragments by reverse transcriptase (RT)-PCR, using Superscript one-step RT-PCR (Invitrogen) and the internal primers described above. The absence of DNA contamination of the RNA was checked by PCR. For mutant verification, bacteria were grown to an optical density at 600 nm (OD600) of 0.6. Primers iap-F (5′-AAAGCAACTATCGCGGCTAC-3′) and iap-R (5′-TCTTGAACAGAAACACCGTA-3′), which amplify the iap gene encoding the p60 protein, were used as a control for RNA amplification. In the semiquantitative PCR assay, bacteria were grown to an OD600 of 0.8, as in the gentamicin survival assay (see below). Primers inlA-1 (5′-CATCACCTTATATGCCCAATATAGC-3′) and inlA-2 (5′-GATTTTTCGTAAATTGAGCGTACAG-3′) and primers gyrA-1 (5′-AACTTTGGTTCGGTTGATGG-3′) and gyrA-2 (5′-TGGCTCACGTTCAGAACC-3′), which amplify the inlA and gyrA genes, respectively, were used as controls for known expressed genes.
Gentamicin survival assay.
A gentamicin survival assay was performed as described previously (25, 51). Briefly, the Listeria strains were grown in BHI medium to an OD600 of 0.8 to 1, washed in phosphate-buffered saline, and diluted in DMEM, so that the multiplicity of infection was about 50 bacteria per cell. Bacterial suspensions were added to mammalian cells for 1 h, the cells were washed, and the noninvasive bacteria were killed by adding 10 μg/ml gentamicin for 2 h. After washing, the cells were lysed in 0.2% Triton X-100, and the number of viable bacteria released from the cells was assessed by plating on agar plates.
In vivo experiments.
Four animals were used for each experiment, and all experiments were reproduced at least once and gave similar results. Prior to oral infection, the animals were starved to prevent variations linked to gastric repletion, which may influence intragastric bacterial survival. Statistical analyses were performed using the Student t test. P values of <0.05 were considered statistically significant. All animals were treated in accordance with Institut Pasteur guidelines for laboratory animal husbandry.
(i) Guinea pig oral infections.
Experiments were performed as described previously (40). Briefly, 300-g male Hartley guinea pigs (Charles River) were starved for 2 days and anesthetized (15 mg/ml ketamine injected intramuscularly). Five milliliters of a 25-mg/ml CaCO3 solution in phosphate-buffered saline was injected intragastrically, followed by 1 ml of a sublethal bacterial inoculum (2.5 × 1010 CFU). After 96 h of infection, spleens, livers, and the whole mesenteric lymph nodes were sterilely dissected. The central 20-cm portion of the guinea pig small intestine was rinsed in DMEM (Gibco) to remove the intestinal contents, incubated at 20°C for 2 h in DMEM containing 100 mg/liter gentamicin (Gibco) to kill the extracellular bacteria from the intestinal lumen, and rinsed three times in DMEM. For bacterial enumeration, the numbers of CFU were determined by plating serial dilutions of organ (intestine, mesenteric lymph node, liver, and spleen) homogenates on BHI agar.
iFABP-hEcad transgenic mouse oral infections.
iFABP-hEcad transgenic mice (40) were starved for 1 day and anesthetized. Three hundred microliters of a 50-mg/ml CaCO3 solution was injected intragastrically along with 200 μl of a sublethal bacterial inoculum (109 CFU). After 72 h of infection, spleens, livers, and the whole mesenteric lymph nodes were sterilely dissected. The central long portion of the mouse small intestine was rinsed in DMEM (Gibco) to remove the intestinal contents, incubated at 20°C for 2 h in DMEM containing 100 mg/liter gentamicin (Gibco) to kill the extracellular bacteria from the intestinal lumen, and rinsed three times in DMEM. For bacterial enumeration, the numbers of CFU were determined by plating serial dilutions of organ (intestine, mesenteric lymph node, liver, and spleen) homogenates on BHI agar.
Mouse intravenous infections.
Bacterial growth in mice was studied by injecting 4- to 6-week-old specific-pathogen-free female BALB/c mice (Charles River) intravenously with a sublethal bacterial inoculum (104 CFU). At 24, 48, and 72 h after infection, the livers and spleens were sterilely dissected, and the numbers of CFU were determined by plating serial dilutions of organ (liver and spleen) homogenates on BHI agar.
RESULTS
Role of sortase A substrates in L. monocytogenes oral infections in guinea pigs.
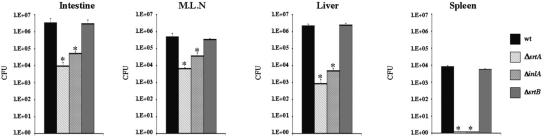
L. monocytogenes translocation across the intestinal barrier occurs efficiently by an InlA-mediated process in guinea pigs (37). To address the global role of LPXTG proteins in the crossing of the intestinal barrier, we analyzed the virulence of an srtA deletion mutant in this animal model and compared it to that of a ΔinlA strain. We also addressed the effect of inactivating the second sortase gene, srtB. Quantification of the level of infection in the small intestine, mesenteric lymph nodes, liver, and spleen was used as a measure of virulence. Guinea pigs were orally infected with the L. monocytogenes wild-type reference strain EGDe or with srtA, inlA, or srtB isogenic deletion mutants. Bacterial counts in the organs were determined 96 h postinfection, which corresponded to the peak for the bacterial load in this animal model (40). For the ΔsrtA strain there were decreases in the bacterial counts of 3 log10 in the intestine and liver and 2 log10 in the mesenteric lymph nodes compared to the wild-type strain and decreases of 1 log10 in both organs compared to the ΔinlA strain (Fig. 1). No ΔsrtA or ΔinlA bacteria were recovered from spleens. In contrast to the ΔsrtA strain, the ΔsrtB strain did not exhibit a virulence defect. These results confirmed that at least one SrtA substrate other than InlA plays a role in the intestinal and hepatic phases of orally acquired listeriosis, which is consistant with our previous results for mice (6). Moreover, they strongly suggest that some LPXTG proteins play an additive role along with InlA in the crossing of the intestinal barrier.
FIG. 1.
srtA is involved in L. monocytogenes oral infections in guinea pigs. The wild-type EGDe strain (wt) and isogenic ΔsrtA, ΔinlA, and ΔsrtB strains were orally inoculated into guinea pigs, as described in Materials and Methods. Animals were euthanized 96 h after infection, and organs were recovered, homonogenized, and plated. The numbers of bacteria able to colonize the intestine, mesenteric lymph nodes (M.L.N), liver, and spleen are expressed in log10 CFU. In each experiment four animal were used for each bacterial strain. The results are representative of two independent experiments. An asterisk indicates a significant difference (P < 0.05) between a mutant strain and the wild-type strain.
Search for new L. monocytogenes virulence factors in the LPXTG protein family.
In order to identify new LPXTG proteins implicated in L. monocytogenes virulence, we exploited the data resulting from a Listeria DNA macroarray analysis (14) to find genes conserved in pathogenic species. Seven genes encoding surface proteins were found as markers for the L. monocytogenes species; these genes were conserved in the genomes of all L. monocytogenes isolates and were absent from all other Listeria species (14). Six of the genes encode proteins belonging to the internalin family (inlA, inlB, inlH, inlE, lmo0333, and lmo2821). All these genes are present in the five L. monocytogenes strains that have been sequenced (14, 28, 53). ΔinlA, ΔinlB, ΔinlH, and ΔinlGHE strains have been shown to be attenuated in virulence (18, 35, 38, 40, 55, 58), while the contributions of the lmo0333 and lmo2821 genes have never been studied. Therefore, we focused on the lmo0333 and lmo2821 genes in this study.
Predicted structures of the internalin-like proteins Lmo0333 (InlI) and Lmo2821 (InlJ).
lmo0333 (5,337 bp) and lmo2821 (2,556 bp) are predicted to encode structurally related proteins consisting of 1,778 and 851 amino acids, respectively. The two proteins possess an N-terminal signal sequence followed by an LRR domain consisting of 28 and 15 LRRs, respectively, a domain characteristic of internalin family members (8, 15). Given this structural characteristic, Lmo0333 and Lmo2821 were desiganted InlI and InlJ. InlI is the largest internalin among the 19 internalin-like proteins present in the EGDe strain (8). As in InlA and in other described internalins (58), the LRR domains of both InlI and InlJ are followed by a conserved immunoglobulin-like domain, a variable region, and a C-terminal sorting signal that includes an LPXTG motif. The variable regions of InlI and InlJ contain specific repeats that are ∼70 residues long reported to be MucBP (mucin binding protein) domains (previously DUF1085; accession number PF06458) (Fig. 2A). MucBP domains are found in conjunction with an LRR domain and an LPXTG sorting signal in several bacterial peptidoglycan-bound proteins, especially in Listeria and Lactobacillis species. The InlI and InlJ proteins contain three and four MucBP domains, respectively. The InlI variable region also contains a PKD (polycystin kidney disease) repeat region (8) related to that found in the human membrane polycystin kidney disease protein (4).
FIG. 2.
InlJ LRR defines a new family of bacterial cysteine-containing LRRs. (A) Amino acid sequence of the inlJ gene product. The signal peptide is indicated by boldface letters. The 15 LRRs are aligned, with leucines, isoleucines, and valines indicated by a red background and cysteines indicated by a blue background. The other conserved residues are indicated by colored letters. The immunoglobulin (Ig)-like domain is indicated by a gray background. The four MucBP repeats are aligned with conserved residues in boldface letters. The LPXTG motif in the C-terminal sorting signal is underlined, and the hydrophobic residues are indicated by italics. (B) Comparison of InlA and InlJ LRR consensus sequences. Residue positions are numbered as described by Schubert et al. (58). The positions of the expected β strand and 310-helix are indicated. Conserved residues are indicated by colored letters (hydrophobic core, red; cysteines, blue). (C) Alignment of LRR consensus sequences of InlJ and proteins from different species. (Top) Alignment of InlJ LRR with LRRs of bacterial or viral proteins. (Bottom) Alignment of InlA and InlJ LRR consensus sequences with LRRs of RNase inhibitor (RI) and cysteine-containing (CC) types from eukaryotes. Conserved residues are indicated by colored letters (leucines, valines, and isoleucines, red; cysteines, dark blue; threonine, light blue; asparagine, green; aspartic acid, orange). O, hydrophobic residues; bCC, bacterial cysteine containing.
Interestingly, while the LRR consensus in InlI is similar to that in InlA and other previously described internalins (58), the consensus in InlJ is slightly different (Fig. 2B). The internalin LRR prototype motif forms a structural unit consisting of a β strand and a 310-helix, which occurs several times in tandem arrrays, and most repeats are 22 residues long (46). InlJ LRRs are in most cases 21 residues long and contain a conserved cysteine at position 7 (or position 9 in repeat 12) in place of a leucine (Fig. 2B). Furthermore, the expected 310-helix region appears to be shorter than that in InlA, with only one residue (usually an aspartate) between the two leucines instead of the two residues in InlA. Lastly, an asparagine residue usually flanks the 310-helix.
Only one other internalin-like protein in L. monocytogenes, Lmo0331, and its ortholog in L. innocua, Lin0354, contain LRRs of the InlJ type. We also found LRRs in proteins from five other bacterial species (Fig. 2C). Two proteins from entomopoxviruses may be related to this subfamily. Finally, several eukaryotic LRRs contain cysteines, but they are not at the same position as in the InlJ LRR consensus. This is the case for the RNase inhibitor LRR type and several proteins of the CC (cysteine-containing) subfamily (Fig. 2C) (33, 36). Thus, InlJ defines a new type of LRR in the internalin family in bacteria.
Effect of inlI or inlJ gene inactivation in the L. monocytogenes cellular infectious process in vitro.
In the genome of L. monocytogenes EGDe, inlI is the first gene of a putative operon comprising seven other genes (28). Six of these genes are predicted to encode proteins with unknown functions, while one, lmo0339, exhibits similarities with a pyrophosphatase-encoding gene. Four genes in this operon, including inlI, are absent in the genome of L. innocua CLIP11262, a sequenced strain. The inlJ gene is also absent from the L. innocua genome, but the surrounding regions are identical in L. innocua and L. monocytogenes. inlJ is in the orientation opposite that of lmo2820, a gene encoding a protein with an N-terminal domain exhibiting similarities with the transcriptional AraC/XylS family of regulators (26). No specific recognition sequence for the Listeria virulence regulator PrfA precedes the inlI and inlJ genes. The expression of inlI and inlJ in bacteria grown in BHI medium in the late exponential phase at 37°C was assessed by RT-PCR and compared to that of inlA and gyrA genes, which encode the invasion protein InlA and gyrase, respectively. Expression products were detected in all cases, indicating that inlI and inlJ are transcribed in these conditions (Fig. 3A).
FIG. 3.
Expression of inlI and inlJ. (A) RT-PCR for total RNAs from the end of the exponential phase (OD600, 0.8) for cultures in BHI medium at 37°C of L. monocytogenes wild-type strain. Serial dilutions of the RNA templates were used. The gyrA and inlA genes were used as controls of known expressed genes. (B) RT-PCR for RNAs from L. monocyogenes wild-type and ΔinlI or ΔinlJ strains. Amplification of the iap gene was used to control RNA amounts. wt, wild type.
In order to study their potential role in virulence, the inlI and inlJ genes were inactivated by in-frame deletion to prevent polar effects. Deletion mutants were obtained by allelic exchange in L. monocytogenes EGDe, as previously described (1). As expected, no RT-PCR products were detected from the mutant strains (Fig. 3B). The ΔinlI and ΔinlJ strains displayed no growth defect in BHI medium at 37°C compared to the wild-type strain (data not shown). We then examined the ability of these strains to adhere to two epithelial cell lines, intestinal Caco-2 and placental JEG-3 cells, and to J774 macrophages, using previously described techniques (7, 15). After 1 h of infection at an initial multiplicity of infection of 50 bacteria per cell, the ΔinlI and ΔinlJ strains displayed no noticeable difference from the wild-type strain in adherence to host cells (data not shown). Then their ability to enter cells was examined by using nine different cell types, including epithelial and endothelial cells, hepatocytes, and macrophages. As shown in Table 2, the entry of the ΔinlI or ΔinlJ strain after 1 h of infection was similar to that of the wild-type strain for all cell types, suggesting that these genes are not involved in the L. monocytogenes internalization process. Finally, these mutants did not appear to be altered in intracellular multiplication, actin-based motility, and cell-to-cell spreading (data not shown). Taken together, these results indicate that the ΔinlI and ΔinlJ strains displayed no detectable defect in infection of tissue-cultured cells.
TABLE 2.
Determination of internalization efficiencies
| Cell linea | Relative % of entry into cell
|
||
|---|---|---|---|
| Wild type | ΔinlI | ΔinlJ | |
| Caco-2 | 100 | 115 ± 27 | 110 ± 32 |
| JEG-3 | 100 | 100 ± 3 | 102 ± 3 |
| Hep-G2 | 100 | 102 ± 8 | 96 ± 11 |
| HUVEC | 100 | 86 ± 8 | 102 ± 8 |
| LoVo | 100 | 115 ± 9 | 93 ± 4 |
| HeLa | 100 | 115 ± 6 | 102 ± 5 |
| HEK293 | 100 | 104 ± 5 | 101 ± 4 |
| J774 | 100 | 86 ± 9 | 127 ± 14 |
| RAW 264.7 | 100 | 125 ± 9 | 107 ± 5 |
Cells were infected with the bacterial strains for 1 h, and invasion frequencies were calculated from the number of bacteria that survived gentamicin treatment compared with the total number of inoculated bacteria. The level of entry of the wild-type strain was arbitrarly defined as 100%, and the levels of entry of the different mutants are relative values.
Effect of inlI and inlJ inactivation on organ colonization.
The contribution of the inlI and inlJ genes in the L. monocytogenes infectious process in vivo was examined first by using an intravenous mouse model of infection. BALB/c mice were infected intravenously with the wild type or with the ΔinlI or ΔinlJ strain. Bacterial counts in the liver and spleen were determined 24, 48, and 72 h postinfection. As shown in Fig. 4A, at 72 h postinfection a 1-log10 decrease in the liver and spleen was observed for the inlJ mutant strain. In contrast, no difference was observed between ΔinlI and the wild-type strain. These results strongly suggest that InlJ contributes to Listeria virulence.
FIG. 4.
inlJ is involved in L. monocytogenes virulence. (A) Mice were inoculated intravenously with 104 CFU of the wild-type EGDe strain or ΔinlI or ΔinlJ strain. Bacterial growth was monitored in the liver and the spleen at 24 h, 48 h, and 72 h. The results are the means of two independent experiments. (B) hEcad mice were orally infected with 109 CFU of the wild-type strain or the ΔinlI, ΔinlJ, ΔinlA, or ΔsrtA strain. The numbers of bacteria able to colonize the intestine, mesenteric lymph nodes (M.L.N), liver, and spleen were determined at 72 h. The data are expressed in log10 CFU (see Materials and Methods). In each experiment four animals were used for each bacterial strain. The results are the means of two (ΔinlI strain) or three (wild-type, ΔinlJ, ΔinlA, and ΔsrtA strains) independent experiments. An asterisk indicates a significant difference (P < 0.05) between a mutant strain and the wild-type strain. wt, wild type.
To address the contribution of these internalins during oral infection, we then performed oral infection experiments with hEcad mice expressing human E-cadherin in enterocytes. hEcad mice are a better model than wild-type mice for studying orally acquired listeriosis as L. monocytogenes efficiently crosses their intestinal barrier by an InlA-mediated process (40). The virulence phenotypes of ΔinlI and ΔinlJ strains were compared to those of wild-type, ΔinlA, and ΔsrtA strains. After 72 h of infection, the ΔinlJ strain displayed 1- and 1.5-log10 decreases in colonization of the mesenteric lymph nodes and spleen, respectively, compared to the wild-type strain (Fig. 4B). This attenuation was comparable to that of ΔinlA and ΔsrtA strains. The bacterial counts of the ΔinlJ strain were reduced 2 log10 in the liver, as observed for ΔinlA, while the ΔsrtA strain displayed a more severe attenuation defect. Finally, ΔinlJ bacteria were recovered from intestines at a level of 3 × 103 CFU, a level intermediate between that of the wild-type strain (3 × 104 CFU) and that of ΔinlA and ΔsrtA strains (3 × 102 CFU), which is at the limit of bacterial detection in this tissue. No significant attenuation in virulence was detected with the ΔinlI strain.
Taken together, these results indicate that the inlJ gene product is involved in several steps of L. monocytogenes infection and validate the hypothesis that LPXTG-anchored proteins other than InlA are implicated in virulence when the oral route is used.
Inactivation of two other LPXTG protein-encoding gene markers for L. monocytogenes serovar 4b.
L. monocytogenes serovar 4b strains are responsible for the majority of listeriosis epidemic cases, suggesting that this serovar expresses specific factors favoring infection in human. However, data generated with the biodiversity array did not reveal any gene that was present only in serovar 4b strains (14). Nevertheless, three genes encoding LPXTG proteins, ORF29, ORF1761, and ORF2568, were specifically detected in lineage II (serovars 1/2b, 4a, 4b, and 4c) and not in the other serovars (14). We therefore investigated whether these genes could be involved in L. monocytogenes serovar 4b infection. Reexamination of the sequence of ORF1761 revealed that this gene fused with ORF2017 and is present in serovar 1/2a. Therefore, we focused on the ORF29 and ORF2568 genes in this study.
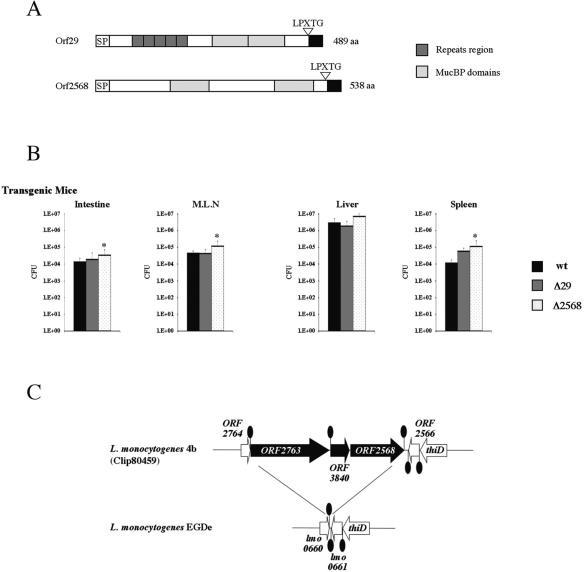
Both ORF29 and ORF2568 are present in the three L. monocytogenes serovar 4b strains that have been sequenced (14, 53). ORF29 (1,470 bp) and ORF2568 (1,617 bp) are predicted to encode LPXTG proteins consisting of 489 and 538 amino acids, respectively, that do not possess an LRR domain but contain MucBP domains (Fig. 5A). Notably, the amino-terminal region encoded by ORF29 contains five hydrophobic repeats consisting of 22 amino acids of unknown structure and two repeated MucBP domains. The protein encoded by ORF2568 exhibits no similarity with known proteins except that it contains two MucBP domains separated by 156 amino acids.
FIG. 5.
(A) Schematic representation of ORF29 and ORF2568 LPXTG proteins specific for serovar 4b. The signal peptide (SP), repeat region, MucBP domains, and C-terminal sorting signal (solid box) are indicated. (B) Effect of ORF29 and ORF2568 inactivation on L. monocytogenes serovar 4b virulence. The wild-type L. monocytogenes serovar 4b strain or ΔORF29 and ΔORF2568 isogenic mutants were orally inoculated into hEcad mice (109 CFU). The numbers of bacteria able to colonize the intestine, mesenteric lymph nodes (M.L.N), liver, and spleen were determined at 72 h and expressed in log10 CFU (see Materials and Methods). In each experiment four animals were used for each bacterial strain. The results are the means of two (ΔORF29 strain) or three (wild-type and ΔORF2568 strains) independent experiments. An asterisk indicates a significant difference (P < 0.05) between a mutant strain and the wild-type strain. wt, wild type. (C) Genetic organization of the ORF2568 region in L. monocytogenes serovar 4b strain CLIP80459 and in L. monocytogenes serovar 1/2a strain EGDe. Open reading frames are indicated according to the genome nomenclature described previously (ORF [14] and lmo [28]). Open reading frames present only in L. monocytogenes serovar 4b are indicated by solid arrows. Other open reading frames are indicated by open arrows. The ovals indicate putative transcription terminators.
To address the contribution of ORF29 and ORF2568 to virulence, the genes were inactivated in the L. monocytogenes serovar 4b CLIP 80459 sequenced strain, and deletion mutants were verified by PCR and RT-PCR, as described previously for the inlI and inlJ genes (data not shown). When tested in vitro in invasion assays for entry, intracellular replication, and motility, ΔORF29 and ΔORF2568 strains behaved like the wild-type strain (data not shown). These mutants were then tested in vivo by oral infection of transgenic hEcad mice, as described above for ΔinlI and ΔinlJ strains. As shown in Fig. 5A, the colonization of the intestine, mesenteric lymph nodes, liver, and spleen by the ΔORF29 strain was similar to the colonization by the wild-type serovar 4b strain. In contrast, strikingly, for the ΔORF2568 strain there was a reproducible and statistically significant slight increase in the number of CFU in the intestine, mesenteric lymph nodes, and spleen (Fig. 5B). Thus, the ΔORF2568 bacteria displayed levels of colonization higher than those of the wild-type serovar 4b strain (0.5 log10 in the intestine, mesenteric lymph nodes, and liver and 1 log10 in the spleen). Together, these results indicate that ORF29 is not required for efficient L. monocytogenes serovar 4b oral infection, at least in the experimental conditions tested here. Moreover, inactivation of ORF2568 increased bacteriemia in tissues, suggesting that expression of this gene may affect L. monocytogenes virulence during host infection. The ORF2568 gene and the adjacent ORF3840 and ORF2763 genes are absent from the L. monocytogenes EGDe genome (Fig. 5C). These genes, whose functions are not known, are located in a chromosomal region which has not previously been associated with virulence or regulation.
DISCUSSION
Role of sortases and LPXTG proteins in orally acquired listeriosis.
In this study we investigated and demonstrated the contribution of sortase A to orally acquired listeriosis, using guinea pigs and hEcad mice in which L. monocytogenes can efficiently cross the intestinal barrier. The invasion protein InlA was the first LPXTG protein reported to be required for efficient L. monocytogenes invasion of the intestinal epithelium (40). We show here that a ΔsrtA strain is significantly more attenuated in virulence than a ΔinlA strain after oral infection in guinea pigs or hEcad mice, confirming the critical role of SrtA in bacterial invasion and/or persistence in deeper organs following oral infection (6). Therefore, it is likely that SrtA substrates other than InlA participate in different steps of the infectious process, from the crossing of the intestinal barrier to the hepatic phase of infection.
The second sortase of L. monocytogenes, SrtB, which does not play any detectable role in virulence in the mouse (5), is not required for L. monocytogenes oral infection in guinea pigs. This suggests that the two SrtB substrates (5, 54) do not play major roles in food-borne listeriosis. In Staphylococcus aureus srtB inactivation moderately attenuates virulence, and detection of this effect requires long-term infection (32, 49).
Identification of a new virulence factor among internalin-LPXTG proteins.
Consistant with the srtA attenuation phenotype, we identified a new virulence factor among the 41 LPXTG proteins encoded by L. monocytogenes EGDe (8, 28), the InlJ protein. Recent work in our laboratory has identified another LPXTG protein involved in orally acquired listeriosis, VIP, encoded by the lmo0320 gene (10). InlJ, in contrast to VIP, belongs to the internalin multigene family, which encompasses 25 genes in EGDe (8, 28). Internalins are characterized by the presence of an N-terminal LRR domain. Nineteen of these proteins, including InlA, have the LPXTG motif that is expected to anchor the protein to the peptidoglycan. One, InlB, is loosely attached to lipoteichoic acids at the bacterial surface. The last five, including InlC (13, 19), do not display any surface-targeting domain and are therefore expected to be secreted into the extracellular medium. InlA, InlH, InlB, and InlC are established virulence factors (18, 35, 58). Thus, InlJ extends the list of internalins involved in virulence.
inlJ is present only in L. monocytogenes among the Listeria species, as shown by PCR analysis of a small pool of virulent and avirulent strains (43) and confirmed on a larger scale by the use of DNA arrays (14). The inlJ mutant displays a virulence defect in both wild-type mice after intravenous infection and hEcad mice after oral infection, suggesting that it may be required at successive steps of listeriosis. Interestingly, ΔinlJ exhibits a 10-fold defect in intestinal colonization of hEcad mice, whereas ΔinlA has at least a 100-fold defect, which is at the limit of detection of L. monocytogenes in this tissue. In contrast, both mutants are similarly attenuated in the liver. InlA is probably the limiting factor among LPXTG proteins for efficient crossing of the intestinal epithelium by bacteria in hEcad mice, as it promotes Listeria internalization into enterocytes (40).
No phenotype could be attributed to the ΔinlJ strain in the cell culture system in vitro, making the function of inlJ elusive. As observed for inlJ, deletions of some other internalin genes, such as inlC or inlGHE, decrease the virulence in a mouse model but do not affect entry into epithelial cells, intracellular multiplication, or cell-to-cell spread (13, 15, 55). The various internalins may cooperate for efficient cell invasion, as proposed previously (3), or may have a totally different role in pathogenicity, such as interaction with the immune system.
Transcription of the inlJ gene is detectable by RT-PCR, similar to the inlA gene in bacteria grown in BHI medium at 37°C at the growth phase used for infection. However, we do not know yet at what level the encoded protein is produced. Using a highly sensitive gel-less method, Calvo et al. recently identified a pool of polypeptides linked to the L. monocytogenes EGDe peptidoglycan from bacteria grown in the same conditions (11). It is interesting that the InlJ protein was not detected in that pool, suggesting that its abundance at the bacterial surface is low. Of the 19 internalin-LPXTG proteins, only 5 were detected in cell wall extracts by this method (InlA, InlG, InlH, Lmo0327, and Lmo0610). This raises the possibility that InlJ synthesis, exportation, or anchoring at the bacterial surface might be low when bacteria are grown in BHI medium at 37°C and used to infect cultured cells. Interestingly, inlJ is adjacent to a gene encoding a putative transcriptional regulator, Lmo2820. Work is in progress to investigate where and when the InlJ protein is expressed and to address its possible regulation by the Lmo2820 protein.
InlJ defines a new subclass family of cysteine-containing LRR proteins.
Sequence comparisons for the large group of LRR proteins suggest that there are several different subfamilies of LRR, which are characterized by different lengths and consensus sequences (33, 36). An LRR in internalins typically contains 22 residues. The LRR domain forms a curved solenoid with conserved leucines and isoleucines forming the hydrophobic core, as described for InlB, InlH, and InlA (44, 47, 58, 59). The InlJ consensus type described here is unusual in that it comprises 21 residues, possibly shortening the 310-helix, and displays a conserved cysteine at position 7 in place of a leucine (Fig. 2C). Cysteines are presumably part of the hydrophobic core of the molecule and may be protected from oxidation and therefore unlikely to form disulfide bonds. Only one cysteine in repeat 12 is not in the conserved position and could be accessible to an external ligand. Other internalin-like proteins containing LRRs of the InlJ type are found in Listeria species and in proteins from only five other bacterial species (Fig. 2C). Interestingly, all of them interact with mammalian hosts and are either commensals or pathogens.
A wide range of functions have been ascribed to LRR proteins, and these functions are related to the ability to bind structurally unrelated protein ligands (34, 36). This is exemplified by the specific interaction of the LRRs of InlA and InlB with at least two different cellular receptors, E-cadherin (39, 51, 59) and the hepatocyte growth factor receptor (60), respectively. In prokaryotes, several LRR proteins of pathogenic bacteria, such as secreted internalins in Listeria ivanovii (20), YopM in Yersinia (41, 50), IpaH in Shigella (22, 62), SspH in Salmonella (52), and Slr in Streptococcus (56), are known to play roles in host-pathogen interactions. LRRs in microorganisms may have been selected during evolution as a consequence of their structural similarities with mammalian LRR proteins, especially those involved in recognition of bacterial pathogens, such as TLR and NOD proteins (2, 29). It will be very interesting to identify the eukaryotic binding partner of InlJ.
Another putative structural feature of InlJ is the presence of MucBP domains, which are repeated four times in the C-terminal part of the protein (Fig. 2A). Identical motifs are present in InlI, ORF29, and ORF2568 and in several proteins bound to peptidoglycan of gram-positive bacteria (accession number PF06458). The function of these domains has not been elucidated.
Inactivation of the ORF2568 gene in L. monocytogenes serovar 4b increases virulence.
Inactivation of two genes, ORF29 and ORF2568, encoding LPXTG proteins present in a subset of L. monocytogenes serovars, including epidemic serovar 4b, does not alter the listerial infectious process in vitro and does not attenuate Listeria virulence following oral infection. However, the bacterial loads of the ORF2568 deletion mutant in hEcad mice organs following oral inoculation, especially spleens, were increased compared to the loads of the wild-type serovar 4b strain. Inactivation of this gene may somehow affect the expression or function of other virulence factors and enhance the bacterial fitness in organs. Future work will address these possibilities.
In conclusion, this study showed the power of the postgenomic approach for successful identification of new virulence determinants. However, this candidate-based approach has its limitations. In contrast to InlA and InlB, whose role in Listeria-induced phagocytosis was identified following screening for noninvasive mutants in tissue-cultured cells (23), the InlJ function has not been determined. The challenge for the future is to identify the eukaryotic binding ligand and the signaling pathways triggered by the interaction with InlJ. In addition, in this study we could not identify any role for inlI or the serovar 4b LPXTG ORF29 gene in vitro or in virulence assays. These genes may not be required for pathogenesis but may function in another host or in specific infection conditions. Finally, the presence of multiple surface LPXTG proteins suggests that some of them may be important for virulence, while others may play a role in survival in food or other environments, which is critical for contamination.
Acknowledgments
We thank M. Doumith and C. Buchrieser for sharing unpublished results, M. Débarbouillé for the gift of plasmid pMAD, D. Heinz and W. D. Schubert for helpful discussions on the LRR structure, and M. Hamon for critical reading of the manuscript.
Work in P. Cossart's laboratory received financial support from the Pasteur Institute (GPH N°9), INRA, INSERM, Ministère de l"Agriculture et de la Pêche (DGAL no. A 03/02), and the Ministère de l'Education Nationale, de la Recherche et de la Technologie (ACI Microbiology Mic0312). P.C. is an international research scholar of Howard Hughes Medical Institute. H.B. is on the INRA staff.
Editor: D. L. Burns
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, J. K., G. E. Mullen, C. A. Leifer, A. Mazzoni, D. R. Davies, and D. M. Segal. 2003. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 24:528-533. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, B., D. Raffelsbauer, M. Kuhn, M. Goetz, S. Hom, and W. Goebel. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol. Microbiol. 43:557-570. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, C., J. Senderek, F. Kupper, F. Schneider, C. Dornia, E. Windelen, T. Eggermann, S. Rudnik-Schoneborn, J. Kirfel, L. Furu, L. F. Onuchic, S. Rossetti, P. C. Harris, S. Somlo, L. Guay-Woodford, G. G. Germino, M. Moser, R. Buttner, and K. Zerres. 2004. PKHD1 mutations in autosomal recessive polycystic kidney disease (ARPKD). Hum. Mutat. 23:453-463. [DOI] [PubMed] [Google Scholar]
- 5.Bierne, H., C. Garandeau, M. G. Pucciarelli, C. Sabet, S. Newton, F. Garcia-del Portillo, P. Cossart, and A. Charbit. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. Garcia-del Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 7.Braun, L., H. Ohayon, and P. Cossart. 1998. The InIB protein of Listeria monocytogenes is sufficient to promote entry into mammalian cells. Mol. Microbiol. 27:1077-1087. [DOI] [PubMed] [Google Scholar]
- 8.Cabanes, D., P. Dehoux, O. Dussurget, L. Frangeul, and P. Cossart. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238-245. [DOI] [PubMed] [Google Scholar]
- 9.Cabanes, D., O. Dussurget, P. Dehoux, and P. Cossart. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51:1601-1614. [DOI] [PubMed] [Google Scholar]
- 10.Cabanes, D., S. Sousa, A. Cebria, M. Lecuit, F. Garcia-Del Portillo, and P. Cossart. 2005. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J. 24:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo, E., M. G. Pucciarelli, H. Bierne, P. Cossart, J. P. Albar, and F. Garcia-Del Portillo. 2005. Analysis of the Listeria cell wall proteome by two-dimensional nanoliquid chromatography coupled to mass spectrometry. Proteomics 5:433-443. [DOI] [PubMed] [Google Scholar]
- 12.Dhar, G., K. F. Faull, and O. Schneewind. 2000. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry 39:3725-3733. [DOI] [PubMed] [Google Scholar]
- 13.Domann, E., S. Zechel, A. Lingnau, T. Hain, A. Darji, T. Nichterlein, J. Wehland, and T. Chakraborty. 1997. Identification and characterization of a novel PrfA-regulated gene in Listeria monocytogenes whose product, IrpA, is highly homologous to internalin proteins, which contain leucine-rich repeats. Infect. Immun. 65:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doumith, M., C. Cazalet, N. Simoes, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dramsi, S., P. Dehoux, M. Lebrun, P. L. Goossens, and P. Cossart. 1997. Identification of four new members of the internalin multigene family of Listeria monocytogenes EGD. Infect. Immun. 65:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dramsi, S. P. Trieu-Cuot, and H. Bierne. 2005. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156:289-297. [DOI] [PubMed] [Google Scholar]
- 17.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchrieser, P. Glaser, and P. Cossart. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 18.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 19.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 20.Engelbrecht, F., C. Dickneite, R. Lampidis, M. Gotz, U. DasGupta, and W. Goebel. 1998. Sequence comparison of the chromosomal regions encompassing the internalin C genes (inlC) of Listeria monocytogenes and L. ivanovii. Mol. Gen. Genet. 257:186-197. [DOI] [PubMed] [Google Scholar]
- 21.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Prada, C. M., D. L. Hoover, B. D. Tall, A. B. Hartman, J. Kopelowitz, and M. M. Venkatesan. 2000. Shigella flexneri IpaH(7.8) facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun. 68:3608-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 24.Gaillard, J. L., P. Berche, J. Mounier, S. Richard, and P. Sansonetti. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaillard, J. L., S. Dramsi, P. Berche, and P. Cossart. 1994. Molecular cloning and expression of internalin in Listeria. Methods Enzymol. 236:551-565. [DOI] [PubMed] [Google Scholar]
- 26.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garandeau, C., H. Reglier-Poupet, I. Dubail, J. L. Beretti, P. Berche, and A. Charbit. 2002. The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect. Immun. 70:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 29.Inohara, N., M. Chamaillard, C. McDonald, and G. Nunez. 2005. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74:355-383. [DOI] [PubMed] [Google Scholar]
- 30.Jacquet, C., M. Doumith, J. I. Gordon, P. M. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 31.Jacquet, C., E. Gouin, D. Jeannel, P. Cossart, and J. Rocourt. 2002. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 68:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonsson, I. M., S. K. Mazmanian, O. Schneewind, T. Bremell, and A. Tarkowski. 2003. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microbes Infect. 5:775-780. [DOI] [PubMed] [Google Scholar]
- 33.Kajava, A. V. 1998. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 277:519-527. [DOI] [PubMed] [Google Scholar]
- 34.Kedzierski, L., J. Montgomery, J. Curtis, and E. Handman. 2004. Leucine-rich repeats in host-pathogen interactions. Arch. Immunol. Ther. Exp. 52:104-112. [PubMed] [Google Scholar]
- 35.Khelef, N., M. Lecuit, C. Buchrieser, D. Cabanes, O. Dussurget, and P. Cossart. 2005. Listeria monocytogenes and the genus Listeria, 3rd ed. Springer, New York, N.Y.
- 36.Kobe, B., and A. V. Kajava. 2001. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11:725-732. [DOI] [PubMed] [Google Scholar]
- 37.Lecuit, M., and P. Cossart. 2002. Genetically-modified-animal models for human infections: the Listeria paradigm. Trends Mol. Med. 8:537-542. [DOI] [PubMed] [Google Scholar]
- 38.Lecuit, M., D. M. Nelson, S. D. Smith, H. Khun, M. Huerre, M. C. Vacher-Lavenu, J. I. Gordon, and P. Cossart. 2004. Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: role of internalin interaction with trophoblast E-cadherin. Proc. Natl. Acad. Sci. USA 101:6152-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lecuit, M., H. Ohayon, L. Braun, J. Mengaud, and P. Cossart. 1997. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 65:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lecuit, M., S. Vandormael-Pournin, J. Lefort, M. Huerre, P. Gounon, C. Dupuy, C. Babinet, and P. Cossart. 2001. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292:1722-1725. [DOI] [PubMed] [Google Scholar]
- 41.Leung, K. Y., and S. C. Straley. 1989. The yopM gene of Yersinia pestis encodes a released protein having homology with the human platelet surface protein GPIb alpha. J. Bacteriol. 171:4623-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, D., A. J. Ainsworth, F. W. Austin, and M. L. Lawrence. 2003. Characterization of virulent and avirulent Listeria monocytogenes strains by PCR amplification of putative transcriptional regulator and internalin genes. J. Med. Microbiol. 52:1065-1070. [DOI] [PubMed] [Google Scholar]
- 44.Machner, M. P., S. Frese, W. D. Schubert, V. Orian-Rousseau, E. Gherardi, J. Wehland, H. H. Niemann, and D. W. Heinz. 2003. Aromatic amino acids at the surface of InlB are essential for host cell invasion by Listeria monocytogenes. Mol. Microbiol. 48:1525-1536. [DOI] [PubMed] [Google Scholar]
- 45.Mackaness, G. B. 1962. Cellular resistance to infection. J. Exp. Med. 116:381-406. [PubMed] [Google Scholar]
- 46.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 2000. A framework for interpreting the leucine-rich repeats of the Listeria internalins. Proc. Natl. Acad. Sci. USA 97:8784-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marino, M., L. Braun, P. Cossart, and P. Ghosh. 1999. Structure of the LnlB leucine-rich repeats, a domain that triggers host cell invasion by the bacterial pathogen L. monocytogenes. Mol. Cell 4:1063-1072. [DOI] [PubMed] [Google Scholar]
- 48.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 49.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDonald, C., P. O. Vacratsis, J. B. Bliska, and J. E. Dixon. 2003. The Yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J. Biol. Chem. 278:18514-18523. [DOI] [PubMed] [Google Scholar]
- 51.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 52.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 53.Nelson, K. E., D. E. Fouts, E. F. Mongodin, J. Ravel, R. T. DeBoy, J. F. Kolonay, D. A. Rasko, S. V. Angiuoli, S. R. Gill, I. T. Paulsen, J. Peterson, O. White, W. C. Nelson, W. Nierman, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, D. H. Haft, J. Selengut, S. Van Aken, H. Khouri, N. Fedorova, H. Forberger, B. Tran, S. Kathariou, L. D. Wonderling, G. A. Uhlich, D. O. Bayles, J. B. Luchansky, and C. M. Fraser. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 32:2386-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pucciarelli, M. G. C., E. Sabet, C. Bierne, H. Cossart, and P. F. Garcia del Portillo. Identification of substrates of the Listeria monocytogenes sortases A and B by a non-gel proteomics analysis. Proteomics, in press. [DOI] [PubMed]
- 55.Raffelsbauer, D., A. Bubert, F. Engelbrecht, J. Scheinpflug, A. Simm, J. Hess, S. H. Kaufmann, and W. Goebel. 1998. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol. Gen. Genet. 260:144-158. [DOI] [PubMed] [Google Scholar]
- 56.Reid, S. D., A. G. Montgomery, J. M. Voyich, F. R. DeLeo, B. Lei, R. M. Ireland, N. M. Green, M. Liu, S. Lukomski, and J. M. Musser. 2003. Characterization of an extracellular virulence factor made by group A Streptococcus with homology to the Listeria monocytogenes internalin family of proteins. Infect. Immun. 71:7043-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlech, W. F., 3rd, P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 58.Schubert, W. D., G. Gobel, M. Diepholz, A. Darji, D. Kloer, T. Hain, T. Chakraborty, J. Wehland, E. Domann, and D. W. Heinz. 2001. Internalins from the human pathogen Listeria monocytogenes combine three distinct folds into a contiguous internalin domain. J. Mol. Biol. 312:783-794. [DOI] [PubMed] [Google Scholar]
- 59.Schubert, W. D., C. Urbanke, T. Ziehm, V. Beier, M. P. Machner, E. Domann, J. Wehland, T. Chakraborty, and D. W. Heinz. 2002. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell 111:825-836. [DOI] [PubMed] [Google Scholar]
- 60.Shen, Y., M. Naujokas, M. Park, and K. Ireton. 2000. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501-510. [DOI] [PubMed] [Google Scholar]
- 61.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694:269-278. [DOI] [PubMed] [Google Scholar]
- 62.Toyotome, T., T. Suzuki, A. Kuwae, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, T. Toyofuku, M. Hori, and C. Sasakawa. 2001. Shigella protein IpaH(9.8) is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 276:32071-32079. [DOI] [PubMed] [Google Scholar]
- 63.World Health Organization. 2001. Programme, Proposed draft guidelines for the control of Listeria monocytogenes in foods. Report of the 34th session of the Codex Committee on Food Hygiene, Bangkok, Thailand, October 2001.