Abstract
Dermatophytes are adapted to infect keratinized tissues by their ability to utilize keratin as a nutrient source. Although there have been numerous reports that dermatophytes like Trichophyton sp. secrete proteolytic enzymes, virtually nothing is known about the patterns of gene expression in the host or even when the organisms are cultured on protein substrates in the absence of a host. We characterized the expression of an aminopeptidase gene, the Trichophyton mentagrophytes homolog of the Trichophyton rubrum Tri r 4 gene. The T. rubrum gene was originally isolated based on the ability of the protein encoded by it to induce immediate and delayed-type hypersensitivity in skin tests. T. mentagrophytes Tri m 4 is closely related to Tri r 4 (almost 94% identity at the protein level). Tri m 4 resembles other protease-encoding genes thought to be virulence factors (for example, DPP V of Aspergillus fumigatus). The Tri m 4 protein was detected immunochemically both in fungal extracts and in the culture medium. Expression of the Tri m 4 gene was induced severalfold when T. mentagrophytes was grown on keratin and elastin. Ex vivo, strong induction was observed after culture on blood plasma, but the use of homogenized skin did not result in a significant increase in Tri m 4 transcript levels. In order to identify additional genes encoding putative virulence factors, differential cDNA screening was performed. By this method, a fungal thioredoxin and a cellulase homolog were identified, and both genes were found to be strongly induced by skin extracellular matrix proteins. Induction by superficial (keratin) and deep (elastin) skin elements suggests that the products of these genes may be important in both superficial and deep dermatophytosis, and models for their function are proposed. Upregulation of several newly identified T. mentagrophytes genes on protein substrates suggests that these genes encode proteins which are relevant to the dermatophyte-skin interaction.
Several opportunistic fungal pathogens of humans have received attention in recent years, including the ascomycetes Candida albicans and Aspergillus fumigatus and the basidiomycete Cryptococcus neoformans. In contrast to these organisms, dermatophyte fungi of the genus Trichophyton infect healthy individuals, causing infections of keratinized structures, including the skin, hair, and nails (20). Dermatophytoses are among the few fungal diseases that are directly communicable from person to person. Trichophyton rubrum and Trichophyton mentagrophytes are distributed worldwide and are the most common cause of athlete's foot and nail infections. Dermatophytes colonize the outermost skin layers (stratum corneum) and can penetrate deeper only if the host is immunocompromised (18). Although not normally life threatening, dermatophyte infections are often difficult to eliminate completely. Despite its importance, little is known about the molecular basis of dermatophyte pathogenesis. This is in marked contrast to C. neoformans, for example, whose sequence has recently been completed (14) and was the subject of a detailed mechanistic investigation (9). Likewise, A. fumigatus has been the subject of much molecular work, yet it is not such a widespread pathogen of healthy individuals.
In host-pathogen interactions, the gene expression of the pathogen is modulated by signals from the host, and understanding the expression patterns may provide insight into the mechanisms of disease. Little or no information is available on the transcription patterns of dermatophyte fungi, in contrast to other fungal pathogens, such as Candida, A. fumigatus, C. neoformans, and pathogens of plants (8, 19, 23-25). In Candida albicans, for instance, activation of individual SAP (secreted aspartyl proteinase) genes depends on the progress of the infection; some members of the gene family are induced immediately after contact with the host, whereas others are expressed only after dissemination into deep organs (8, 23). C. neoformans cells isolated from rabbit cerebrospinal fluid exhibited expression of fungal genes belonging to several functional classes. The high level of expression of these genes suggested that they might be required for C. neoformans to survive and proliferate in the host environment; temporal, host-specific signals were postulated to regulate these transcriptional patterns (24). A. fumigatus is an important opportunistic fungal pathogen that can cause acute invasive disease in neutropenic hosts. In this species, two genes encoding the regulatory subunit of cyclic AMP-dependent protein kinase were identified (12); furthermore, a member of the RAS family provided a new approach for finding virulence-related genes induced by the interaction with host cells (19).
One route to the identification of fungal genes associated with disease is the study of fungus-derived antigens. Chronic dermatophytosis has been associated with atopic disease (30). Furthermore, dermatophyte antigens seem to contribute to allergic disease, and indeed asthma symptoms in dermatophyte-sensitized patients were improved after antifungal treatment (28, 29). The genes encoding two antigens derived from T. rubrum were cloned and characterized previously (32). One of these genes, Tri r 2, encodes a 29-kDa protein which is a member of the subtilisin protease family. The protein encoded by the other gene, a 83-kDa antigen encoded by Tri r 4, has limited sequence identity to the prolyl oligopeptidase family of serine proteinases. In A. fumigatus the closest homolog of Tri r 4 is designated DPP V (3), and the T. rubrum homolog encodes a secreted aminopeptidase (15). The expression of these genes might be regulated by the host and host-derived molecules. A second approach to identify such genes is to screen for fungal transcripts that are regulated by growth on skin extracellular matrix proteins or skin explants (5). Although dermatophytes do not normally penetrate beyond the epidermis, deeper penetration and systemic infections can occur in immunocompromised hosts (17). This suggests that dermatophytes might have evolved the ability to detect other host proteins, including those in plasma, so we also tested whether growth on plasma could induce our candidate genes. In this study, using both approaches, we identified several genes that have potential as molecular markers for host-induced gene expression in dermatophytosis.
MATERIALS AND METHODS
Organism and culture conditions.
A clinical isolate of T. mentagrophytes var. mentagrophytes was maintained on Sabouraud dextrose agar (Difco) containing 0.05 mg/liter chloramphenicol at 30°C for 14 to 21 days. Fungal microconidia were isolated as described previously (5) and incubated on mineral medium (MM) (11) with 0.5% (wt/vol) keratin (Sigma) or 0.25% elastin (Sigma) to study gene expression on these proteins as sole substrates. Minimal medium, referred to as “conditional” medium (CM) [glucose, 2.5 g liter−1; (NH4)2HPO4, 6.6 g liter−1; KH2PO4, 0.46 g liter−1; K2HPO4 · 3H2O, 1.3 g liter−1; MgSO4 · 7H2O, 0.49 g liter−1], was used for comparisons between induced and noninduced cultures. To obtain a homogenized human skin suspension, 3 to 5 mg of human skin sections obtained as a by-product from plastic surgery (5) was added to 100 ml of MM or CM and homogenized three to five times. Microconidia were incubated at 30°C with the homogenized skin suspension for 21 to 28 days or with human plasma type AB for 14 days.
Nucleic acid isolation and analysis.
Crude genomic DNA for PCR analysis was isolated from mycelium ground in liquid nitrogen as described previously for plant tissue (6). For isolation of total RNA, mycelial samples were ground in liquid nitrogen and extracted with Tri reagent (Molecular Research Center). Total RNA from keratin- or elastin-induced and noninduced mycelia was electrophoresed and transferred to a Hybond membrane (Amersham).
Construction and verification of the subtracted library.
Poly(A) mRNA was isolated from total RNA of keratin-induced fungal mycelium (tester population) and noninduced fungal mycelium (driver population) using the PolyATtract mRNA isolation system (Promega), and a suppression subtractive library was prepared according to the manufacturer's instructions (PCR-Select cDNA subtraction kit; Clontech). Clones were ligated into the vector pGEM-T Easy (Promega) and transformed into Escherichia coli XL-1 Blue (Stratagene). For colony hybridization (on Biotrace NT filters [Gelman Sciences]), labeling of probes by random priming with [α-32P]dCTP (Amersham), and other nucleic acid techniques we used standard procedures (2, 21). Plasmid DNA for sequencing was prepared with a Wizard Plus SV Minipreps kit (Promega). Probes for hybridization of Northern blots were prepared by PCR amplification (standard T7 and SP6 primers) from the plasmid clones, gel purified, and labeled.
PCR and RT-PCR analysis.
PCRs were performed with BioTaq DNA polymerase (Bioline). For amplification prior to direct sequencing of PCR products, we used the high-fidelity BIO-X-ACT polymerase (Bioline). Total RNA was treated with RNase-free DNase (QIAGEN) and amplified with the Access reverse transcription (RT)-PCR system (Promega). Isolation of a fragment of the Tri m 4 gene was accomplished with primers M4#4452 (5′-CGTCAGGGCTTGCATTCAC-3′) and M4#3544 (5′-CTTGAGGCCAGAGAACGCTC-3′), and isolation of a fragment of the Tri m 2 gene was accomplished with primers M2#7990 (5′-CAGGGCCTACTCTGGCGAG-3′) and M2#2897 (5′-GTTACCAGCAGCGACAGC-3′). The following primers were used for RT-PCR to complete the 3′ portion of the cDNA sequence: M4#7388 (5′-CTTCCTCCAGCCCAGTCTTCTC-3′) and M4#9477 (5-CGAAGTAAGAGTGAGCCTCATGGTC-3′).
PCR-based genome walking.
Libraries were constructed from the genomic DNA of T. mentagrophytes with a Universal Genome Walker kit (Clontech). PCR-based genome walking with this kit was used to obtain sequences upstream and downstream of the coding region of Tri m 4. The PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced (Hylabs, Rehovot, Israel).
The following primers were used for genome walking: M4-5′-GSP1 (5′-GAATGCAAGCCCTGACGAGGCAAACG-3′), M4-5′-GSP2 (5′-CCAAGGAGGCAATCAACCATTTGGCT-3′), M4-3′-GSP1 (5′-CAGCAGGTCGGATGGATCAACAAG-3′), and M4-3′-GSP2 (5′-TGACCATGAGGCTCACTCTTACTTCG-3′). Extension was continued from the ends of the PCR products with the following primers: M4-5-WALK2 (5′-CTGTTCTCCGTACTCAAACTCAGG-3′), TM4-3-WALK2 (5′-CCAGTATACCAATGGACTAGATTTGC-3′), TM4-5-WALK3 (5′-CAAGTGTTGGTGCGGGTGCCT-3′), and TM4-3-WALK3 (5′-CCAATGTATCGAAAGGGCCTGG-3′).
Real-time PCR detection.
Total RNA (3 μg) was treated with RNase-free DNase (QIAGEN) for 15 min at 25°C, followed by 15 min at 65°C. First-strand cDNA was synthesized using the ImProm-II reverse transcription system (Promega) according to the manufacturer's protocol. Quantitative PCR for Tri m 4 induction in the presence of keratin, elastin, and homogenized skin was carried out by performing real-time PCR. For Tri m 4, the following primers were synthesized: M4#1804F (5′-CACGACTTCAACGGAACCTTCT-3′) and M4#1859R (5′-CAATCCCAGCGGTCATAGTTCT-3′). The “housekeeping” gene was the actin gene, and the primers used were actin#219F (5′-CGAGCGTGGCTACAGCTTCT-3′) and actin#279R (5′-CTCCTTGATGTCACGGACGAT-3′). The reaction mixtures contained 4 μl of cDNA, 10 μl of SYBR green ROX mixture (ABGene, Surrey, United Kingdom), and 1 pmol of each primer, and the volume was brought to 20 μl with nuclease-free water. The reaction protocol was 50°C for 2 min, 95°C for 15 min, and 40 cycles of 95°C for 15 s and annealing and synthesis at 60°C for 1 min. This was followed by a dissociation protocol, and the dissociation curves verified that the signal corresponded to a single PCR product. Reactions were performed in duplicate with an Applied Biosystems 7000 cycler. Data were analyzed using the 2−ΔΔCT method (13). The cycle threshold values for the duplicate PCRs for each RNA sample were averaged, and then 2−ΔΔCT values were calculated (actin was used as the reference); this was followed by normalization to the value for RNA samples from CM.
Protein extraction, purification, and analysis.
For extraction of cellular protein, mycelia were ground in liquid nitrogen, and about 0.5 mg of ground mycelia was transferred into 50 μl of Laemmli buffer (2× concentrated) and 50 μl of water. After vortexing, the suspension was centrifuged at 10,000 rpm for 5 min, and the supernatant was heated at 95°C for 5 min and used for protein analysis. Thirty microliters of each sample was run on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (10% polyacrylamide running gel and 4% polyacrylamide stacking gel) and electrotransferred onto a Hybond ECL nitrocellulose membrane (Amersham Biosciences). The membrane was blocked and incubated with the first antibody against protein V (a homolog of Tri r 4) (31) (diluted 1:1,000), and after washing it was incubated with the rabbit anti-mouse second antibody labeled with horseradish peroxidase (diluted 1:25,000; ECL Plus Western blotting reagents; Amersham Biosciences). The membrane was incubated with developing reagent (ECL Plus reagent; Amersham Biosciences) and exposed to Hyperfilm ECL (Amersham Biosciences) for 1 to 30 min. The gel was stained with Page Blue staining solution (Fermentas) for 1 h and then washed with water. To obtain secreted proteins, the medium was filtered through a 0.22-μm filter (Stericup; Millipore) and then concentrated by ultrafiltration at 2,000 × g through a Centriprep YM-10 (Millipore) filter with a cutoff of 10,000 Da. Protein concentrations were measured with the Bradford reagent (Sigma) with bovine serum albumin as the standard. Forty nanograms of protein per lane was separated by electrophoresis and probed with antibody as described above for the cellular proteins. The gel was stained according to the instructions for the silver staining kit (Amersham Biosciences).
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in the GenBank database under accession number AY929335.
RESULTS
T. mentagrophytes DPP V homolog.
Proteases are strong candidates for host- or substrate-regulated genes. An initial search for subtilisin family members in T. mentagrophytes was carried out by PCRs with degenerate primers designed on the basis of subtilisin conserved active site regions. One of the products obtained from this search was a homolog of Tri r 2 (32). We also isolated the T. mentagrophytes homolog of the second dermatophyte antigen, Tri r 4, using primers designed from the database sequences (GenBank accession numbers AFO82514 and AJ430836). The predicted coding sequences of T. rubrum and T. mentagrophytes are 95% identical at the nucleotide level; the four introns are at the same locations in the two species (32). Five kilobases of sequence (Fig. 1B), including the complete coding sequence and upstream and downstream regions, has been deposited in the GenBank database (see above). A BLAST (1) search of the translated databases with the predicted Tri m 4 protein sequence revealed that this gene is conserved in a large number of eukaryotic and prokaryotic organisms, many of which are pathogenic to plants or animals. The phylogenetic relationships between these DPP V-related genes are shown in Fig. 1A, and a map of the genomic sequence is shown in Fig. 1B.
FIG. 1.
T. mentagrophytes DPP V-related gene Tri m 4. (A) Phylogenetic relationships between DPP V-related proteins. The tree was constructed using CLUSTALW and was plotted using TREEVIEW. Hypothetical proteins are indicated by number signs. Bar = 0.1 nucleotide substitution/site. (B) Map of the Tri m 4 locus. The predicted start and stop codons are indicated, as are the four introns starting at bp 1627, 1745, 2044, and 3574. Intron positions were confirmed by sequencing of cDNA. PCR primers (for sequences, see Materials and Methods) are indicated by arrows.
We then asked whether expression of Tri m 4 is indeed regulated. Tri m 4 was expressed on MM (salts only; see Materials and Methods) with keratin, elastin, or skin as the sole carbon and nitrogen source (Fig. 2A). In CM, a synthetic minimal medium with glucose as a carbon source (see Materials and Methods), the presence of keratin resulted in strong induction compared with the level on glucose alone (Fig. 2A). Primers were designed to detect the transcripts of several other candidate genes, including the predicted protease Tri m 2 and Alp1 genes and the metalloprotease Mep2 and Mep3 genes (10). The primer pairs gave no signals in RT-PCR with 40 cycles of PCR amplification (data not shown) or signals that were at the limit of detection.
FIG.2.
Detection of transcripts of Tri m 4 and other candidate genes by RT-PCR. (A) Total RNAs from mycelia grown on different nutrient sources were used as templates in RT-PCRs with primers specific for Tri m 4, Tri m 2, or dermatophyte actin, as indicated above the lanes. Lanes DNA show the results for amplification with the same primers from the genomic DNA template. MM contained salts only and was supplemented with keratin or elastin. CM contained salts and glucose, with or without keratin. (B) Expression of Tri m 4 and other genes encoding proteolytic enzymes in T. mentagrophytes grown on blood plasma. RT-PCR was performed with primer pairs designed to detect the following transcripts (predicted sizes are indicated in parentheses): dipeptidyl peptidases encoded by Tri m 4 (362 bp); alkaline proteases encoded by Tri m2 (629 bp) and Alp1 (680 bp); and metalloproteases encoded by Mep2 (804 bp) and Mep3 (907 bp). (C) Expression of Tri m 4 and Tri m2 on homogenized skin explant tissue as a sole carbon and nitrogen source (upper panel). RT-PCR amplifications were performed starting with 1, 0.5, and 0.1 μg total RNA (decreasing amounts of template are indicated above each set). The photographs in the lower panel show growth of the dermatophyte (arrow in right photograph) on homogenized skin in MM (salts only).
For Tri m 2 weak bands consistent with the expected size (629 bp) were detected in some experiments (for example, the CM + keratin sample shown in Fig. 2A). The stronger band at 250 bp amplified by the Tri m 2 primers (Fig. 2A) was not predicted from the sequence and was apparently nonspecific. Using the same primer pairs, Tri m 4 gave a strong signal, and the metalloprotease Mep3 gene could also be clearly detected by RT-PCR from RNA isolated from cultures grown on blood plasma (Fig. 2B). Tri m 2, Alp1, and Mep2 expression, however, could not be detected under these conditions (Fig. 2B). On homogenized skin as the sole nutrient source, the dermatophyte grew slowly but was clearly visible on the surface of the skin fragments; Tri m 4 was clearly detectable (Fig. 2C), while only a faint Tri m 2 band was visible (Fig. 2C) following 40 cycles of PCR amplification.
Of the candidate protease-related genes considered (Fig. 2), Tri m 4 was the only gene that was expressed at sufficient levels and showed evidence of induction by protein substrates. This gene was therefore chosen for further quantitative analysis of transcript levels. Real-time quantitative RT-PCR amplifications were performed with the fungal actin gene as the reference gene. In CM with glucose as the carbon source, addition of keratin or elastin resulted in severalfold induction of Tri m 4 transcript levels compared to the levels on CM alone (Fig. 3). Addition of homogenized skin explant tissue to CM did not result in a significant increase compared with the control level (Fig. 3).
FIG. 3.
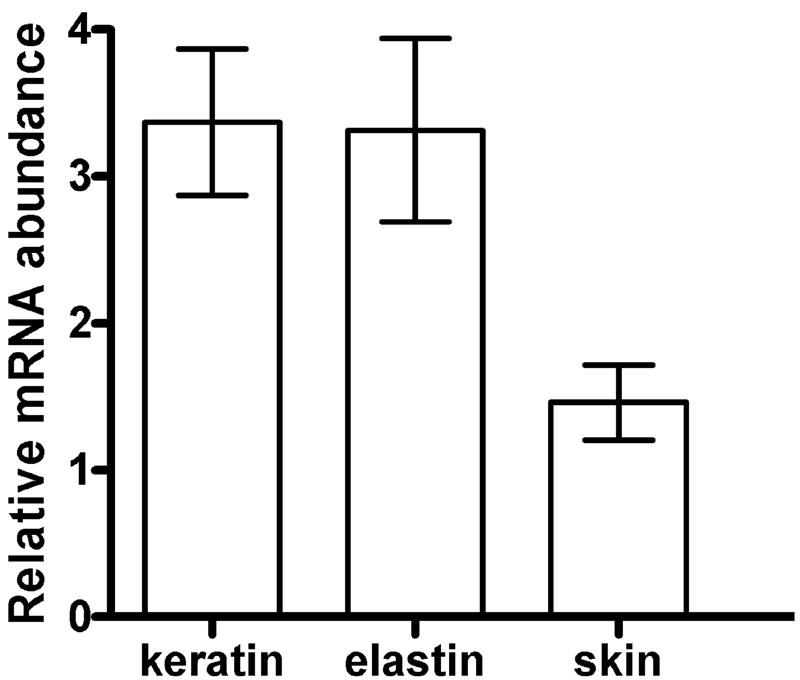
Detection of induction of Tri m 4 expression by real-time quantitative PCR amplification. RNA samples were used as templates for synthesis of cDNA, which was followed by amplification using Tri m 4-specific and fungal actin-specific primers. The signal is relative to actin and normalized to the signal on CM (indicated by a value of 1.0 on the y axis). The error bars indicate standard errors of the means of three independent experiments.
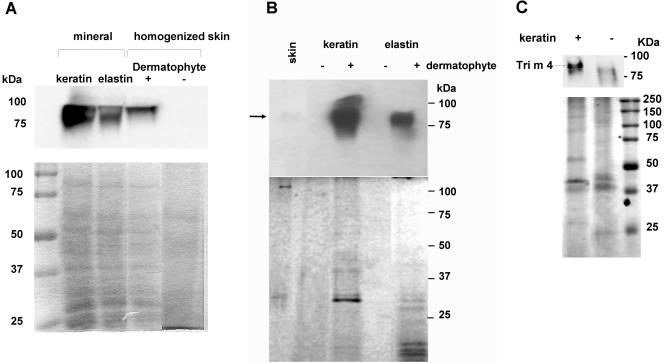
By using sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis a protein with a molecular mass consistent with the mass of a homolog of the Tri r 4-encoded protein was detected in extracts from mycelia grown on MM with keratin, elastin, or skin explant tissue (Fig. 4A), as well as from culture media under the same conditions (Fig. 4B). On CM, the fungus could be grown without a protein substrate (as shown in Fig. 3). In this case, Tri m 4 was clearly induced, at the protein level, by addition of keratin to the medium (Fig. 4C). Without keratin, some weaker cross-reacting bands were visible at lower molecular weights (Fig. 4C). The level of the Tri m 4 protein signal on keratin and elastin compared to that on skin reflected the pattern of the transcript levels (Fig. 2 to 4). The major regulation of the Tri m 4 gene thus appears to be at the level of transcription.
FIG. 4.
Immunochemical detection of the Tri m 4-encoded protein in intracellular (A) and extracellular (B and C) protein samples with monoclonal antibody to T. rubrum Tri r 4-encoded protein. The lower panels show Coomassie blue staining. In panel B, which shows extracellular protein from experiments on mineral medium, note that without the dermatophyte (lanes −), only some diffuse weak protein staining is visible (probably degraded protein substrates), while with the dermatophyte (lanes +) several distinct extracellular polypeptide bands are visible. In panel C induction by keratin on CM is shown. A plus sign indicates CM with keratin, while a minus sign indicates CM without keratin; in both cases, dermatophyte cultures were present.
Screening for differential gene expression.
Proteases like that encoded by Tri m 4 provided a logical starting point to identify regulated genes in dermatophytes, but little is known about the mechanisms of dermatophyte virulence. Thus, direct identification of relevant target genes for study is not possible. We therefore adopted the nonbiased differential cDNA screening approach, using the suppression-subtractive hybridization (SSH) technique. This PCR-based method enriches for sequences that are differentially expressed in two mRNA populations, referred to as “tester” and “driver.” Abundant transcripts, as well as transcripts present at similar levels in both populations, are selected against, while transcripts that are more abundant in the tester population than in the driver population are selected for. An SSH cDNA library was constructed for T. mentagrophytes grown on CM with keratin (tester mRNA population) and compared to the library obtained with CM alone (with glucose as the carbon source; driver mRNA population). A sample of 20 clones was initially sequenced. A majority of these clones corresponded to a fungal thioredoxin gene. We therefore screened the library for additional clones that did not correspond to this overrepresented clone. After colony hybridization with the thioredoxin probe, clones that did not hybridize were sequenced. A second relatively abundant clone exhibited high homology to a fungal cellobiohydrolase. Both thioredoxin and the cellobiohydrolase were differentially expressed, as shown by Northern analysis of total mycelial RNA (Fig. 5). Although differentially expressed (Fig. 2 and 3), Tri m 4 was not recovered from the initial SSH screening. This was perhaps not unexpected, considering (i) that the complexity of the library was low and (ii) that Tri m 4 was differentially expressed threefold (Fig. 3), while the abundant transcripts shown in Fig. 5 were more strongly induced (no transcript was detectable on CM alone [Fig. 5]).
FIG. 5.
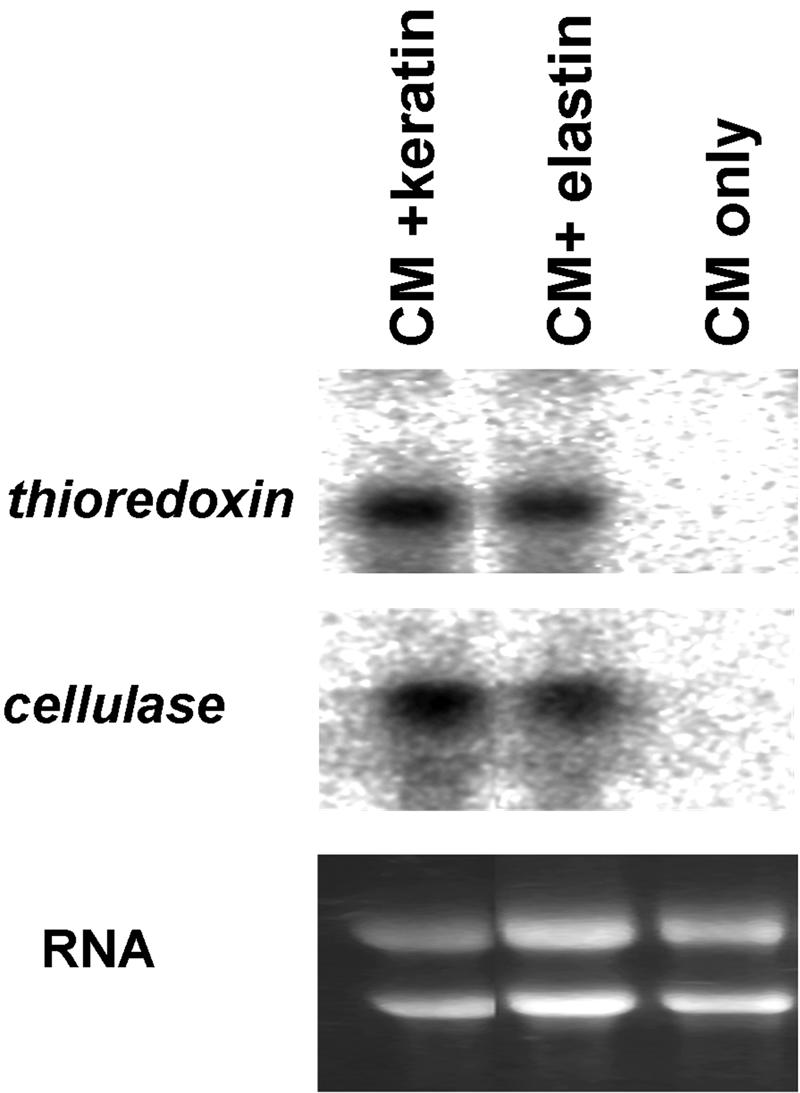
Expression of two differentially regulated genes on protein substrates. Northern blots of total mycelial RNA were hybridized with the probes indicated. The bottom panel shows ethidium bromide staining for equal loading.
DISCUSSION
The dipeptidyl peptidase class to which the Tri m 4-encoded protein belongs is conserved among a variety of fungal, bacterial, and protozoan species, suggesting that it has a general role. Some of the microorganisms are pathogenic, and the gene is upregulated while the organism is infecting the host. This is true for the tripeptidyl peptidase of Beauveria (Cordyceps) bassiana (26), DPP IV and V of A. fumigatus (3, 15), and proteases that are virulence factors in the bacterium Porphyromonas gingivalis (16), for example. The presence of related proteins in dermatophytes suggests that there is a similar invasive mechanism. The existence of members of the dipeptidyl peptidase family in nonpathogenic fungi, however, suggests that there are proteins that have roles unrelated to virulence. Genes for hydrolytic enzymes that could be virulence related are found in saprophyte genomes, however, perhaps reflecting the pathogenic potential of filamentous fungi in general.
The T. mentagrophytes DPP V homolog is very similar to Tri r 4 of T. rubrum (7), as expected since these species are very closely related phylogenetically (Fig. 1A). Like the T. rubrum homolog, the Tri m4-encoded protein is predicted to have a signal peptide for secretion, with an identical sequence. Indeed, the protein was detected on Western blots of total cellular protein, as well as in the culture medium (Fig. 4). The 3′ untranslated region (UTR) is very similar (90% identity), while in the 5′ UTR there is no obvious homology between T. rubrum and T. mentagrophytes; the reason for this is not yet clear, and this finding might suggest that there are different patterns of regulation in the two species. In both species the 3′ UTR includes what is apparently the 3′ end of a dynein gene, whose coding region is oriented in the direction opposite that of DPP V. Since the Tri m 4-encoded protein is not a very abundant secreted protein, it might have a role in detecting, rather than degrading, protein substrates. Keratin is encountered by the pathogen immediately upon adherence and in the early stages of growth, while elastin occurs in the dermis. Dermatophyte infections penetrate deeply in immunocompromised hosts (17, 22) and occasionally cause systemic infections (27). Profuse growth, as well as strong expression of Tri m 4, was observed on blood plasma, which is consistent with the view that dermatophytes have the potential to cause deep mycoses and are stopped from doing so only by the immune system. In contrast to growth on plasma, homogenized skin was not an efficient substrate for induction of Tri m 4 under the conditions tested.
Differential library screening was initiated to identify other regulated genes. Two candidate proteins with highly differential expression were identified (Fig. 5). One of these, a cellobiohydrolase, seems unlikely to have a direct role in pathogenicity on skin. The other, thioredoxin, could provide defense against oxidative stress. Another role for thioredoxin could be activation of proteases. It was recently reported, for example, that thioredoxin secreted following UV irradiation modulates activity of a matrix metalloproteinase in human dermal fibroblasts (4). Thioredoxin has also been reported to be a virulence factor in Helicobacter pylori and in the opportunistic fungal pathogen Pneumocystis carinii.
In conclusion, we examined, for the first time, expression and upregulation of potential pathogenicity genes in dermatophytes, using extracellular matrix substrates and tissues which are likely to be infected by T. mentagrophytes. In this study we identified three dermatophyte genes whose expression is regulated by protein substrates and can serve as molecular markers of growth and differentiation during infection.
Acknowledgments
We are grateful to Yehuda Ullman for providing the skin samples.
This work was supported in part by the Technion V.P.R. Fund.
Editor: T. R. Kozel
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kinston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1987. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
- 3.Beauvais, A., M. Monod, J. P. Debeaupuis, M. Diaquin, H. Kobayashi, and J. P. Latge. 1997. Biochemical and antigenic characterization of a new dipeptidyl-peptidase isolated from Aspergillus fumigatus. J. Biol. Chem. 272:6238-6244. [DOI] [PubMed] [Google Scholar]
- 4.Didier, C., I. Kerblat, C. Drouet, A. Favier, J. C. Beani, and M. J. Richard. 2001. Induction of thioredoxin by ultraviolet-A radiation prevents oxidative-mediated cell death in human skin fibroblasts. Free Radic. Biol. Med. 31:585-598. [DOI] [PubMed] [Google Scholar]
- 5.Duek, L., G. Kaufman, Y. Ulman, and I. Berdicevsky. 2004. The pathogenesis of dermatophyte infections in human skin sections. J. Infect. 48:175-180. [DOI] [PubMed] [Google Scholar]
- 6.Edwards, K., C. Johnstone, and C. Thompson. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19:1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, J., and A. Takashima. 2004. Cloning and characterization of Trichophyton rubrum genes encoding actin, Tri r2, and Tri r4. J. Clin. Microbiol. 42:3298-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hube, B., M. Monod, D. A. Schofield, A. J. Brown, and N. A. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 9.Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557-615. [DOI] [PubMed] [Google Scholar]
- 10.Jousson, O., B. Lechenne, O. Bontems, S. Capoccia, B. Mignon, J. Barblan, M. Quadroni, and M. Monod. 2004. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology 150:301-310. [DOI] [PubMed] [Google Scholar]
- 11.Kaufman, G., B. A. Horwitz, R. Hadar, Y. Ullmann, and I. Berdicevsky. 2004. Green fluorescent protein (GFP) as a vital marker for pathogenic development of the dermatophyte Trichophyton mentagrophytes. Microbiology 150:2785-2790. [DOI] [PubMed] [Google Scholar]
- 12.Liebmann, B., S. Gattung, B. Jahn, and A. A. Brakhage. 2003. cAMP signaling in Aspergillus fumigatus is involved in the regulation of the virulence gene pksP and in defense against killing by macrophages. Mol. Genet. Genomics 269:420-435. [DOI] [PubMed] [Google Scholar]
- 13.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 14.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monod, M., B. Lechenne, O. Jousson, D. Grand, C. Zaugg, R. Stocklin, and E. Grouzmann. 2005. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology 151:145-155. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nir-Paz, R., H. Elinav, G. Pierard, D. Walker, A. Maly, M. Shapiro, R. Barton, and I. Polacheck. 2003. Deep infection by Trichophyton rubrum in an immunocompromised patient. J. Clin. Microbiol. 41:5298-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa, H., R. C. Summerbell, K. V. Clemons, T. Koga, Y. P. Ran, A. Rashid, P. G. Sohnle, D. A. Stevens, and R. Tsuboi. 1998. Dermatophytes and host defence in cutaneous mycoses. Med. Mycol. 36(Suppl. 1):166-173. [PubMed] [Google Scholar]
- 19.Rhodes, J. C., B. G. Oliver, D. S. Askew, and T. W. Amlung. 2001. Identification of genes of Aspergillus fumigatus up-regulated during growth on endothelial cells. Med. Mycol. 39:253-260. [DOI] [PubMed] [Google Scholar]
- 20.Richardson, M., and S. Aljabre. 1993. Pathogenesis of dermatophytosis. Curr. Top. Med. Mycol. 5:49-77. [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Sommer, S., R. C. Barton, S. M. Wilkinson, W. J. Merchant, E. G. Evans, and M. K. Moore. 1999. Microbiological and molecular diagnosis of deep localized cutaneous infection with Trichophyton mentagrophytes. Br. J. Dermatol. 141:323-325. [DOI] [PubMed] [Google Scholar]
- 23.Staib, P., M. Kretschmar, T. Nichterlein, G. Kohler, S. Michel, H. Hof, J. Hacker, and J. Morschhauser. 1999. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol. Microbiol. 32:533-546. [DOI] [PubMed] [Google Scholar]
- 24.Steen, B. R., S. Zuyderduyn, D. L. Toffaletti, M. Marra, S. J. Jones, J. R. Perfect, and J. Kronstad. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straney, D., R. Khan, R. Tan, and S. Bagga. 2002. Host recognition by pathogenic fungi through plant flavonoids. Adv. Exp. Med. Biol. 505:9-22. [DOI] [PubMed] [Google Scholar]
- 26.Tartar, A., and D. G. Boucias. 2004. A pilot-scale expressed sequence tag analysis of Beauveria bassiana gene expression reveals a tripeptidyl peptidase that is differentially expressed in vivo. Mycopathologia 158:201-209. [DOI] [PubMed] [Google Scholar]
- 27.Tateishi, Y., H. Sato, M. Akiyama, M. Abe, H. Kobayashi, S. Umehara, J. Yamaguchi, H. Shibaki, and H. Shimizu. 2004. Severe generalized deep dermatophytosis due to Trichophyton rubrum (trichophytic granuloma) in a patient with atopic dermatitis. Arch. Dermatol. 140:624-625. [DOI] [PubMed] [Google Scholar]
- 28.Ward, G. W., Jr., G. Karlsson, G. Rose, and T. A. Platts-Mills. 1989. Trichophyton asthma: sensitisation of bronchi and upper airways to dermatophyte antigen. Lancet i:859-862. [DOI] [PubMed] [Google Scholar]
- 29.Ward, G. W., Jr., J. A. Woodfolk, M. L. Hayden, S. Jackson, and T. A. Platts-Mills. 1999. Treatment of late-onset asthma with fluconazole. J. Allergy Clin. Immunol. 104:541-546. [DOI] [PubMed] [Google Scholar]
- 30.Woodfolk, J. A., and T. A. Platts-Mills. 1998. The immune response to dermatophytes. Res. Immunol. 149:436-445. [DOI] [PubMed] [Google Scholar]
- 31.Woodfolk, J. A., J. B. Slunt, B. Deuell, M. L. Hayden, and T. A. Platts-Mills. 1996. Definition of a Trichophyton protein associated with delayed hypersensitivity in humans. Evidence for immediate (IgE and IgG4) and delayed hypersensitivity to a single protein. J. Immunol. 156:1695-1701. [PubMed] [Google Scholar]
- 32.Woodfolk, J. A., L. M. Wheatley, R. V. Piyasena, D. C. Benjamin, and T. A. Platts-Mills. 1998. Trichophyton antigens associated with IgE antibodies and delayed type hypersensitivity. Sequence homology to two families of serine proteinases. J. Biol. Chem. 273:29489-29496. [DOI] [PubMed] [Google Scholar]