Abstract
Borrelia hermsii is the primary cause of tick-borne relapsing fever in North America. When its tick vector, Ornithodoros hermsi, acquires these spirochetes from the blood of an infected mammal, the bacteria switch their outer surface from one of many bloodstream variable major proteins (Vmps) to a unique protein, Vtp (Vsp33). Vtp may be critical for successful tick transmission of B. hermsii; however, the gene encoding this protein has been described previously in only one isolate. Here we identified and sequenced the vtp gene in 31 isolates of B. hermsii collected over 40 years from localities throughout much of its known geographic distribution. Seven major Vtp types were found. Little or no sequence variation existed within types, but between them significant variation was observed, similar to the pattern of diversity described for the outer surface protein C (OspC) gene in Lyme disease spirochetes. The pattern of sequence relatedness among the Vtp types was incongruent in two branches compared to two genomic groups identified among the isolates by multilocus sequence typing of the 16S rRNA, flaB, gyrB, and glpQ genes. Therefore, both horizontal transfer and recombination within and between the two genomic groups were responsible for some of the variation observed in the vtp gene. O. hermsi ticks were capable of transmitting spirochetes in the newly identified genomic group. Therefore, given the longevity of the tick vector and persistent infection of spirochetes in ticks, these arthropods rather than mammals may be the likely host where the exchange of spirochetal DNA occurs.
Tick-borne relapsing fever associated with the vector Ornithodoros hermsi is endemic throughout the higher elevations of western North America (10, 29). This tick specifically transmits Borrelia hermsii but is incapable of transmitting other species of spirochetes associated with other species of ticks (24, 25). One early investigation demonstrated that O. hermsi transmitted B. hermsii by bite (39), and our study with immunofluorescence antibodies and microscopy demonstrated B. hermsii in the salivary glands of every experimentally infected O. hermsi examined (66).
In 1967, Coffey and Eveland demonstrated the ability of B. hermsii to generate a series of distinct serotypes during single infections in rats (18, 19). In 1982, Stoenner et al. showed that a single cell of B. hermsii HS1 could give rise to 24 serotypes in mice (81). When four different serotypes of spirochetes were transferred from mice to Kelly's medium (45), a novel and common serotype emerged with passage in vitro, and the authors of that study referred to these spirochetes as “culture adapted” (81). Barbour et al. identified a unique 19,000 molecular weight protein (pIC) produced by the culture-adapted spirochetes, which these authors renamed “C serotype” (7). pIC and other serotype-specific surface proteins of B. hermsii were later named variable major proteins (VMPs) (7), with the protein specific for serotype C designated VMPC (8). As the genes encoding the VMPs in B. hermsii were sequenced and analyzed, the gene encoding VMPC was also sequenced, and the protein was renamed Vmp33 (16) and then Vsp33 (5, 15). Through all of these nomenclatural changes, the biological significance of Vsp33 remained unclear, and until the present study the gene has been sequenced in only one B. hermsii isolate (HS1) (16).
In 1998, we reported that B. hermsii produces Vsp33 in ticks, and that the switch from a bloodstream Vmp to Vsp33 is accelerated in vitro by lowering the cultivation temperature (66). The high prevalence of spirochetes expressing Vsp33 while persistently infecting salivary glands in these ticks that feed in minutes suggested that this surface protein may be important in the tick transmission of spirochetes. This hypothesis is supported by the observations that the Lyme disease spirochete, Borrelia burgdorferi, upregulates outer surface protein (Osp) C, a Vsp33 ortholog, when transmitted by its vector Ixodes scapularis (69, 71). vsp33 is unique from all of the other vmp genes in that there is only a single copy (in the one isolate examined previously), it is under the control of a promoter different from the other vmps (5), and it is persistently expressed in ticks rather than mammals (66). These differences led Barbour to change the name of Vsp33 to Vtp for variable tick protein (3). To further address the biological significance of Vtp in the life cycle of B. hermsii, we identified the vtp gene in all isolates of this bacterium and compared the sequences to a phylogenetic analysis based on four other highly conserved genes. Here we show that all isolates of B. hermsii examined have vtp and that the sequences exhibit significant heterogeneity. We also identify two genomic groups of B. hermsii that overlap in their distribution within a wide geographic area and most likely share the same species of tick vector for their transmission and natural reservoir.
MATERIALS AND METHODS
Borrelia strains and cultivation.
Thirty-one isolates of B. hermsii collected over nearly 40 years were studied (Table 1). Twenty-eight of the isolates originated from the blood of patients who acquired the infection in the western United States or the Okanagan Valley of southern British Columbia, Canada. Two isolates originated from ticks, including the type strain HS1 (ATCC 35209) recovered near Spokane, Wash. (84), and SIS that was from northeastern California (35). One isolate (EST-7) was cultured from the blood of a Unita chipmunk (Tamias umbrinus), bled during an investigation of an outbreak of relapsing fever near Estes Park, Colo. (86). Borrelia parkeri RML, Borrelia turicatae 91E135, and Borrelia anserina BA2 were isolated from Ornithodoros parkeri, Ornithodoros turicata, and a domestic chicken, respectively. Borrelia crocidurae CR2A was provided by Sven Bergström, Umeå University, Umeå, Sweden. Borrelia coriaceae Co53 was isolated from Ornithodoros coriaceus in California (49).
TABLE 1.
Origin of B hermsii isolates examined
| Genomic group and isolatea | Date | Sourceb | Localityc | Source or reference |
|---|---|---|---|---|
| GGI | ||||
| HS1 | 1968 | Tick | Spokane Co., Wash. | 84 |
| MAN | 1960s | Human | Sierra Nevada Mtns, Calif. | 46 |
| CON | 1960s | Human | Sierra Nevada Mtns, Calif. | 46 |
| FRO | 1987 | M/8 yr | Eastern Washington | 74 |
| DAH | 1991 | F/Adult | Spokane Co., Wash. | 73 |
| FRE | 1996 | M/10 yr | Pend Oreille Co., Wash. | 73 |
| MIL | 1996 | F/Unkn | Kootenai Co., Idaho | This study |
| BRO | 1996 | M/Unkn | Kootenai Co., Idaho | This study |
| SWA | 1996 | M/Unkn | Kootenai Co., Idaho | This study |
| CAR | 1996 | F/46 yr | Benewah Co., Idaho | This study |
| EST-7 | 1996 | Chipmunk | Larimer Co., Colo. | 86 |
| BAK | 1997 | F/50 yr | Okanogan Co., Wash. | This study |
| BYM | 1997 | M/Unkn | Kootenai Co., Idaho | This study |
| ALL | 1997 | M/39 yr | Duchesne Co., Utah | This study |
| RAL | 1997 | F/39 yr | Siskiyou Co., Calif. | 35 |
| SIS | 1998 | Tick | Siskiyou Co., Calif. | 35 |
| WAD | 1998 | M/2 yr | Placer Co., Calif. | 35 |
| HAL | 1998 | M/73 yr | Kootenai Co., Idaho | This study |
| GAR | 2001 | M/42 yr | Okanagan Valley, BC | This study |
| GGII | ||||
| YOR | 1964 | M/50 yr | Siskiyou Co., Calif. | 46 |
| HAN | 1990 | M/1 mo | Boundary Co., Idaho | 73 |
| REN | 1992 | F/37 yr | Okanogan Co., Wash. | 73 |
| OKA-1 | 1995 | F/58 yr | Okanagan Valley, BC | 2 |
| OKA-2 | 1996 | M/Adult | Okanagan Valley, BC | 2 |
| OKA-3 | 1996 | M/Adult | Okanagan Valley, BC | 2 |
| GMC | 1997 | M/42 yr | Stevens Co., Wash. | This study |
| CMC | 1997 | F/36 yr | Stevens Co., Wash. | This study |
| RUM | 1997 | M/4 yr | Stevens Co., Wash. | This study |
| SIL | 2002 | F/40 yr | Boundary Co., Idaho | This study |
| LAK-1 | 2002 | F/5 yr | Lake Co., Mont. | 72 |
| LAK-2 | 2002 | M/43 yr | Lake Co., Mont. | 72 |
GGI and GGII were as defined in this study.
M, male; F, female; Unkn, unknown age.
Co., County; BC, British Columbia, Canada; Mtns, Mountains.
The new isolates of B. hermsii were established in pure culture by inoculating laboratory mice (Mus musculus) with blood from spirochetemic patients. Mouse inoculation was used because most attempts to isolate the organisms in Complete BSK-H medium (Sigma-Aldrich, St. Louis, Mo.) directly from the patients' blood failed. Adult mice (outbred strain RML) were inoculated intraperitoneally with 0.25 ml of human blood in EDTA. A drop of peripheral blood obtained daily from the tip of the mouse tail vein was smeared on a microscope slide, air dried, stained with Giemsa, and examined by bright-field microscopy at ×970 magnification and oil immersion. On the second or third day of the spirochetemia, 0.25 ml of the infected blood was collected by cardiac puncture and passaged into a second mouse by intraperitoneal inoculation, which generally resulted in a higher level of spirochetemia. Infected blood was collected from the second mouse by cardiac puncture, and 100 μl was inoculated into a tube containing 9 ml of Complete BSK-H medium, sealed, and incubated at 34°C. Spirochetes were harvested and examined as uncloned isolates after two to four passages. The use of animals for this research was approved by the Rocky Mountain Laboratories Animal Care and Use Committee.
Polyacrylamide gel electrophoresis.
Whole-cell lysates of spirochetes were prepared as described previously (68). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Laemmli buffer (48) and a vertical gel electrophoresis system (Bethesda Research Laboratories/Gibco, Gaithersburg, Md.) were used to separate proteins according to the manufacturer's instructions. Proteins were stained with Coomassie brilliant blue.
Western blot analysis.
Whole-cell lysates were electrophoresed in one-dimensional acrylamide gels and blotted onto nitrocellulose membranes using Towbin buffer (85) and a Trans-Blot Cell (Bio-Rad Laboratories, Hercules, Calif.) according to the manufacturer's instructions. Membranes were blocked overnight at room temperature with TSE-Tween (50 mM Tris [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.05% Tween 20) and subsequently incubated with anti-flagellin monoclonal antibodies H9724, specific for all members of the genus Borrelia (6), and H9826, specific for B. hermsii (65). Bound antibodies were detected with 125I-labeled protein A and autoradiography.
DNA purification and analysis.
Genomic DNA was purified from 100- or 500-ml stationary-phase cultures of spirochetes (77). Total genomic DNA samples were electrophoresed in 1% agarose gels with 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 10 mM EDTA) without ethidium bromide. Plasmids were resolved in a reverse-pulse electrical field provided with a PPI-200 Programmable Power Inverter (MJ Research, Watertown, Mass.). DNA was electrophoresed at 100 V for 15 min and run on Program 3 for 18 h with recirculation of the buffer in ice. Program 3 was set by the manufacturer for resolving linear plasmids in the 10 to 100 kb size. The gels were stained with ethidium bromide and visualized with UV transillumination.
PCR and DNA sequence analysis.
DNA was quantified by UV spectroscopy and diluted to ca. 0.1 μg for each 100-μl PCR. Taq enzyme and reaction constituents were added as suggested in the manufacturer's instructions (Perkin-Elmer, Roche Molecular Systems, Inc., Branchburg, N.J.). The DNA sequence was determined for five genes in all 31 isolates of B. hermsii, including vtp, 16S rRNA, flaB, gyrB, and glpQ. Preliminary analysis of the 16S rRNA gene sequences in GenBank for some of the species of relapsing fever spirochetes indicated possible errors in the database. Therefore, we determined the DNA sequence of the 16S rRNA and flaB genes of B. coriaceae, B. crocidurae, B. anserina, B. parkeri, and B. turicatae. The primers (Invitrogen, Carlsbad, Calif.) used to amplify DNA fragments for sequencing the five genes are shown in Table 2. PCRs were performed under mineral oil for 25 cycles by using a Perkin-Elmer thermocycler. Each cycle consisted of denaturation at 94°C for 1 min, annealing at 50°C for 30 s, and extension at 72°C for 2 min. After the 25th cycle, an additional 7-min extension was done at 72°C.
TABLE 2.
Oligonucleotide primers used for gene amplification and DNA sequencing
| Gene and primera | Sequence (5′ to 3′)b or description | Base positions |
|---|---|---|
| vtp | Amplicon size for DAH, 836 bp; ORF, 627 | |
| Vtp sp-7 | TGATAATATTTTTGTTTTGTAAAATTATTTACG | −115 to −83 |
| Vtp sp-12 | GCTTTCTATTTATTGACTTTATTTTTCCAG | +91 to +62 |
| 16S rRNA | Amplicon size1489 bp; Trimmed: 1273 bp | |
| FD3* | AGAGTTTGATCCTGGCTTAG | −89 to −70 |
| T50* | GTTACGACTTCACCCTCCT | +127 to +109 |
| Rec4* | ATGCTAGAAACTGCATGA | 533 to 550 |
| Rec9* | TCGTCTGAGTCCCCATCT | 1052 to 1035 |
| 16s (−) | TAGAAGTTCGCCTTCGCCTCTG | 641 to 620 |
| 16s (+) | TACAGGTGCTGCATGGTTGTCG | 939 to 960 |
| flaB | Amplicon size, 1,285 bp; ORF, 1,002 bp | |
| Bh fla 5′ | AATCTTTGAATTTACAGCGACAAAACAGG | −155 to −127 |
| Bh fla 3′ | AAACTCCAATGCGAAAACATTACAATCC | +125 to +98 |
| Fla +1 | AGAGCTTGGAATGCAACCCG | 447 to 466 |
| Fla −1 | TGCCTCATCCTGATTTGCG | 552 to 534 |
| gyrB | Amplicon size, 2,141 bp; ORF, 1902 bp | |
| gyrB 5′ A-1 | TTTATTGGTTTTAAGTCAAGTTGAATATGTC | −120 to −90 |
| gyrB 3′ | GGCTCTTGAAACAATAACAGACATCGC | +116 to +90 |
| gyrB 5′ | GGTTTATGAGTTATGTTGCTAGTAATATTCAAGTGC | −5 to 31 |
| gyrB 5′+1 | TTATCAAAGAGACTTAGGGAACTTGC | 547 to 572 |
| gyrB 5′+2 | GAAAGATGTTCCAAGTCTTACATTAGATG | 906 to 934 |
| gyrB 5′+3 | GCTGATGCTGATGTTGATGG | 1480 to 1499 |
| gyrB 3′+1 | TGCCCATTCTCAATTAACTCCC | 1568 to 1547 |
| gyrB 3′+2 | CATCATGCACAATAGTTTCAACG | 1060 to 1038 |
| gyrB 3′+3 | TTCTCTTTTCCCGATCTCCTATC | 629 to 607 |
| glpQ | Amplicon size, 1,396 bp; ORF, 1,020 and 1,026 bp | |
| glpQ F+1 | GGGGTTCTGTTACTGCTAGTGCCATTAC | −252 to −225 |
| Rev-2 | CAATACTAAGACCAGTTGCTCCTCCGCC | +121 to +94 |
| Rev-1 | GCACAGGTAGGAATGTTGGAATTTATCCTG | 482 to 511 |
| glpQ F-1 | CAATTTTAGATATGTCTTTACCTTGTTGTTTATGCC | 565 to 530 |
*, For flaB, gyrB, and glpQ, the minus (−) numbers represent positions upstream of the A in the ATG start codon; positive (+) numbers represent positions downstream of the last base in the stop codon; numbers with no “+” or “−” are within the ORF, beginning with the A in the start codon. For 16S rRNA, minus (−) and positive (+) positions flank the sequence used in the analysis.
PCR amplification products were first visualized by agarose gel electrophoresis. If primer dimer products or heterogeneous secondary bands were present, the total reaction mixture was electrophoresed in an agarose gel and the band of interest was excised. DNA was purified from gel fragments with Minus EtBr spin columns (Supelco, Inc., Bellefonte, Pa.) according to the manufacturer's instructions. PCRs resulting in a single fragment of the predicted size were purified in Centricon 100 concentrators (Millipore, Bedford, Mass.) according to the manufacturer's instructions. All DNA samples were quantified by UV spectroscopy and diluted to the appropriate concentration recommended for automated DNA sequencing.
DNA sequencing reactions were performed with Model 370 and 3700 Automated DNA Sequencers (Applied Biosystems, Inc., Foster City, Calif.) and ABI Prism Dye Terminator Cycle Sequencing Ready Reaction sequencing kits according to the manufacturer's instructions (Applied Biosystems). Nucleotide and deduced amino acid sequences were analyzed with the MacVector version 6.0 software package (Oxford Molecular, Beaverton, Oreg.). DNA sequences were first aligned with the CLUSTAL V program in the Lasergene software package (DNASTAR, Madison, Wis.). The alignments were transferred into the MacClade program (52a) for manual correction. MacClade output files were opened in PAUP (81a), and maximum-likelihood neighbor-joining trees were created with a paraphyletic outgroup. The robustness of clade designations was tested with a full heuristic search and 1,000 bootstrap replicates. DNA polymorphism data, DNA divergence between the genomic groups, and recombination analysis via R = 4Nr were all performed with the DNasp package of algorithms (http://www.ub.es/dnasp). Sawyer's test was performed with GENECONV (http://www.math.wustl.edu/∼sawyer/mbprogs/).
vtp gene mapping.
Total genomic DNA samples were electrophoresed in 1% agarose gels as described above and transferred onto GeneScreen Plus membranes (Perkin-Elmer Life and Analytical Sciences, Shelton, Conn.) by the method of Southern (78). DNA hybridization probes specific for the vtp gene were produced after the DNA sequences were determined in all isolates. Primer sequences were chosen to amplify an internal, variable region to prevent hybridization of these DNA fragments with other vsp genes. Pairs of primers were made (Invitrogen) (Table 3) to PCR amplify DNA from genomic DNA purified from B. hermsii BAK, DAH, FRO, HAN, and YOR. The predicted size of the amplification products ranged from 260 to 327 bp. The DNA probes were labeled directly with horseradish peroxidase for detection by enhanced chemiluminescence with the ECL Direct Nucleic Acid Labeling and Detection System (Amersham Life Science) according to the manufacturer's instructions.
TABLE 3.
Primer sequences to amplify vtp-specific probes and ORF fragments lacking the signal sequence for cloning into the BamHI site of pET-15b
| Isolate | Sequence (5′ to 3′) | Size of probe (bp) |
|---|---|---|
| For probes | ||
| BAK-1 | GGTAAGAAGATAGTTGCTGGTGGTGC | 260 |
| BAK-2 | TGGCTTTTTTTGCATTTGCATCAG | |
| DAH-1 | GATGAGCTTGCTAAAGCTATTGGACAG | 324 |
| DAH-2 | TGTATTTAACTTACCAAGTTCTTCAGCTCCC | |
| FRO-1 | CGCTATTGGAAAGAAAATTAAAGAAGATG | 265 |
| FRO-2 | GCTTTTTGTGCATTCTCATCAGTAGC | |
| HAN-1 | GCTATTAAAAAGAAAATTCAAGCAGATGGTC | 277 |
| HAN-2 | ATCTATGGCTTCCTTTGCATTAGCAC | |
| YOR-1 | GCTAAAGCTATTGGGAAAAAAATTGATC | 327 |
| YOR-2 | TTCAACTTACCAAGCTCTTCAGCTCC | |
| For cloning | ||
| Vtp1C | GGATCCGTGTAATAATGGAGGCCCAGAG | |
| Vtp1G | GGATCCGTGTAATAATGGAGGGCCAGAG | |
| Vtp2A | GGATCCTTAAGGTTTAACAGGGGTCGC | |
| Vtp2G | GGATCCTTAAGGTTTAGCAGGGGTCGC |
Production of Vtp monoclonal antibodies.
Five isolates of B. hermsii were chosen (DAH, YOR, BRO, FRO, and BYM). The vtp coding region minus the signal sequence was amplified with genomic DNA by PCR with primers that included BamHI sites on both ends of the amplicons (Table 3). Primers Vtp1C and Vtp 2A were used with YOR and DAH, primers Vtp1C and Vtp2G were used with BRO and FRO, and primers Vtp1G and Vtp2A were used with BYM. The methods used for PCR amplification, cloning amplicons into the pET-15b vector (Novagen, Inc., Madison, Wis.), purification, and quantification of the heterologous His-tagged fusion proteins were done as described previously (58). Monoclonal antibodies H1131, H3548, and H4337 were products of fusions of RML mouse spleen and NS1 myeloma cells as described previously (8, 65), except that the immunogens were the purified heterologous proteins, and no antibiotics were administered to the mice. Hybridoma supernatants were examined for reactivity with methanol-fixed spirochetes and an indirect fluorescent antibody (IFA) test (65). The isotype for each monoclonal antibody was determined by IFA with methanol-fixed spirochetes, affinity-purified antigen-specific rabbit antibodies to mouse immunoglobulin M (IgM), IgG1, IgG2A, and IgG2B (Zymed Laboratories, Inc., South San Francisco, Calif.), and goat anti-rabbit immunoglobulin labeled with rhodamine (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.).
Tick infection and transmission.
The ability of O. hermsi to transmit spirochetes identified as B. hermsii in the newly identified genomic group II (the present study) was studied. To initiate the transmission cycle, one mouse each was infected with B. hermsii REN or HAN. A 0.7-ml frozen aliquot of each culture was thawed and inoculated in total by intraperitoneal injection. Mice were examined daily for spirochetemia by collecting blood from the tail vein, preparing a thick drop of blood on a microscope slide, staining the samples with Giemsa, and examining them with a bright-field microscope at ×970 magnification and oil immersion. The numbers of circulating spirochetes were 2.4 × 107 (HAN, 2 days postinoculation) and 7.2 × 107 (REN, 7 days postinoculation) bacteria per ml. These mice were used to infect nymphal and adult O. hermsi from a colony of uninfected ticks maintained at Rocky Mountain Laboratories. Approximately 40 ticks fed on each mouse at the time of the spirochetemias cited above. Bacterial acquisition by ticks was confirmed immediately after feeding by dissecting the midgut from two ticks in each group (HAN- and REN-infected ticks) and examining wet mounts with a dark-field microscope. The remaining ticks were held at 25°C and 85% relative humidity.
Tick transmission of B. hermsii REN and HAN was attempted by allowing groups of two ticks to feed on individual 10-day-old mice or allowing groups of 8 to 10 ticks to feed on individual adult mice. Tick transmission was attempted 53 to 122 days after the ticks had ingested their previous infectious blood meal. Together, 61 ticks exposed previously to B. hermsii were fed on 19 mice, which were then examined daily for infection as described above.
Nucleotide sequence accession numbers.
Nucleotide sequences (151 of 155) of the five loci from the 31 isolates of B. hermsii have been deposited in the GenBank database under accession numbers AY597656 to AY597806. Three sequences for the type strain HS1 were already determined and identical to our results: glpQ (U65980) (76), flaB (M86838) (57), and gyrB (AF098862), as was the glpQ sequence we determined previously for B. hermsii DAH (U40762) (73); therefore, these sequences were not duplicated in the database. Nucleotide sequences of 16S rRNA and flaB for the other Borrelia species have also been deposited in the GenBank database under the accession numbers AY604974 to AY604982. The glpQ sequences for these species were deposited previously (AF247151, AF247152, AF247156 to AF247158, and AY368276 [58] and AY368276 [1]).
RESULTS
Geographic distribution of B. hermsii isolates examined.
The 31 isolates of B. hermsii came from localities throughout a major part of the known distribution where relapsing fever has been associated with the occurrence of the tick vector, O. hermsi (Table 1). The geographic range of the isolates spanned the Okanagan Valley in southern British Columbia, south to Placer County (Co.), Calif., and east to Estes Park in Larimer Co., Colo. All of the isolates, except the well-studied type strain HS1, were tentatively identified as B. hermsii based on their ecological or epidemiological association with their probable tick vector and locality of origin.
Protein and plasmid analysis.
Whole-cell lysates of the 31 isolates of B. hermsii examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis demonstrated a homogeneous protein profile except for the heterogeneity in the Vmps. Their identities as B. hermsii were confirmed by immunoblot analysis and positive reactivity with monoclonal antibodies H9724 and H9826. Protein profiles and positive immunoblots with H9826 have been presented elsewhere for 12 of the isolates (HS1, MAN, CON, YOR, DAH, OKA-1, OKA-2, OKA-3, WAD, RAL, SIS, and FRO) (2, 35, 65). Similar results for the other 19 isolates are not shown.
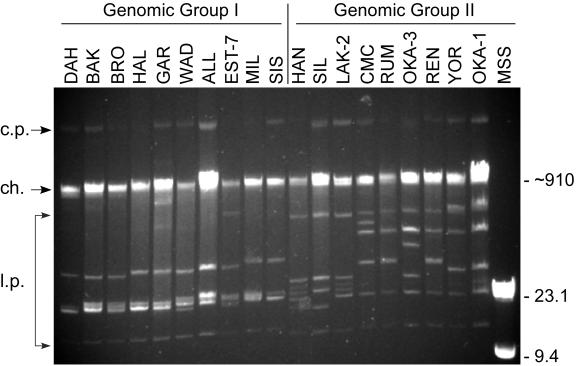
Three general patterns of the linear plasmids were observed among the isolates (Fig. 1). The largest linear plasmid of 180 to 200 kb in these spirochetes (40) was not seen as it comigrated with the chromosomal DNA, and the 30-kb circular plasmids described previously (80) migrated untrue to their size, well above the chromosome (Fig. 1). Nineteen isolates represented by DAH contained a very similar plasmid profile with an estimated six plasmids ranging in size from approximately 16 to 55 kb (Fig. 1). The 12 other isolates contained linear plasmids that varied more in size and included one or more plasmids significantly larger than 55 kb. The difference in plasmid profiles correlated with the DNA sequence data below that separated the isolates into two genomic groups. Isolates HAN, SIL, and LAK-2 had very similar plasmid profiles unique from the other isolates in their group, and these three isolates originated from localities not far apart in northern Idaho and western Montana.
FIG. 1.
Plasmid profiles of representative isolates of B. hermsii demonstrating patterns associated with the two genomic groups based on DNA sequence analysis (see subsequent figures). Isolate designations are shown above each lane, and DNA size estimates are shown on the right in kilobases. Arrows on the left show the positions of the circular plasmids (c.p.), chromosome (ch.), and various-sized linear plasmids (l.p.).
DNA sequence analysis of four conserved chromosomal loci.
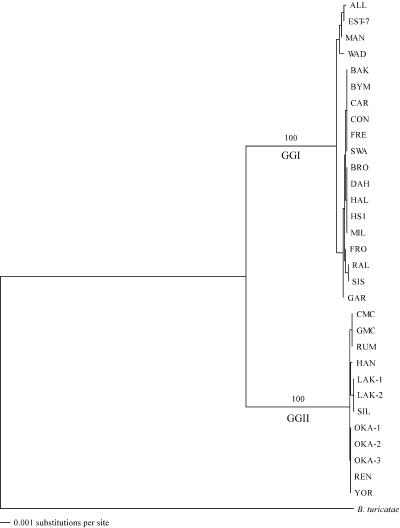
Further efforts to characterize the B. hermsii isolates at the genomic level focused on multilocus sequence typing. This approach can provide a genome-wide, subsample measure of evolutionary parameters including recombination frequency (59). DNA sequence was determined for most (1,273 bp) of the 16S rRNA gene and the complete open reading frames (ORFs) of three highly conserved, protein-encoding chromosomal loci, flaB (1,002 bp), gyrB (1,902 bp), and glpQ (1,020 or 1,026 bp). The 16S rRNA phylogram rooted with B. burgdorferi B31 supported the cluster formation of B. hermsii into two genomic groups designated genomic group I (GGI) and genomic group II (GGII) (data not shown). Analysis of the 16S rRNA alignment for all isolates identified five (0.39%) segregating or polymorphic sites between the two groups (Table 4). Mean nucleotide diversity at each aligned position (π) of the 16S rRNA gene was low (0.00193) between the groups, and the sequences were identical within each group (Table 4). GGI contained 19 isolates, including the type strain HS1, and GGII contained 12 isolates, exemplified by isolates YOR, OKA-1, OKA-2, and OKA-3 described in less detail elsewhere (2, 64).
TABLE 4.
Descriptive statistics and Sawyer's test for recombination of five loci in B. hermsii GGI and GGIIa
| Group | Locus | No. of:
|
Aligned characters
|
Sawyer's test
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples | Alleles | Bp | No. of:
|
π | Maximum score | SD | P | BC | No. of fragments | |||
| Gaps | Polymorphisms (%) | |||||||||||
| All isolates | 16S rRNA | 31 | 2 | 1,273 | 0 | 5 (0.39) | 0.0019 | |||||
| GGI | 16S rRNA | 19 | 1 | 1,273 | 0 | 0 | 0 | |||||
| GGII | 16S rRNA | 12 | 1 | 1,273 | 0 | 0 | 0 | |||||
| All isolates | flaB | 31 | 5 | 1,002 | 0 | 16 (1.6) | 0.0064 | 1.67 | 0.87 | 0.17 | 0 | |
| GGI | flaB | 19 | 3 | 1,002 | 0 | 5 (0.5) | 0.0014 | |||||
| GGII | flaB | 12 | 2 | 1,002 | 0 | 1 (0.1) | 0.0005 | |||||
| All isolates | gyrB | 31 | 5 | 1,902 | 0 | 40 (2.1) | 0.0099 | 0.86 | −0.48 | 0.74 | 0 | |
| GGI | gyrB | 19 | 4 | 1,902 | 0 | 3 (0.16) | 0.0005 | |||||
| GGII | gyrB | 12 | 1 | 1,902 | 0 | 0 | 0 | |||||
| All isolates | glpQ | 31 | 7 | 1,026 | 6 | 37 (3.6) | 0.0172 | 2.17 | 0.66 | 0.18 | 0 | |
| GGI | glpQ | 19 | 4 | 1,026 | 6 | 3 (0.29) | 0.0010 | |||||
| GGII | glpQ | 12 | 3 | 1,026 | 6 | 2 (0.19) | 0.0009 | |||||
| All isolates | Concat | 31 | 3,930 | 6 | 93 (2.4) | 0.0109 | ||||||
| GGI | Concat | 19 | 3,924 | 6 | 11 (0.28) | 0.0009 | ||||||
| GGII | Concat | 12 | 3,930 | 6 | 3 (0.08) | 0.0004 | ||||||
| All isolates | vtp | 31 | 13 | 654 | 44 | 268 (44) | 0.1745 | 21.30 | 14.72 | <10−5 | 0.020 | 57 |
| GGI | vtp | 19 | 10 | 654 | 44 | 243 (40) | 0.1790 | 23.98 | 17.43 | <10−5 | 0.017 | 23 |
| GGII | vtp | 12 | 3 | 654 | 13 | 196 (30) | 0.1124 | 5.19 | 3.46 | 0.007 | 0 | |
π, Mean nucleotide at each aligned position; SD, standard deviations above the mean; P, P value with Bonferroni correction for multiple samples; BC, Bonferroni corrected.
Phylograms for flaB, gyrB, and glpQ, each segregated the B. hermsii isolates into the same two genomic groups (data not shown). Therefore, DNA sequences of the three genes from each isolate were concatenated with the order flaB, glpQ, and gyrB (3,924 or 3,930 bp per isolate). These sequences were compared to the orthologous concatenated sequence for B. turicatae 91E135 as the outgroup, in which the phylogram had 100% of the bootstrap replicates again supporting the two genomic groups (Fig. 2). Alignments of the individual gene sequences showed a higher degree of polymorphism and mean nucleotide diversity in these three loci in GGI than in GGII (Table 4).
FIG. 2.
Phylogram of the concatenated sequences (flaB-glpQ-gyrB) of B. hermsii isolates and B. turicatae 91E135 used for the outgroup. The tree was constructed with CLUSTAL V and the neighbor-joining method with 1,000 bootstrap replicates. Numbers at the nodes are the percentages of bootstraps that supported this pattern. The scale bar for the branch lengths represents the number of substitutions per site.
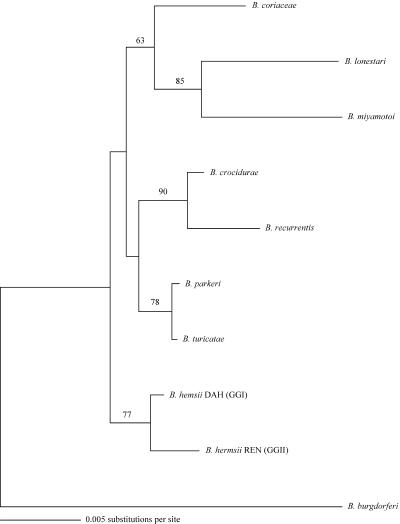
Single isolates from the two genomic groups of B. hermsii (DAH in GGI and REN in GGII) were compared to single isolates of other Borrelia species in a phylogenetic analysis with 16S rRNA, flaB, and glpQ genes. The phylogram for 16S rRNA demonstrated that the two genomic groups of B. hermsii partitioned with longer branch lengths than did B. parkeri and B. turicatae (Fig. 3). Phylograms for flaB and glpQ also clustered the two genomic groups of B. hermsii with similar branch patterns in relation to the other species (data not shown).
FIG. 3.
Phylogram of the16S rRNA sequences of one member of each genomic group of B. hermsii, other representative species of relapsing fever spirochetes, and B. burgdorferi used for the outgroup. The tree was constructed and analyzed as described for Fig. 2.
Sequence analysis of vtp.
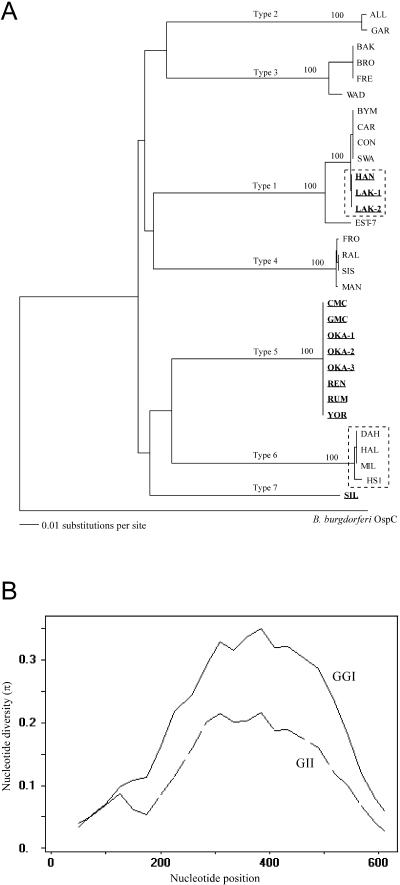
The DNA sequence of vtp was determined previously for one isolate (HS1) of B. hermsii (GenBank L24911) (16). We also determined the DNA sequence for HS1 and the 30 other isolates described above. Our vtp sequence for HS1 varied at four positions from what was reported. Therefore, we PCR amplified and sequenced the gene again, obtained an identical result, and used our sequence for further analysis. Unlike the four highly conserved chromosomal loci, the vtp gene was highly polymorphic (44%) and clustered into seven distinct Vtp types (Fig. 4A) (Table 4). An unrooted tree generated a star-shaped pattern (data not shown), similar to that described for ospC in B. burgdorferi sensu lato (12).
FIG. 4.
(A) Tree of the vtp sequences of the B. hermsii isolates and ospC of B. burgdorferi B31 used for the outgroup. The tree was constructed and analyzed as described for Fig. 2. All isolates in GGII are distinguished from GGI isolates with bold and underlined names. Probable examples of horizontal transfer of vtp sequences between spirochetes in the two genomic groups are shown within dashed-line boxes. (B) Distribution of polymorphic sites in the vtp gene for all isolates in each genomic group. A sliding window of 100 with a step size of 25 bases was used for same length alignments of the vtp genes. The mean nucleotide diversity (π) is on the y axis in relation to the nucleotide position on the x axis.
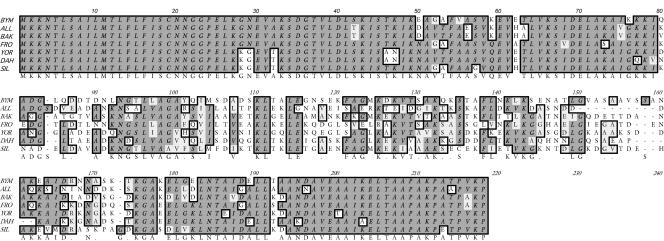
All 31 vtp genes encoded an identical signal sequence of 18 amino acids (MKKNTLSAILMTLFLFIS) preceding a cysteine, and the entire predicted, unprocessed peptides varied from 207 to 216 amino acids. The estimated molecular masses after cleavage of the signal peptide ranged from 19,568 to 20,388 Da. An amino acid alignment with one member of each of the seven Vtp types (Fig. 5) demonstrated, as did the DNA sequences (Fig. 4B), that most of the heterogeneity in this locus was in the internal hypervariable region, whereas the amino- and carboxy-terminal regions were highly conserved.
FIG. 5.
Alignment of deduced amino acid sequences for single isolates in each of the seven Vtp types identified in B. hermsii. The consensus is shaded and shown below the alignment. BYM (type 1), All (type 2), BAK (type 3), FRO (type 4), YOR (type 5), DAH (type 6), SIL (type 7). BYM, ALL, BAK, FRO, and DAH are in GGI; YOR and SIL are in GGII.
Vtp amino acid sequences within each type were identical or nearly so, but among the seven types they were highly divergent, with identity values ranging from 60.2 to 74.2% (mean = 66.8%) (Table 5). DNA sequences in each Vtp type were also identical or varied by only 0.2 to 2.3%; among the seven Vtp types, sequences varied from 75.9 to 82%. Thirteen alleles were found among the seven Vtp types (ten alleles in GGI and only three alleles in GGII), and the mean nucleotide diversity was greater in GGI than in GGII (Fig. 4B) (Table 4). Some isolates with different Vtp types came from the same geographic region, such Kootenai Co, Idaho, where five isolates in three Vtp types originated (BYM and SWA in type 1, BRO in type 2, and MIL and HAL in type 6). In contrast, isolates with identical amino acid and DNA sequences were also found from far ranging localities, such as the Vtp type 5 isolates that came from southern British Columbia, eastern Washington, and northern California.
TABLE 5.
Vtp amino acid sequence identity values between single isolates in each Vtp typea
| Isolate (type) | Sequence identity (%) of isolates of B. hermsii representing the seven Vtp typesb
|
||||||
|---|---|---|---|---|---|---|---|
| BYM | GAR | BAK | RAL | YOR | DAH | SIL | |
| BYM (1) | 100 | 66.0 | 66.4 | 68.2 | 67.6 | 70.8 | 66.7 |
| GAR (2) | 100 | 68.9 | 64.6 | 64.6 | 63.4 | 60.2 | |
| BAK (3) | 100 | 65.0 | 66.4 | 66.0 | 66.4 | ||
| RAL (4) | 100 | 68.2 | 67.0 | 65.1 | |||
| YOR (5) | 100 | 74.2 | 68.7 | ||||
| DAH (6) | 100 | 69.4 | |||||
| SIL (7) | 100 | ||||||
Percent identity values were determined with the GCG-Lite sequence comparison tool at http://molbio.info.nih.gov/molbio/gcglite/compare.html.
The lowest and highest identity values are underlined.
Recombination at the vtp locus.
The tree with seven vtp types in B. hermsii had long, internal branch lengths (Fig. 4A). This result suggested that recombination may have caused some of the diversity in this locus, thus further analyses were performed. There was a 10- to 90-fold greater mean nucleotide diversity in vtp compared to the conserved chromosomal loci (Table 4). Sawyer's test identified numerous recombination sites in the GGI vtp genes but none in GGII (Table 4). Hudson's formula (42) and the four-gamete test (43) gave the same results in that recombination was identified only in the GGI vtp genes (data not shown).
Horizontal gene transfer at the vtp locus.
Spirochetes in Vtp type 1 included seven isolates with identical amino acid sequences (BYM, CAR, CON, SWA, HAN, LAK-1, and LAK-2). However, part of this branch of the Vtp tree was incongruent with the tree for the two genomic groups (Fig. 2). Isolates BYM, CAR, CON, and SWA belonged to GGI, whereas isolates HAN, LAK-1, and LAK-2 were in GGII. Thus, in Vtp type 1 isolates, identical amino acid sequences were shared among multiple members of both genomic groups. This difference in branch pattern suggested that the Vtp gene in HAN, LAK-1, and LAK-2 represented a horizontal transfer of some part or all of the vtp sequence from a donor in GGI, that is, these sequences were xenologous (33). Analysis of the vtp DNA sequence in these seven isolates showed two alleles that differed in only one synonymous substitution at position 30 in the tenth codon (TTG versus TTA; both code for leucine in the signal peptide). All GGII isolates had an A in position 30, whereas in GGI only six of the isolates had an A and the remaining thirteen isolates had a G. If the entire gene had been transferred, we would expect identical characters in the 30th position; however, this was not observed. This suggests that most but not all of the vtp sequence downstream of position 30 was transferred from a GGI type 1 to GGII spirochete and therefore that one recombination event had occurred that was not detectable by the statistical tests used above.
Another incongruity in the branch pattern of the Vtp tree (Fig. 4A) compared to the two genomic groups (Fig. 2) was the greater similarity of Vtp type 6 isolates (in GGI) to Vtp type 5 isolates (in GGII) than to the other GGI types. The DNA sequences in these two types were 82% identical, which was greater than the identity values among other types. We aligned Vtp DNA sequences from one isolate of each of the seven types, which included YOR (GGII, Vtp type 5) and DAH (GGI, Vtp type 6). We divided the ORFs into three regions (bases 1 to 200 with no gaps, bases 201 to 521 with gaps, and bases 522 to 654 with no gaps) and constructed separate trees for each region (not shown). DAH grouped closely with YOR in the tree made with the first 200 bases but grouped with other GGI isolates in the trees based on the other two regions. Thus, isolates in GGI Vtp type 6 had a vtp sequence that may have evolved, in part, from the horizontal transfer of the upstream region of the vtp gene from a GGII spirochete.
Mapping the vtp gene.
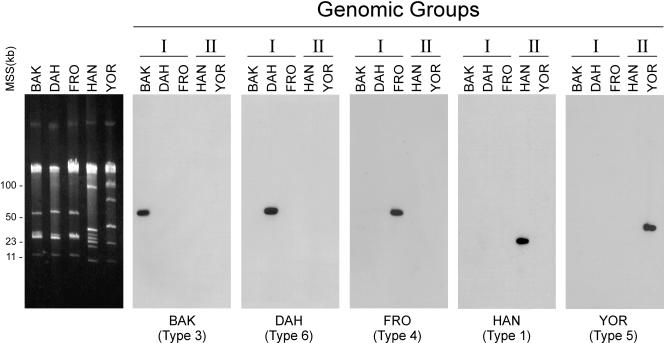
A single copy of this gene was mapped previously to a 53-kb linear plasmid in B. hermsii HS1 (5). We mapped the location of vtp in HS1 and 21 additional isolates that represented both genomic groups. We utilized the DNA sequences from the internal, hypervariable region to develop hybridization probes that were specific to five Vtp types that included 28 of the 31 isolates studied. These probes were specific to each vtp type and did not hybridize to vsp sequences on the smaller linear plasmids (Fig. 6). These probes hybridized to linear plasmids of approximately 53 kb in GGI isolates (Fig. 6) and to smaller linear plasmids of approximately 35 to 40 kb in GGII isolates (data not shown).
FIG. 6.
Mapping the location of the vtp gene with type-specific DNA probes. Plasmids from B. hermsii representing five of the Vtp types are shown in the agarose gel in the left panel. Southern blots containing these DNAs probed with different vtp probes are shown in the other panels, demonstrating their specificity. The isolate source for the DNA is shown above each lane along with its genomic group. The origins of the probes and Vtp types are shown below each panel. Molecular size standards (MSS) are shown on the left in kilobases.
Vtp-specific monoclonal antibodies.
Previously, we demonstrated that the DAH isolate of B. hermsii switched its outer surface from a bloodstream Vmp to Vtp when infecting ticks (66). For that study, we used monoclonal antibody H4825 (4) to identify Vtp-positive spirochetes in ticks; however, this antibody recognized only type 6 Vtps. Therefore, to study antigenic switching in ticks in the future with other isolates of B. hermsii, we produced three additional antibodies that recognize other Vtps. Antibody H3548 reacted only with type 5 Vtps, antibody H4337 reacted only with type 4 Vtps, and antibody H1131 was broadly reactive with Vtps in types 1, 3, 4, and 5. The three new antibodies were isotype IgG2A, while H4825 was IgG2B.
Tick infection and transmission.
Nymphal and adult O. hermsi transmitted GGII spirochetes (REN and HAN) to 74% (14 of 19) of the mice when 2 to 10 ticks fed on single mice 53 to 122 days after acquiring the spirochetes. Therefore, O. hermsi was a competent host and efficient transmitter of these spirochetes in the laboratory, which suggests that this tick is the probable vector of these spirochetes in nature.
DISCUSSION
One goal of our laboratory is to identify adaptations of B. hermsii that allow this spirochete to infect both ticks and mammals and to understand how this bacterium successfully alternates between these two very different types of hosts. Vtp may be critical for B. hermsii to infect ticks or to be infectious in mammals at the time of delivery by tick bite (66). Therefore, we sought additional isolates to examine the vtp and other loci to gain an understanding of the genetic structure at the species level. Multilocus sequence typing analysis of four chromosomal loci demonstrated two genomic groups of spirochetes that are highly clonal in structure, as has been demonstrated previously for the Lyme disease spirochete Borrelia burgdorferi (32). Both genomic groups met the criteria for bacterial clones (75) in that they contained isolates that were identical or nearly so throughout the single and concatenated gene phylogenies, they originated from locations over a wide geographic area, and they were isolated over a time span of 40 years. In addition, the GGI isolates had greater nucleotide diversity (π) and more polymorphisms in all of the loci examined than did the GGII isolates (Table 4), which suggests that GGI may be ancestral to GGII. The evidence for recombination and greater nucleotide diversity observed in vtp compared to the other loci suggests that this gene is under greater selective pressure (59).
Recently, Bunikis et al. described four genotypes among nine isolates of B. hermsii based on 685 bp of sequence in the 16S-23S rRNA intergenic spacer (IGS) region (13). The IGS may be more susceptible to accumulating mutations than are the protein-coding loci we examined, and therefore the IGS may be useful in identifying local geographic variants of this spirochete. Our GGI contained a cluster of four isolates (ALL, EST-7, MAN, and WAD) (Fig. 2) that originated from Utah, Colorado, and California but not from Washington and Idaho, where most of the other GGI isolates were found. Future efforts are needed to compare genotypes of B. hermsii based on the IGS with the genomic groups and geographic clusters described here to determine the utility of the IGS region for phylogenetic studies.
The vtp locus in B. hermsii demonstrated a pattern of diversity similar to what has been described for ospC in B. burgdorferi sensu lato. Numerous investigators have examined the ospC locus with PCR-RFLP, PCR-SSCP, and DNA sequencing (12, 31, 44, 52, 82, 83, 87, 88). The results of these studies identified 21 major ospC groups. Within each group the sequence varies little (>99% identity), but among the groups the sequences are quite different (average of 80% identity) (88). Also, sequence divergence in ospC is as great within a local population of spirochetes as it is within an entire species over large geographic areas (88). For example, eight major ospC groups were found among 20 isolates of B. burgdorferi sensu lato collected near Vienna, Austria, while identical ospC sequences were found in spirochetes from the United States and France (52). ospC sequences also group more closely within a species of Lyme disease spirochete (44, 52, 82, 83). However, many comparisons of ospC sequences have not agreed with phylogenetic analyses. This has allowed investigators to identify probable examples of horizontal transfer and recombination of part of this gene both within and between species (31, 44, 52, 87).
Two studies of sequence variation in ospC in B. burgdorferi led investigators to conclude that the major groups in this locus are maintained by frequency-dependent selection, a form of balancing selection, driven by the vertebrate host's immune system (31, 88). OspC is a major outer surface protein produced by Lyme disease spirochetes when these bacteria are transmitted by the bite of slow-feeding Ixodes ticks (21, 69, 71); hence, this protein is a dominant antigen when the spirochetes first enter the mammalian host. After infection by tick bite, humans make an early and strong antibody response to OspC (28, 36, 56). Mice immunized with OspC from one strain are protected from infection when challenged with the homologous but not a heterologous strain of B. burgdorferi (37, 60), and the ospC sequence does not vary during mammalian infection (79). Therefore, anti-OspC antibodies kill OspC-positive spirochetes that contain the same protein to which antibodies are produced; thus, the continued presence of OspC during infection could be lethal for the spirochetes. Interestingly, spirochetes rapidly downregulate this protein when grown continuously in vitro at 37°C (69), and one study showed that this gene is no longer expressed in mice after 17 days after needle inoculation of cultured spirochetes (51). Vtp is upregulated when B. hermsii persists in the salivary glands of its fast-feeding tick vector O. hermsi until the spirochetes are transmitted to the next mammalian host. Once back in mammals, B. hermsii downregulates the synthesis of Vtp within the first few days of infection in blood, whether infected by tick bite (66) or needle inoculation (5). How much specific immunity is produced to Vtp during the early and brief exposure in mammals is not known, although hyperimmune sera produced to this protein in rabbits kills Vtp-positive B. hermsii in vitro (T. G. Schwan, unpublished data).
OspC in B. burgdorferi is antigenically related and orthologous to Vtp and other Vsps produced by B. hermsii (16, 53, 54, 82, 89). Both OspC and Vtp are produced by their respective species of spirochete when transmitted by tick bite, although the dynamics and temporal events of spirochetal infection, localization for their persistence in the ticks, and feeding behaviors of ixodid and argasid ticks are very different (63, 66, 70, 71). These proteins are also members of a family of proteins that are likely shared among most if not all species of Borrelia (16, 53). OspC and Vtp all have an identical amino acid sequence in the signal peptide that differs from the other Vsps (16), but the significance of this is unknown.
The Lyme disease and relapsing fever spirochetes differ in their repertoire of these small, outer surface lipoproteins. B. burgdorferi has only 1, OspC (17, 34), while B. hermsii has at least 11 (Vtp and Vsp1, -2, -3, -6, -8, -11, -13, -22, -24, and -26) (3, 40). Therefore, a single B. burgdorferi infection will stimulate an immune response only to OspC, whereas B. hermsii may stimulate a stronger immunological response to this family of proteins due to the cyclical boosts with related proteins produced during the relapses that follow the initial infection with Vtp. The Vtp amino acid sequence of B. hermsii HS1 shares 52.6 to 58% identity with the other Vsps in this isolate (determined with the GCG-Lite sequence comparison tool [http://molbio.info.nih.gov/molbio/gcglite/compare.html] and data in GenBank). Possibly, this could increase the selective pressure for Vtp to change in immune hosts and result in the greater amino acid sequence divergence among the B. hermsii Vtp types (mean of 66.8% identity) compared to the B. burgdorferi OspC groups (70.7 to 78.8% identity) (44). The lower identity of the amino acid sequences compared to the DNA sequences among both the B. burgdorferi OspC groups and the B. hermsii Vtp types presented here also suggests that these proteins are under selective pressure from host immunity (3).
The ecological parameters that maintain Lyme disease and relapsing fever spirochetes in nature are vastly different, and yet cumulatively they result in a genetic structure for the transmission-associated proteins, OspC and Vtp, that are strikingly similar. The primary reservoir for B. burgdorferi in the eastern United States is the white-footed mouse, Peromyscus leucopus (27), which once infected probably remains so for life (41, 67) with no apparent mortality as a result (14, 41). However, these mice are relatively short-lived, and wild populations usually have a complete turnover and replacement with young each year (47), although rare individuals may live long enough to overwinter into the next spring (14, 62). The primary tick vector associated with these mice, I. scapularis, has a 2-year life cycle (23). However, only the relatively short-lived nymphal ticks that became infected as larvae the previous year are responsible for transferring spirochetes to the next generation of naive mice (14, 50). The primary vertebrate hosts for B. hermsii are chipmunks (Tamias spp.) and tree squirrels (Tamiasciurus spp.) (9, 30). Pine squirrels may live up to 9 years in the wild (55), while yellow-pine chipmunks may live 3 to 4 years (11). However, in spite of their longevity, these animals are poor long-term reservoirs for spirochetes. These wild rodents and experimental mice may be repeatedly spirochetemic over the course of weeks to a few months; however, B. hermsii, unlike some other species of relapsing fever spirochetes, rarely persists in the brain (20, 22). O. hermsi is a relatively long-lived tick with multiple nymphal stages and adults that feed repeatedly and all stages are capable of transmitting B. hermsii during one or more blood meals (26). Therefore, the dynamics of immune selective pressure on B. burgdorferi OspC includes short-lived mice and short-lived ixodid ticks in contrast to B. hermsii Vtp that includes long-lived squirrels and long-lived argasid ticks.
The spatial distribution and density of ticks and vertebrate hosts in the wild, in conjunction with the prevalence of spirochete infection, are also critical for how intense the selective pressure may be for spirochetes to change if they are to persist. Many studies have investigated the ecology and seasonal dynamics of B. burgdorferi in ticks and mice (38). In a recent study in Connecticut, nearly all of the white-footed mice became infected with B. burgdorferi during each of two consecutive transmission seasons, and a high percentage of the mice were seropositive with antibodies to OspC (14). The exposure of white-footed mice to infected ticks may be intense, and these rodents may be exposed repeatedly to spirochetes with different OspC groups in one season. Unfortunately, no such studies exist for tracking the incidence and distribution of B. hermsii in their vertebrate hosts and soft ticks. Thus, we can only speculate that the longer-lived squirrels and soft ticks, while probably less abundant, provide spirochetal infections that cumulatively over time result in the same selective pressure that drives the diversity in Vtp.
OspC and Vtp appear to be under selective pressure from the vertebrate hosts' immune system, with horizontal transfer between spirochetes and recombination accounting for some of the variability. Given the long-lived nature of O. hermsi and persistent infection of B. hermsii in the tick's salivary glands compared to the transient spirochetemias in rodent hosts, ticks are the true reservoir for the long-term perpetuation of these relapsing fever spirochetes in nature. Therefore, dual infections in ticks may provide the greatest opportunity for the exchange of DNA between genetically diverse spirochetes, although recombination between vtp and vls/vsp loci in one spirochete to generate vtp variants cannot be ruled out. For ticks to be susceptible to superinfection, the colonization of the tick's salivary glands with the first population of spirochetes should not make these tissues refractory to colonization with additional spirochetes acquired during subsequent infectious blood meals. Such experiments in ticks are now possible with the arsenal of diverse B. hermsii isolates and specific diagnostic reagents and sequences described here.
Acknowledgments
We thank Robert Karstens for technical assistance; Gary Hettrick and Anita Mora for help with the figures and manuscript preparation; Larry Bronson, Charles Smith, James Tucker, Curtis Fritz, Jane Wong, Satyendra Banerjee, and Ken Gage for field material used in the study; James Musser, Philip Stewart, and Michael Chaussee for reviewing an early draft of the manuscript, and Jonas Bunikis and Alan Barbour for reviewing the penultimate draft.
Editor: J. T. Barbieri
REFERENCES
- 1.Bacon, R. M., M. A. Pilgard, B. J. B. Johnson, S. J. Raffel, and T. G. Schwan. 2004. Glycerophosphodiester phosphodiesterase gene (glpQ) of Borrelia lonestari identified as a target for differentiating Borrelia species associated with hard ticks (Acari: Ixodidae). J. Clin. Microbiol. 42:2326-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, S. N., M. Banerjee, K. Fernando, W. Burgdorfer, and T. G. Schwan. 1998. Tick-borne relapsing fever in British Columbia, Canada: first isolation of Borrelia hermsii. J. Clin. Microbiol. 36:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, A. G. 2003. Antigenic variation in Borrelia: relapsing fever and Lyme borreliosis, p. 319-356. In A. Craig, and A. Scherf (ed.), Antigenic variation. Academic Press, Ltd., London, England.
- 4.Barbour, A. G. 1987. Immunobiology of relapsing fever, p. 125-137. In J. M. Cruse and R. E. Lewis, Jr. (ed.), Contributions to microbiology and immunology, vol. 8. Karger, Basel, Switzerland. [PubMed] [Google Scholar]
- 5.Barbour, A. G., C. J. Carter, and C. D. Sohaskey. 2000. Surface protein variation by expression site switching in the relapsing fever agent Borrelia hermsii. Infect. Immun. 68:7114-7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour, A. G., S. F. Hayes, R. A. Heiland, M. E. Schrumpf, and S. L. Tessier. 1986. A Borrelia-specific monoclonal antibody binds to a flagellar epitope. Infect. Immun. 52:549-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbour, A. G., S. L. Tessier, and H. G. Stoenner. 1982. Variable major proteins of Borrelia hermsii. J. Exp. Med. 156:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barstad, P. A., J. E. Coligan, M. G. Raum, and A. G. Barbour. 1985. Variable major proteins of Borrelia hermsii: epitope mapping and partial sequence analysis of CNBr peptides. J. Exp. Med. 161:1302-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck, M. D. 1937. California field and laboratory studies on relapsing fever. J. Infect. Dis. 60:64-80. [Google Scholar]
- 10.Beck, M. D. 1942. Present distribution of relapsing fever in California, p. 20-25. In F. R. Moulton (ed.), A symposium on relapsing fever in the Americas. American Association for the Advanced Sciences, Washington, D.C.
- 11.Broadbrooks, H. E. 1958. Life history and ecology of the chipmunk, Eutamias amoenus, in eastern Washington. Misc. Publ. Mus. Zool. Univ. Mich. 103:1-42. [Google Scholar]
- 12.Bunikis, J., U. Garpmo, J. Tsao, J. Berglund, D. Fish, and A. G. Barbour. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741-1755. [DOI] [PubMed] [Google Scholar]
- 13.Bunikis, J., J. Tsao, U. Garpmo, J. Berglund, D. Fish, and A. G. Barbour. 2004. Typing of Borrelia relapsing fever group strains. Emerg. Infect. Dis. 10:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunikis, J., J. Tsao, C. J. Luke, M. G. Luna, D. Fish, and A. G. Barbour. 2004. Borrelia burgdorferi infection in a natural population of Peromyscus leucopus mice: a longitudinal study in an area where Lyme borreliosis is highly endemic. J. Infect. Dis. 189:1515-1523. [DOI] [PubMed] [Google Scholar]
- 15.Cadavid, D., P. M. Pennington, T. A. Kerentseva, S. Bergström, and A. G. Barbour. 1997. Immunologic and genetic analyses of VmpA of a neurotropic strain of Borrelia turicatae. Infect. Immun. 65:3352-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter, C. J., S. Bergström, S. J. Norris, and A. G. Barbour. 1994. A family of surface-exposed proteins of 20 kilodaltons in the genus Borrelia. Infect. Immun. 62:2792-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 18.Coffey, E. M., and W. C. Eveland. 1967. Experimental relapsing fever initiated by Borrelia hermsi. I. Identification of major serotypes by immunofluorescence. J. Infect. Dis. 117:23-28. [DOI] [PubMed] [Google Scholar]
- 19.Coffey, E. M., and W. C. Eveland. 1967. Experimental relapsing fever initiated by Borrelia hermsi. II. Sequential appearance of major serotypes in the rat. J. Infect. Dis. 117:29-34. [DOI] [PubMed] [Google Scholar]
- 20.Coleman, G. E. 1934. Relapsing fever in California. III. The carrier condition: epidemiology. J. Infect. Dis. 54:282-294. [Google Scholar]
- 21.Coleman, J. L., J. A. Gebbia, J. Piesman, J. L. Degen, T. H. Bugge, and J. L. Benach. 1997. Plasminogen is required for efficient dissemination of Borrelia burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 89:1111-1119. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham, J. 1937. Further observations on Indian relapsing fever. Part III. Persistence of spirochaetes in the blood and organs of infected animals. Ind. J. Med. Res. 24:571-580. [Google Scholar]
- 23.Dammin, G. J. 1986. Lyme disease: its transmission and diagnostic features. Lab. Manag. 24:33-38. [Google Scholar]
- 24.Davis, G. E. 1956. The identification of spirochetes from human cases of relapsing fever by xenodiagnosis with comments on local specificity of tick vectors. Exp. Parasitol. 5:271-275. [DOI] [PubMed] [Google Scholar]
- 25.Davis, G. E. 1942. Species unity or plurality of the relapsing fever spirochetes, p. 41-47. In F. R. Moulton (ed.), A symposium of relapsing fever in the Americas. American Association for the Advanced Sciences, Washington, D.C.
- 26.Davis, G. E., and M. E. Walker. 1940. Ornithodoros hermsi: feeding and molting habits in relation to the acquisition and transmission of relapsing fever spirochetes. Public Health Rep. 55:492-504. [Google Scholar]
- 27.Donahue, J. G., J. Piesman, and A. Spielman. 1987. Reservoir competence of white-footed mice for Lyme disease spirochetes. Am. J. Trop. Med. Hyg. 36:92-96. [DOI] [PubMed] [Google Scholar]
- 28.Dressler, F., J. A. Whalen, B. N. Reinhardt, and A. C. Steere. 1993. Western blotting in the serodiagnosis of Lyme disease. J. Infect. Dis. 167:392-400. [DOI] [PubMed] [Google Scholar]
- 29.Dworkin, M. S., D. E. Anderson, Jr., T. G. Schwan, P. C. Shoemaker, S. N. Banerjee, B. O. Kassen, and W. Burgdorfer. 1998. Tick-borne relapsing fever in the northwestern United States and southwestern Canada. Clin. Infect. Dis. 26:122-131. [DOI] [PubMed] [Google Scholar]
- 30.Dworkin, M. S., T. G. Schwan, and D. E. Anderson. 2002. Tick-borne relapsing fever in North America. Med. Clin. N. Am. 86:417-433. [DOI] [PubMed] [Google Scholar]
- 31.Dykhuizen, D. E., and G. Baranton. 2001. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 32.Dykhuizen, D. E., D. S. Polin, J. Dunn, B. Wilske, V. Preac-Mursic, R. J. Dattwyler, and B. J. Luft. 1993. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc. Natl. Acad. Sci. USA 90:10163-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitch, W. M. 2000. Homology: a personal view on some of the problems. Trends Genet. 16:227-231. [DOI] [PubMed] [Google Scholar]
- 34.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. V. Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 35.Fritz, C. L., L. R. Bronson, C. R. Smith, M. E. Schriefer, J. R. Tucker, and T. G. Schwan. 2004. Isolation and characterization of Borrelia hermsii associated with two foci of tick-borne relapsing fever in California. J. Clin. Microbiol. 42:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fung, B. P., G. L. McHugh, J. M. Leong, and A. C. Steere. 1994. Humoral immune response to outer surface protein C of Borrelia burgdorferi in Lyme disease: role of the immunoglobulin M response in the serodiagnosis of early infection. Infect. Immun. 62:3213-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilmore, R. D., K. J. Kappel, M. C. Dolan, T. R. Burkot, and B. J. B. Johnson. 1996. Outer surface protein C (OspC), but not P39, is a protective immunogen against a tick-transmitted Borrelia burgdorferi challenge: evidence for a conformational protective epitope in OspC. Infect. Immun. 64:2234-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray, J. S., O. Kahl, R. S. Lane, and G. Stanek (ed.). 2002. Lyme borreliosis: biology, epidemiology and control. CABI Publishing, Oxford, England.
- 39.Herms, W. B., and C. M. Wheeler. 1935. Tick transmission of California relapsing fever. J. Econ. Entomol. 28:846-855. [Google Scholar]
- 40.Hinnebusch, B. J., A. G. Barbour, B. I. Restrepo, and T. G. Schwan. 1998. Population structure of the relapsing fever spirochete Borrelia hermsii as indicated by polymorphism of two multigene families that encode immunogenic outer surface lipoproteins. Infect. Immun. 66:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmeister, E. K., B. A. Ellis, G. E. Glass, and J. E. Childs. 1999. Longitudinal study of infection with Borrelia burgdorferi in a population of Peromyscus leucopus at a Lyme disease-enzootic site in Maryland. Am. J. Trop. Med. Hyg. 60:598-609. [DOI] [PubMed] [Google Scholar]
- 42.Hudson, R. R. 1987. Estimating the recombination parameter of a finite population model without selection. Genet. Res. Cambridge 50:245-250. [DOI] [PubMed] [Google Scholar]
- 43.Hudson, R. R., and N. L. Kaplan. 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jauris-Heipke, S., G. Liegl, V. Preac-Mursic, D. Rossler, E. Schwab, E. Soutschek, G. Will, and B. Wilske. 1995. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J. Clin. Microbiol. 33:1860-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelly, R. 1971. Cultivation of Borrelia hermsi. Science 173:443-444. [DOI] [PubMed] [Google Scholar]
- 46.Kurashige, S., M. Bissett, and L. Oshiro. 1990. Characterization of a tick isolate of Borrelia burgdorferi that possesses a major low-molecular-weight surface protein. J. Clin. Microbiol. 28:1362-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lackey, J. A., D. G. Huckaby, and B. G. Ormiston. 1985. Peromyscus leucopus. Mammalian Species 247:1-10. [Google Scholar]
- 48.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 49.Lane, R. S., W. Burgdorfer, S. F. Hayes, and A. G. Barbour. 1985. Isolation of a spirochete from the soft tick, Ornithodoros coriaceus: a possible agent of epizootic bovine abortion. Science 230:85-87. [DOI] [PubMed] [Google Scholar]
- 50.Lane, R. S., J. Piesman, and W. Burgdorfer. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36:587-609. [DOI] [PubMed] [Google Scholar]
- 51.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Livey, I., C. P. Gibbs, R. Schuster, and F. Dorner. 1995. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol. Microbiol. 18:257-269. [DOI] [PubMed] [Google Scholar]
- 52a.Maddison, D. R., and W. P. Maddison. 2003. MacClade 4: analysis of phylogeny and character evolution, version 4.06. Sinauer Associates, Sunderland, Mass.
- 53.Marconi, R. T., D. S. Samuels, T. G. Schwan, and C. T. Garon. 1993. Identification of a protein in several Borrelia species which is related to OspC of the Lyme disease spirochetes. J. Clin. Microbiol. 31:2577-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margolis, N., D. Hogan, W. Cieplak, Jr., T. G. Schwan, and P. A. Rosa. 1994. Homology between Borrelia burgdorferi OspC and members of the family of Borrelia hermsii variable major proteins. Gene 143:105-110. [DOI] [PubMed] [Google Scholar]
- 55.McAdam, A. G., and S. Boutin. 2003. Variation in viability selection among cohorts of juvenile red squirrels (Tamiasciurus hudsonicus). Evolution 57:1689-1697. [DOI] [PubMed] [Google Scholar]
- 56.Padula, S. J., A. Sampieri, F. Dias, A. Szczepanski, and R. W. Ryan. 1993. Molecular characterization and expression of p23 (OspC) from a North American strain of Borrelia burgdorferi. Infect. Immun. 61:5097-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picken, R. N. 1992. Polymerase chain reaction primers and probes derived from flagellin gene sequences for specific detection of the agents of Lyme disease and North American relapsing fever. J. Clin. Microbiol. 30:99-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porcella, S. F., S. J. Raffel, M. E. Schrumpf, M. E. Schriefer, D. T. Dennis, and T. G. Schwan. 2000. Serodiagnosis of louse-borne relapsing fever with glycerophosphodiester phosphodiesterase (GlpQ) from Borrelia recurrentis. J. Clin. Microbiol. 38:3561-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 60.Probert, W. S., M. Crawford, R. B. Cadiz, and R. B. LeFebvre. 1997. Immunization with outer surface protein (Osp) A, but not OspC, provides cross-protection of mice challenged with North American isolates of Borrelia burgdorferi. J. Infect. Dis. 175:400-405. [DOI] [PubMed] [Google Scholar]
- 61.Ras, N. M., B. Lascola, D. Postic, S. J. Cutler, F. Rodhain, G. Baranton, and D. Raoult. 1996. Phylogenesis of relapsing fever Borrelia spp. Int. J. Syst. Bacteriol. 46:859-865. [DOI] [PubMed] [Google Scholar]
- 62.Schug, M., S. Vessey, and A. Korytko. 1991. Longevity and survival in a population of white-footed mice (Peromyscus leucopus). J. Mamm. 72:360-366. [Google Scholar]
- 63.Schwan, T. G. 1996. Ticks and Borrelia: model systems for investigating pathogen-arthropod interactions. Infect. Agents Dis. 5:167-181. [PubMed] [Google Scholar]
- 64.Schwan, T. G., K. L. Gage, and B. J. Hinnebusch. 1995. Analysis of relapsing fever spirochetes from the western United States. J. Spirochetal Tick-Borne Dis. 2:3-8. [Google Scholar]
- 65.Schwan, T. G., K. L. Gage, R. H. Karstens, M. E. Schrumpf, S. F. Hayes, and A. G. Barbour. 1992. Identification of the tick-borne relapsing fever spirochete Borrelia hermsii by using a species-specific monoclonal antibody. J. Clin. Microbiol. 30:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwan, T. G., and B. J. Hinnebusch. 1998. Bloodstream- versus tick-associated variants of a relapsing fever bacterium. Science 280:1938-1940. [DOI] [PubMed] [Google Scholar]
- 67.Schwan, T. G., R. H. Karstens, M. E. Schrumpf, and W. J. Simpson. 1991. Changes in antigenic reactivity of Borrelia burgdorferi, the Lyme disease spirochete, during persistent infection in mice. Can. J. Microbiol. 37:450-454. [DOI] [PubMed] [Google Scholar]
- 68.Schwan, T. G., K. K. Kime, M. E. Schrumpf, J. E. Coe, and W. J. Simpson. 1989. Antibody response in white-footed mice (Peromyscus leucopus) experimentally infected with the Lyme disease spirochete (Borrelia burgdorferi). Infect. Immun. 57:3445-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwan, T. G., and J. Piesman. 2002. Vector interactions and molecular adaptations of Lyme disease and relapsing fever spirochetes associated with transmission by ticks. Emerg. Infect. Dis. 8:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwan, T. G., P. F. Policastro, Z. Miller, R. L. Thompson, T. Damrow, and J. E. Keirans. 2003. Tick-borne relapsing fever caused by Borrelia hermsii, Montana. Emerg. Infect. Dis. 9:1151-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwan, T. G., M. E. Schrumpf, B. J. Hinnebusch, D. E. Anderson, and M. E. Konkel. 1996. GlpQ: an antigen for serological discrimination between relapsing fever and Lyme borreliosis. J. Clin. Microbiol. 34:2483-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwan, T. G., W. J. Simpson, M. E. Schrumpf, and R. H. Karstens. 1989. Identification of Borrelia burgdorferi and B. hermsii using DNA hybridization probes. J. Clin. Microbiol. 27:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Selander, R. K., and J. M. Musser. 1990. Population genetics of bacterial pathogens, p. 11-36. In B. H. Iglewski and V. L. Clark (ed.), Molecular basis of bacterial pathogenesis. Academic Press, Inc., San Diego, Calif.
- 76.Shang, E. S., J. T. Skare, H. Erdjument-Bromage, D. R. Blanco, P. Tempst, J. N. Miller, and M. A. Lovett. 1997. Sequence analysis and characterization of a 40-kilodalton Borrelia hermsii glycerophosphodiester phosphodiesterase homolog. J. Bacteriol. 179:2238-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simpson, W. J., C. F. Garon, and T. G. Schwan. 1990. Analysis of supercoiled circular plasmids in infectious and noninfectious Borrelia burgdorferi. Microb. Pathog. 8:109-118. [DOI] [PubMed] [Google Scholar]
- 78.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 79.Stevenson, B., and S. W. Barthold. 1994. Expression and sequence of outer surface protein C among North American isolates of Borrelia burgdorferi. FEMS Microbiol. Lett. 124:367-372. [DOI] [PubMed] [Google Scholar]
- 80.Stevenson, B., S. F. Porcella, K. L. Oie, C. A. Fitzpatrick, S. J. Raffel, L. Lubke, M. E. Schrumpf, and T. G. Schwan. 2000. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect. Immun. 68:3900-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stoenner, H. G., T. Dodd, and C. Larsen. 1982. Antigenic variation in Borrelia hermsii. J. Exp. Med. 156:1297-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81a.Swofford, D.L. 1998. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 82.Theisen, M., M. Borre, M. J. Mathiesen, B. Mikkelsen, A.-M. Lebech, and K. Hansen. 1995. Evolution of the Borrelia burgdorferi outer surface protein OspC. J. Bacteriol. 177:3036-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Theisen, M., B. Frederiksen, A.-M. Lebech, J. Vuust, and K. Hansen. 1993. Polymorphism in ospC gene of Borrelia burgdorferi and immunoreactivity of OspC protein: implications for taxonomy and for use of OspC protein as a diagnostic antigen. J. Clin. Microbiol. 31:2570-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thompson, R. S., W. Burgdorfer, R. Russell, and B. J. Francis. 1969. Outbreak of tick-borne relapsing fever in Spokane County, Washington. JAMA 210:1045-1050. [PubMed] [Google Scholar]
- 85.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trevejo, R. T., M. E. Schriefer, K. L. Gage, T. J. Safranek, K. A. Orloski, W. J. Pape, J. A. Montenieri, and G. L. Campbell. 1998. An interstate outbreak of tick-borne relapsing fever among vacationers at a Rocky Mountain cabin. Am. J. Trop. Med. Hyg. 58:743-747. [DOI] [PubMed] [Google Scholar]
- 87.Wang, G., A. P. van Dam, and J. Dankert. 1999. Evidence for frequent OspC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol. Lett. 177:289-296. [DOI] [PubMed] [Google Scholar]
- 88.Wang, I.-N., D. E. Dykhuizen, W. Qiu, J. J. Dunn, E. M. Bosler, and B. J. Luft. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilske, B., V. Preac-Mursic, S. Jauris, A. Hofmann, I. Pradel, E. Soutschek, E. Schwab, G. Will, and G. Wanner. 1993. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect. Immun. 61:2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]