Abstract
Currently, paratuberculosis vaccines are comprised of crude whole-cell preparations of Mycobacterium avium subsp. paratuberculosis. Although effective in reducing clinical disease and fecal shedding, these vaccines have severe disadvantages as well, including seroconversion of vaccinated animals and granulomatous lesions at the site of vaccination. DNA vaccines can offer an alternative approach that may be safer and elicit more protective responses. In an effort to identify protective M. avium subsp. paratuberculosis sequences, a genomic DNA expression library was generated and subdivided into pools of clones (∼1,500 clones/pool). The clone pools were evaluated to determine DNA vaccine efficacy by immunizing mice via gene gun delivery and challenging them with live, virulent M. avium subsp. paratuberculosis. Four clone pools resulted in a significant reduction in the amount of M. avium subsp. paratuberculosis recovered from mouse tissues compared to mice immunized with other clone pools and nonvaccinated, infected control mice. One of the protective clone pools was further partitioned into 10 clone arrays of 108 clones each, and four clone arrays provided significant protection from both spleen and mesenteric lymph node colonization by M. avium subsp. paratuberculosis. The nucleotide sequence of each clone present in the protective pools was determined, and coding region functions were predicted by computer analysis. Comparison of the protective clone array sequences implicated 26 antigens that may be responsible for protection in mice. This study is the first study to demonstrate protection against M. avium subsp. paratuberculosis infection with expression library immunization.
For more than a century, paratuberculosis (Johne's disease) has been recognized as a progressive, chronic enteritis of ruminant animals worldwide. Paratuberculosis is caused by the acid-fast bacterium Mycobacterium avium subsp. paratuberculosis. Transmission of infection is generally thought to be oral as the bacteria are shed into the feces, colostrum, and milk of infected animals, although in utero transmission has also been documented (15, 48). Calves appear to be the stage most susceptible to infection (33), but perhaps this is because their risk of exposure is greater due to confinement with infected dams within the maternity pen or consumption of contaminated milk or colostrum.
Paratuberculosis vaccine studies have demonstrated that both cellular and humoral immune responses are induced (29, 40), yet vaccination is not effective for preventing infection or transmission of the disease to other animals. Under the best conditions, vaccines have been reported to reduce fecal shedding and delay the onset of clinical disease (23, 27, 30, 32, 53, 54, 56). The heat-killed whole-cell vaccine that is approved for use in the United States is now known to be a strain of Mycobacterium avium subsp. avium, not M. avium subsp. paratuberculosis (14). Use of this vaccine does not produce favorable results because of adverse reactions, including severe inflammation and granuloma formation at the injection site. In addition, vaccination interferes with bovine tuberculosis skin testing and serologic detection of M. avium subsp. paratuberculosis-infected cattle (19, 40). The vaccine also poses a health risk to veterinarians who administer the vaccine, as adverse reactions to accidental self-injection have been documented (44).
Although M. avium subsp. paratuberculosis is not considered a zoonotic agent, it has been isolated from intestinal biopsy tissue of human patients with Crohn's disease, a severe inflammatory enteritis whose clinical signs mimic some of those noted in cattle with clinical paratuberculosis. Furthermore, DNA-DNA hybridization has demonstrated a level of homology of 91 to 100% between M. avium subsp. paratuberculosis and M. avium subsp. avium (18). More recently, a large-scale sequence analysis of the two genomes demonstrated that there is more than 97% nucleotide homology between the two subspecies (7). This high degree of homology suggests that immunogens or vaccine candidates that may be useful for the control of paratuberculosis in livestock may also provide potential benefits for human health if they are used to prevent M. avium-associated pulmonary infections in human immunodeficiency virus-infected individuals. A recent study demonstrated the presence of the IS900 insertion sequence typically associated with M. avium subsp. paratuberculosis in 15 isolates of M. avium from human immunodeficiency virus patients (41), further corroborating the level of identity between the two subspecies and lending credence to the hypothesis that there is an overlap in protective sequences that could potentially be exploited for human vaccines as well.
A number of putative antigens have been identified from M. avium subsp. paratuberculosis, and their immunogenicity has been examined by serodiagnostic and lymphocyte stimulation assays (4, 5, 20, 37-39, 43). However, only one protein, a 65-kDa heat shock protein, has been evaluated for use as a potential vaccine (55). In that study, a DNA vaccine encoding the M. avium subsp. paratuberculosis 65-kDa antigen failed to protect mice after challenge with live M. avium. Additionally, several DNA vaccines encoding a single antigen have been developed and tested against Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium leprae, and M. avium (26). Nearly one-half of these mycobacterial DNA vaccines elicited strong levels of protection and exhibited the potential for use as safe, effective DNA vaccines that limit mycobacterial infection and disease progression.
DNA vaccines offer the advantage of being inexpensive and simple to produce, are stable at a variety of temperatures, and do not contain elements that induce allergic or hypersensitivity responses, which are often associated with conventional vaccines (3). There are a number of DNA delivery methods, but gene gun-mediated DNA vaccination is needle free and nanogram quantities of DNA can effectively induce both humoral and cell-mediated immune responses. One technique to identify novel protective antigens of bacterial pathogens that was previously described is expression library immunization (11). This technique enables all of the proteins encoded by a pathogen to be expressed by the host and simultaneously screened for protection in an infection model.
In the present study, a representative genomic library of M. avium subsp. paratuberculosis DNA was cloned into a mammalian expression vector and tested for its protective effects in vivo against challenge with live, virulent M. avium subsp. paratuberculosis in mice.
MATERIALS AND METHODS
Bacterial strains.
M. avium subsp. paratuberculosis strain 19698-1974 (originally isolated in 1974 from a clinical cow at the National Animal Disease Center [NADC], Ames, IA) and strain 6112 (recently isolated from a cow with clinical infection at the NADC) were grown in Middlebrook 7H9 liquid medium (pH 6.0) supplemented with 10% oleic acid-albumin-dextrose complex (Becton Dickinson Microbiology, Sparks, MD), 0.5% Tween 80 (Becton Dickinson Microbiology), and 2 mg/ml mycobactin J (Allied Monitor Inc., Fayette, MO). M. avium subsp. paratuberculosis cultures were grown to the log phase at an optical density at 540 nm of 0.4 at 37°C without shaking. All bacterial strains were minimally passaged to maintain virulence. Bacteria were pelleted by centrifugation at 10,000 × g for 30 min and washed twice with phosphate-buffered saline (PBS; 150 mM NaCl, 10 mM NaPO4; pH 7.4). The bacteria were resuspended in PBS, and the concentration was adjusted to 109 CFU/ml, as determined by the optical density at 540 nm. The final concentration of bacteria was confirmed by serial dilution onto agar slants of Herrold's egg yolk medium containing 2 mg/liter of mycobactin J (Allied Monitor). Escherichia coli ElectroMAX (Gibco BRL, Rockville, MD) cells were routinely grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates supplemented with kanamycin (50 μg/ml) for selection.
Mice.
Six-week-old BALB/cJ female mice were purchased from Jackson Laboratories (Bar Harbor, ME). The mice were housed in a temperature- and humidity-controlled room at the NADC and maintained in disposable plastic cages with free access to water and standard mouse chow during the course of this study. The NADC Animal Care and Use Committee approved all animal procedures described below.
Expression vector.
The mammalian expression vector pVAX1 (Invitrogen Corporation, Carlsbad, CA) was modified for use in this study. In order to enhance mammalian translation initiation and expression, a Kozak translation initiation sequence (consensus sequence, ANNATGG, with ATG initiation codon) was inserted into the multiple cloning site of pVAX1. Briefly, oligonucleotides Koz-1 and Koz-2 (see Table S1 in the supplemental material) were denatured by heating to 85°C for 5 min and then annealed to each other by slow cooling to 37°C. The resulting double-stranded DNA and pVAX1 vector were double digested for 1 h with HindIII and BamHI restriction endonucleases (New England Biolabs, Beverly, MA) at 37°C. Digested products were purified from agarose gels using Geneclean III (Bio 101, Inc., Carlsbad, CA) and ligated with T4 DNA ligase (Gibco BRL) to create pVAX-Koz. Following ligation, pVAX-Koz was transformed into competent E. coli cells, clones were selected for plasmid purification (Midi kit; QIAGEN, Inc., Valencia, CA), and the correct orientation of the Kozak sequence was confirmed by DNA sequencing.
Genomic DNA library.
Genomic DNA was extracted from M. avium subsp. paratuberculosis strain 19698-1974 as previously described (4). A genomic expression library was constructed by partially digesting 55 μg of M. avium subsp. paratuberculosis genomic DNA with 4 U of Sau3A I restriction endonuclease (New England Biolabs) for 1 h at 37°C to obtain 1- to 3-kb fragments. The resulting genomic fragments were gel purified using Geneclean III as described above. pVAX-Koz was digested with BamHI and gel purified, and the 5′ and 3′ ends were dephosphorylated with calf intestinal alkaline phosphatase (Gibco BRL) to prevent vector self-ligation. Ligation reactions with digested vector and inserts were performed overnight at 16°C with T4 DNA ligase. The transformed cells were plated on LB-kanamycin agar so that an average of 1,500 colonies grew per 50-mm plate. A total of 11 plates, termed “clone pools,” were selected for this study and numbered sequentially. Colonies from each clone pool were harvested from each plate and frozen at −80°C in LB storage medium containing 20% glycerol. Plasmids were purified from each clone pool using an EndoFree plasmid maxi kit (QIAGEN, Inc.) and were quantitated by spectrophotometry at 260 nm.
Clone pool analysis.
PCR analysis was performed in order to determine the percentage of clones that contained genomic DNA inserts. Two clone pools, clone pools 3 and 11, were randomly selected, and 50 clones from each of these two pools were randomly selected for plasmid purification. PCRs were designed to amplify across the multiple-cloning site using primers pVAX-MCS5′ and pVAX-MCS3′ (see Table S1 in the supplemental material). Amplicons were visualized on 1.2% agarose gels, and sizes were estimated based on a 100-bp DNA ladder size standard (MBI Fermentas, Hanover, MD). Additionally, all clone pools were surveyed by PCR for the presence of nine known M. avium subsp. paratuberculosis genes, alkA (13), csp1 (4), groES (17), gsbB (51), hsp65 (20), hspX (22), IS900 (24), mig (45), and pks (4). The DNA sequences for each primer set used to detect these nine genes are listed in a table in the supplemental material (Table S1).
DNA vaccine trial I.
BALB/cJ mice were obtained and randomly assigned to immunization groups containing 10 mice as follows: group 1, no DNA, noninfected; group 2, no DNA, infected; group 3, microcarrier, infected; group 4, vector DNA, infected; and group 5, clone pool DNA, infected. For control group 3 (microcarrier, infected), mice were immunized with 0.5 mg of gold microcarrier beads without any plasmid DNA. For control group 4 (vector DNA, infected), mice were immunized with 0.5 mg of gold microcarrier beads carrying 1 μg of pVAX-Koz. Eleven clone pools were tested in mice (10 mice per pool) in this study. DNA was prepared for Helios gene gun immunization as described by the manufacturer (Bio-Rad Laboratories, Hercules, CA). Briefly, 15 μg of pVAX-Koz or pVAX plasmid DNA was precipitated onto 7.5 mg of 1-μm gold beads and loaded into tubing for preparation of vaccine “bullets.” Each resulting DNA vaccine bullet contained 2 μg of DNA that was delivered by 0.5 mg of gold microcarrier beads. In order to increase the efficiency of DNA transfection, the abdomens of the mice were shaved with surgical clippers and wiped with 70% ethanol prior to immunization. Mice were anesthetized by inhalation of isofluorane prior to immunization to aid in handling.
Each group of mice received 2 μg of DNA from the assigned clone pool and then was boosted with the same DNA vaccine 3 weeks after the initial immunization. Two weeks after boosting, mice were inoculated intraperitoneally with live, virulent M. avium subsp. paratuberculosis strain 6112 (log phase, 108 bacteria per mouse in 100 μl). No-DNA, noninfected control mice were mock challenged by intraperitoneal injection of 100 μl sterile PBS. After 3 months of infection, mice were anesthetized by inhalation of isoflurane and were decapitated with a guillotine. The spleen and mesenteric lymph nodes were removed from each mouse, weighed, homogenized in a 0.75% hexadecylpyradinium chloride solution by use of a stomacher for 1 min, and allowed to sit overnight for decontamination. Tenfold serial dilutions (100 to 104) of each tissue homogenate from individual mice were inoculated (100 μl per slant) onto Herrold's egg yolk medium slants and incubated at 37°C for 8 weeks. The number of CFU per slant was determined for each dilution, and the average recovery of bacteria was calculated for each mouse tissue based on the tissue wet weight. The culture data (CFU/mg of tissue) were averaged for each immunization group for comparison and statistical analysis.
Additionally, blood was immediately collected into sterile glass tubes and pooled according to immunization treatment. The blood was allowed to clot overnight at 4°C, and then serum was harvested and frozen at −20°C.
Electrophoresis and immunoblot analysis.
The methods used for preparation of M. avium subsp. paratuberculosis sonicate, polyacrylamide gel electrophoresis, and electrophoretic transfer of M. avium subsp. paratuberculosis proteins onto pure nitrocellulose membranes have been described previously (8). Pooled sera (according to immunization group) from vaccinated mice were individually diluted 1:500 with PBS plus 2% bovine serum albumin and 0.1% Tween 20. Diluted sera were sequentially added to individual rows of a slot blot apparatus (Bio-Rad Laboratories) containing a blot of M. avium subsp. paratuberculosis whole-cell proteins and a molecular weight standard (Precision plus protein standard; Bio-Rad Laboratories). Immunoblots were probed essentially as described by Bannantine et al. (8).
Nucleotide sequence analysis of protective clone pools.
The DNA vaccination and challenge results from trial I demonstrated that four clone pools significantly protected mice from M. avium subsp. paratuberculosis challenge. In order to determine the coding sequence of each clone in a protective clone pool, DNA sequence analysis was performed. Two protective clone pools were retrieved from the frozen stocks, and the clone pools were grown overnight on LB-kanamycin medium plates. Individual clones in each pool were picked and transferred to 96-well plates containing LB-kanamycin medium using a BioRobotics BioPick colony picker (Genomic Solutions, Ann Arbor, MI). Plates of clones were incubated overnight at 37°C in a GeneMachines HiGro incubator (Genomic Solutions) at 480 orbits per min with oxygen supplementation. From each overnight culture, 10 μl was inoculated into fresh, sterile, 96-well, 1.5-ml, deep-well plates containing LB-kanamycin medium. The original culture plates were preserved by adding 20% glycerol to each well and storing the plates at −80°C, while the new deep-well plates were incubated overnight as described above. Plasmids were extracted from deep-well overnight cultures using QIAprep 96 turbo miniprep kits (QIAGEN Inc., Valencia, CA). Plasmid DNA was quantitated using a pico-green double-stranded DNA kit (Molecular Probes, Eugene, OR), and sequencing reactions were performed using Big Dye terminator chemistry v2.0 (Applied Biosystems, Foster City, CA). The insert sequences for each clone were obtained by sequencing across the vector multiple-cloning site using primers T7 (5′-AATACGACTCACTATAG-3′) and Rev (5′-TAGAAGGCACAGTCGAGG-3′). Nucleotide sequence data were obtained with an ABI Prism 3700 DNA analyzer (Applied Biosystems). DNA sequence data were edited, aligned, and analyzed using MacVector and AssemblyLIGN (Genetics Computer Group, Madison, WI). A comparison of clone coding sequences to the M. avium subsp. paratuberculosis genome was performed using Artemis (http://www.sanger.ac.uk/Software/Artemis/).
Clone arrays.
Nonrepeat clones from one protective clone pool were sequentially numbered on the basis of open reading frame size, from the smallest (clone 1) to the largest (clone 536), and they were individually transferred and ordered in new 96-well master plates. The nonrepetitive clones were incubated as described above, and clone growth was visually confirmed the following morning. From the master plates of nonrepetitive clones, 10 new clone pools containing 108 clones each were generated. The 10 new clone pools, termed “clone arrays,” were specifically designed so that each clone array contained an equal distribution of open reading frame sizes. This was accomplished by a complex clone selection method in which clone arrays 1 to 5 contained every fifth clone, while clone arrays 6 to 10 contained every sixth clone. Briefly, clone array 1 contained clones 1, 6, 11, 16, …, 536, clone array 2 contained clones 2, 7, 12, 17, …, 532, clone array 3 contained clones 3, 8, 13, 18, …, 533, etc. By comparison, clone array 6 contained clones 1, 7, 13, 19, …, 535, clone array 7 contained clones 2, 8, 14, 20 …, 536, clone array 8 contained clones 3, 9, 15, 21, …, 531, etc. This configuration of clones resulted in 17 clones shared by any two clone arrays, while it ensured that open reading frame sizes were equally distributed among the new clone arrays. Each clone array was grown overnight in 96-well plates, and culture growth was visually confirmed for each clone. The 108 clones from each clone array were pooled and grown overnight for EndoFree plasmid DNA purification as described above. In addition, the parent protective clone pool (1,500 clones that offered protection in our previous study) and the nonrepetitive clones larger than 7 amino acids (536 clones) were grown, and plasmid DNA was purified.
DNA vaccine trial II.
In DNA vaccine trial II, 40 μg of plasmid DNA was precipitated onto 10 mg of 1-μm gold beads and loaded into tubing for preparation of 20 vaccine “bullets.” Each resulting DNA vaccine bullet contained 2 μg of DNA that was delivered by 0.5 mg of gold microcarrier beads. BALB/cJ mice were obtained and randomly partitioned into immunization groups containing 10 mice as follows: group 1, no DNA, noninfected; group 2, no DNA, infected; group 3, parent protective clone pool DNA, infected; group 4, nonrepetitive clones DNA, infected; group 5, clone array DNA, infected. Ten clone arrays were tested with mice (10 mice per pool) in this experiment. The protocol for vaccination, boosting, challenge with live M. avium subsp. paratuberculosis, and collection and processing of tissues at necropsy was identical to that described above for vaccine trial I.
Statistical analysis.
A two-way analysis of variance was performed to compare differences between the mean values for control infected mice and immunization groups. Differences were considered significant when the P was <0.05.
RESULTS
Clone pool analysis.
An M. avium subsp. paratuberculosis genomic DNA expression library was cloned into the modified pVAX-Koz mammalian expression vector, and a total of 11 clone pools were selected for this study, with an average of 1,500 clones per pool. PCR analysis of 50 clones from each of two randomly selected clone pools revealed that 80.4% (clone pool 3) and 75% (clone pool 11) of the clones contained inserts (data not shown). Given the percentage of recombinant clones, the average insert size, and the number of clone pools, we calculated that all genes are represented in the pVAX-Koz library at least once. This calculation took into account the finding that one-third of the inserts were in frame and that of these, one-half were in the correct orientation. A crude survey was performed, in which we analyzed the 11 clone pools for nine known M. avium subsp. paratuberculosis genes by PCR amplification. Genes were present at different percentages across the clone pools, as follows: alkA, 9.1%; csp1, 0%; groES, 0%; gsbB, 18.2%; hsp65, 45.5%; hspX, 45.5%; IS900, 100%; mig, 9.1%; and pks, 54.5%. No single clone pool contained all nine of the genes assayed. The highest number of selected genes present in a single clone pool was five, and only one clone pool contained five of the genes (data not shown). As expected, the IS900 insertion sequence, which is present at a level of 17 copies in the M. avium subsp. paratuberculosis genome, was detected most frequently among the clone pools (100%). In contrast, neither the csp1 nor groES sequence was detected in any of the clone pools. Taken together, these data demonstrated that the clone pools had some overlaps, as well as some differences, compared with other clone pools.
DNA vaccine trial I.
The protective effect of each DNA vaccine pool was evaluated by comparing the number of viable M. avium subsp. paratuberculosis CFU recovered from the spleens and mesenteric lymph nodes of immunized mice to number of viable M. avium subsp. paratuberculosis CFU recovered from the spleens and mesenteric lymph nodes of no-DNA, infected control mice after challenge. PBS mock-infected (no DNA, noninfected) mice were negative for M. avium subsp. paratuberculosis infection, as expected. The no-DNA, infected mouse positive control treatment group had an average of 2,047 CFU/mg of spleen and 1,240 CFU/mg of mesenteric lymph node (Table 1). Two other control groups (microcarrier, infected; vector DNA, infected) also had high levels of bacterial infection (Table 1), indicating that intraperitoneal infection of BALB/cJ mice with bacterial isolate 6112 was very effective and reproducible. Immunization with clone pools 1, 2, and 10 substantially reduced the numbers of bacteria recovered from the spleens of mice (27 CFU/mg, 11 CFU/mg, and 144 CFU/mg of spleen, respectively). Statistical analysis indicated that these three clone pools resulted in a significant (P < 0.001) reduction in M. avium subsp. paratuberculosis colonization (Table 2). Although less dramatic, clone pools 7 (P < 0.07) and 8 reduced (P < 0.05) bacterial infection in the spleens at least twofold compared to no-DNA, infected control mice (Table 1). By comparison, clone pools 1, 2, 8, and 10 significantly reduced (P < 0.001) the number of bacteria isolated from the mesenteric lymph nodes of immunized mice (Table 1). In contrast, immunization of mice with clone pool 11 resulted in an increase (P < 0.05) in the viable bacteria recovered from mesenteric lymph nodes.
TABLE 1.
Viable bacteria recovered from mice that were vaccinated with clone pool DNA (trial 1) and challenged with live M. avium subsp. paratuberculosisa
| Immunization group | Spleen
|
Mesenteric lymph nodes
|
||||
|---|---|---|---|---|---|---|
| CFU/mgb | SEM | P valuec | CFU/mgb | SEM | P valuec | |
| No DNA, noninfected | 0.0 | 0.0 | 0.0 | 0.0 | ||
| No DNA, infected | 2,047.3 | 478.2 | 1,240.1 | 218.0 | ||
| Microcarrier, infected | 1,422.6 | 264.6 | 0.2680 | 1,538.5 | 298.5 | 0.4295 |
| Vector DNA, infected | 1,604.6 | 268.0 | 0.4299 | 1,246.5 | 270.2 | 0.9855 |
| Clone pool 1 | 27.2 | 26.3 | 0.0001 | 63.5 | 62.4 | 0.0001 |
| Clone pool 2 | 11.4 | 8.9 | 0.0001 | 41.2 | 33.1 | 0.0001 |
| Clone pool 3 | 1,470.0 | 253.6 | 0.3004 | 948.7 | 231.1 | 0.3712 |
| Clone pool 4 | 2,377.7 | 628.2 | 0.6804 | 911.4 | 270.4 | 0.3565 |
| Clone pool 5 | 1,436.7 | 452.9 | 0.3662 | 714.6 | 201.0 | 0.0933 |
| Clone pool 6 | 1,578.9 | 224.2 | 0.3869 | 1,543.7 | 392.5 | 0.5075 |
| Clone pool 7 | 793.6 | 432.2 | 0.0679 | 1,057.2 | 885.6 | 0.8433 |
| Clone pool 8 | 724.4 | 385.5 | 0.0451 | 281.5 | 162.7 | 0.0024 |
| Clone pool 9 | 1,130.3 | 416.7 | 0.1655 | 1,441.9 | 771.9 | 0.8024 |
| Clone pool 10 | 144.2 | 93.7 | 0.0010 | 247.4 | 174.7 | 0.0023 |
| Clone pool 11 | 2,754.2 | 399.5 | 0.2714 | 3,239.0 | 729.4 | 0.0171 |
BALB/c mice were immunized with clone pool DNA (2 μg), boosted 3 weeks later, and challenged intraperitoneally with live M. paratuberculosis (strain 6112; 108 CFU). Mice were necropsied after 3 months of infection, and tissues were processed for culture.
Mean CFU/mg tissue (n = 10).
The values in boldface type are P values for groups that were significantly different than the no-DNA, infected control group (P < 0.05).
TABLE 2.
Sequencing analyses of two clone pools that provided protection against colonization of M. avium subsp. paratuberculosis in BALB/c mice
| Parameter | Clone pool 1 | Clone pool 2 |
|---|---|---|
| No. of Plates | 26 | 24 |
| No. of reactions | 2,496 | 2,304 |
| No. of clones with no data | 187 (7.5)a | 119 (5.2) |
| No. of clones with no insert | 543 (21.8) | 596 (26.0) |
| No. of usable data | 1,767 (70.8) | 1,580 (68.8) |
| No. of nonrepeat clones | 581 (32.8) | 378 (23.9) |
| Average size of clones (bp) | 285 | 342 |
| Total bp | 174,350 | 127,526 |
| % of total genome | 3.9 | 2.8 |
The values in parentheses are percentages.
Serologic analysis of immunized mice.
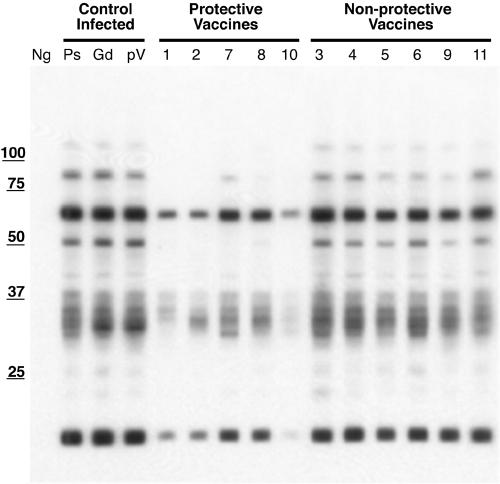
Immunoblot analysis was performed to confirm that the mice mounted an immune response against M. avium subsp. paratuberculosis genes after DNA immunization and bacterial challenge and to assess antibody reactivity between protective and nonprotective clone pools (Fig. 1). Serum from the no-DNA, noninfected group did not recognize any M. avium subsp. paratuberculosis proteins, as expected. Although subtle differences in reactivity profiles were observed for each clone pool, sera from all groups of infected mice reacted strongly with 15-kDa and 65-kDa proteins. Sera from the control infected groups (no DNA, infected; microcarrier, infected; vector DNA, infected) reacted in a manner similar to that of sera from the nonprotective clone pools (clone pools 3, 4, 5, 6, 9, and 11). Interestingly, mice immunized with the clone pools that reduced tissue colonization (clone pools 1, 2, 7, 8, and 10) exhibited reduced reactivity to proteins in the 35-kDa, 50-kDa, and 90-kDa regions (Fig. 1).
FIG. 1.
Immunoblots from control, vaccinated, and challenged mice from DNA vaccine trial I. Sera from mice were blotted against M. avium subsp. paratuberculosis proteins from a whole-cell sonicate to assay for reactivity. Lane Ng, no DNA, noninfected; lane Ps, no DNA, infected; lane Gd, microcarrier, infected; lane pV, vector DNA, infected. Clone pool numbers are indicated above the other lanes. The lines above the lane designations cluster the vaccination groups into infected control groups, protected groups, and nonprotected groups (from left to right). The positions of molecular mass standards (in kDa) are indicated on the left.
Nucleotide sequencing of protective clone pools 1 and 2.
Characterization of sequencing data from protective clone pools suggested that approximately 70.8 and 68.8% of the clones from clone pools 1 and 3, respectively, produced usable data (Table 2). Sequence alignment revealed that there were 581 and 378 nonrepetitive sequences in these clone pools, respectively. Analysis of coding regions revealed that the open reading frame sizes in frame with the cytomegalovirus promoter of pVAX-Koz ranged from 6 to 1,269 bp, with a mean of 120 bp. A total of 174,350 and 127,526 bp of M. avium subsp. paratuberculosis DNA were present in the protective clone pools, representing 3.9 and 2.8% of the genome, respectively.
Generation of clone arrays.
Nucleotide sequence analysis revealed that 581 of the sequenced clones in clone pool 1 contained genomic inserts that were not repeated in the clone pool. Because this clone pool had a much higher number of nonrepeated clones than clone pool 2, it was selected for further assessment of protection in a clonal array format. Of the 581 nonrepeat clones, 536 contained an M. avium subsp. paratuberculosis sequence that coded for an open reading frame larger than 7 amino acids. Clones that contained open reading frames smaller than 8 amino acids (n = 45) were excluded from further analysis because they cannot be loaded onto major histocompatibility complex molecules. The resulting 536 clones were arrayed and retested for protection in the mouse challenge model.
DNA vaccine trial II.
As in the experiment described above, PBS mock-infected (no DNA, noninfected) mice were negative for M. avium subsp. paratuberculosis infection, whereas no-DNA, infected mice had an average of 242 CFU/mg of spleen and 804 CFU/mg of mesenteric lymph node (Table 3). Immunization of mice with clone arrays 1 and 2 significantly (P < 0.001) reduced the number of bacteria isolated from the spleens of mice compared to the no-DNA, infected control group (average, <5 CFU/mg tissue) (Table 3). Altogether, 7 of 10 clone arrays (clone arrays 1, 2, 4, 5, 8, 9, and 10) provided significant (P < 0.05) levels of protection from bacterial colonization in the spleen (Table 3). Clone arrays 2 and 4 were the most effective at preventing bacterial colonization in the mesenteric lymph nodes of mice (<10 CFU/mg of tissue), and 4 of 10 clone arrays (clone arrays 1, 2, 4, and 9) provided significant levels of protection (P < 0.05) (Table 3). Higher levels of bacteria were recovered from the spleens of mice that were immunized with either the parent protective clone pool or the nonrepetitive clone pool than from the spleens of mice in the no-DNA, infected group (723 CFU/mg and 764 CFU/mg of spleen, respectively). Additionally, protection was not observed in the mesenteric lymph nodes of mice immunized with the parent protective clone pool or the nonrepetitive clone pool, although the nonrepetitive clones resulted in a fivefold reduction in bacterial colonization (Table 3). Colonization data for the spleens and mesenteric lymph nodes of immunized mice indicated that clone arrays 1, 2, 4, and 9 provided good protection against M. avium subsp. paratuberculosis infection, while clone array 2 provided almost complete protection of mice from both spleen and mesenteric lymph node infection, as there was negligible recovery of organisms from these tissues.
TABLE 3.
Recovery of viable bacteria from tissues of mice vaccinated with clone array DNA (trial II) and challenged with live M. avium subsp. paratuberculosisa
| Immunization group | Spleen
|
Mesenteric lymph nodes
|
||||
|---|---|---|---|---|---|---|
| CFU/mgb | SEM | P valuec | CFU/mgb | SEM | P valuec | |
| No DNA, noninfected | 0 | 0 | 0 | 0 | ||
| No DNA, infected | 242.49 | 93.51 | 804.21 | 638.61 | ||
| Parent clone pool | 723.41 | 199.83 | 0.1257 | 649.91 | 228.32 | 0.5896 |
| Nonrepetitive clones | 764.88 | 363.74 | 0.6272 | 142.52 | 38.31 | 0.0776 |
| Clone array 1 | 2.74 | 0.68 | 0.0007 | 15.89 | 8.23 | 0.0106 |
| Clone array 2 | 1.34 | 0.33 | 0.0003 | 0.28 | 0.13 | 0.0003 |
| Clone array 3 | 453.08 | 411.48 | 0.5599 | 39.29 | 36.13 | 0.0548 |
| Clone array 4 | 28.88 | 27.76 | 0.0144 | 10.89 | 3.91 | 0.0044 |
| Clone array 5 | 10.91 | 10.5 | 0.0029 | 49.9 | 32.69 | 0.0636 |
| Clone array 6 | 422.96 | 182.77 | 0.8027 | 700.37 | 471.77 | 0.4425 |
| Clone array 7 | 407.82 | 240.27 | 0.6697 | 336.98 | 83.79 | 0.4665 |
| Clone array 8 | 61.47 | 32.78 | 0.0379 | 46.94 | 27.11 | 0.0534 |
| Clone array 9 | 13.43 | 11.83 | 0.0024 | 18.65 | 12 | 0.0116 |
| Clone array 10 | 31.61 | 15.53 | 0.0118 | 89.95 | 54.76 | 0.1212 |
BALB/c mice were immunized with clone pool DNA (2 μg), boosted 3 weeks later, and challenged intraperitoneally with live M. paratuberculosis (strain 6112; 108 CFU). Mice were necropsied after 3 months of infection, and tissues were processed for culture.
Mean CFu/mg tissue (n = 10).
The values in bold-face type are P values for groups that were significantly different than the no-DNA, infected control group (P < 0.05).
Potential protective genes within clone pool arrays.
In order to determine the genes that provided protection from infection, the open reading frames from each nonrepetitive clone (n = 536) were compared to the M. avium subsp. paratuberculosis genome, and predicted gene functions were assigned to each clone. As indicated above, clone arrays 1, 2, 4, and 9 significantly protected mice from bacterial infection in both the spleens and mesenteric lymph nodes; therefore, the analyses focused on these protective clone arrays. Predicted gene functions were separated into seven major categories: transport/binding proteins, membrane proteins, macromolecular metabolism (i.e., macromolecule degradation and synthesis, nucleotide-associated enzymes, polysaccharide degradation and synthesis), IS-like elements, virulence proteins, small-molecule metabolism (i.e., amino acids, cofactors, regulators, fatty acids, energy), and mycobactin/polyketide synthase. Each of the coding regions from the four protective clone arrays was separated into categories for comparison. Genes involved in small-molecule metabolism were most prevalent in all four of the clone arrays. The individual genes present in more than one protective clone array are shown in Table 4. Most notable are members of the proline-rich antigens, PPE family proteins, macrophage cell entry proteins, and mycobactin/exochelin proteins, which were identified in three of four protective clone arrays.
TABLE 4.
Genes present in four clone array pools that provided protection against colonization of mouse tissues with M. avium subsp. paratuberculosis
| General category | Open reading frame | Predicted function | Protective clone arrays
|
|||
|---|---|---|---|---|---|---|
| Array 1 | Array 2 | Array 4 | Array 9 | |||
| Transport/binding proteins | MAP0448 | FisH membrane chaperone | X | X | ||
| MAP1301 | ChaA calcium/proton antiporter | X | X | |||
| MAP1308 | Lgt lipoprotein transferase | X | X | |||
| MAP2491 | OppA oligopeptide transport | X | X | |||
| MAP3498c | Ctpl magnesium transport | X | X | |||
| Membrane proteins | MAP1239c | MmpL4.2 membrane protein | X | X | ||
| MAP1493c | Integral protein | X | X | |||
| MAP1912 | Membrane protein | X | X | |||
| MAP2239 | Membrane protein | X | X | |||
| MAP3049c | Mmpl.2 large membrane protein | X | X | |||
| MAP3131 | Large membrane protein | X | X | |||
| MAP3171c | FisX membrane protein | X | X | |||
| Virulence proteins | MAP0047c | Proline-rich antigen | X | X | X | |
| MAP1003c | PE/PPE family protein | X | X | |||
| MAP2191 | Mce4 macrophage cell entry | X | X | |||
| MAP2192 | Mce4 macrophage cell entry | X | X | X | ||
| MAP3737 | PPE family protein | X | X | X | ||
| Mycobactin/polyketide synthase | MAP1796c | Pks12 polyketide synthase | X | X | ||
| MAP1871c | Polyketide synthase | X | X | |||
| MAP2171c | MbtF mycobactin/exochelin | X | X | |||
| MAP2174c | MbtD mycobactin/exochelin | X | X | X | ||
| MAP2175c | MbtC mycobactin/exochelin | X | X | |||
| MAP2230c | Polyketide synthase | X | X | X | ||
| MAP2604c | Polyketide synthase | X | X | |||
| MAP3742 | Polyketide synthase | X | X | |||
| MAP3764c | Pks2 polyketide synthase | X | X | X | ||
DISCUSSION
In contrast to the present study, DNA vaccines for M. tuberculosis have largely been based on one or more specific genes encoding proteins that induce gamma interferon secretion (31, 34, 47). It has been demonstrated that a single antigen, mycobacterial heat shock protein Hsp65, was effective in protecting mice against challenge with M. tuberculosis, reducing infection in the lung by 2 log10 (35). Other studies have shown that there is reduced tissue infection after challenge in mice or guinea pigs immunized either with Hsp65, Apa, or a combination of ESAT-6 and Ag85A coding sequences (31, 35, 47). The advantage of expression library immunization is that all antigens encoded in a pathogen's genome can be screened simultaneously to identify protective sequences.
Using expression library immunization (10), a high-throughput DNA immunization technique, thousands of bacterial genes can be screened simultaneously to identify protective sequences. This immunization technique, as described above, proceeds through successive rounds of immunization and library segregation to ultimately isolate a single or select group of protective sequences. Sequencing a protective clone pool provided information about potential protective sequences in the pool. Because high-throughput sequencing is performed in a 96-well format, every sequence can be traced back to its original clone for further manipulation. The in silico screening accomplished by nucleotide sequencing resulted in two important results. First, a 64% reduction in clone number was achieved in less than 2 weeks. This level of clone reduction would have otherwise required an 8-month immunization and challenge experiment with a group of 150 mice. Second, because we obtained the nucleotide sequence of every clone in a protective clone pool, subsequent experiments could be directed with specific knowledge of which genes offered protection. Additionally, the 10 new clone arrays were generated so that any two clone arrays shared 17 clones. This was essential to account for the possible additive effects of multiple clones in providing protection. Ideally, expression library immunization aims to identify one protective sequence; however, it is more likely that multiple coding regions provide protection, and their combined use may provide even greater levels of protection.
Our preliminary studies (data not shown) demonstrated that we could deliver and express the β-galactosidase gene in mice. Expression was observed in all tissues examined, including those peripheral to the transfection site (i.e., spleen, inguinal lymph node, and mesenteric lymph node). This suggests that antigen-presenting cells were directly transfected with plasmid and migrated throughout the body, an essential component for generating a systemic immune response. In addition, detection of β-galactosidase up to 140 days postimmunization demonstrated that antigens encoded by DNA vaccines are not immediately cleared but are available for immune stimulation for prolonged periods of time. These data are supported by a previous study, which reported that plasmid-encoded luciferase was expressed for at least 19 months after intramuscular injection in mice (57). Taken together, the systemic and prolonged antigen expression demonstrated that the pVAX-Koz vector and gene gun delivery method was effective for expressing a cloned gene in mice.
Using the protocol for DNA immunization and bacterial challenge described above, we were able to demonstrate that mice were protected against challenge with M. avium subsp. paratuberculosis when they were immunized with four different clone pools of naked DNA. Further partitioning of a protective clone pool into clone arrays resulted in effective protection as well. Bacterial infection was dramatically reduced in both the spleens and mesenteric lymph nodes of mice immunized with DNA from these clone pools compared to the infected control groups and other mouse clone pool groups. Surprisingly, the parent protective clone pool did not reduce bacterial infection in mice compared to the no-DNA, infected group in the second vaccine trial. After resampling of the parent clone pool, it is likely that a different mixture of clones was obtained from the heterogeneous population of clones present. Regardless, results from this study demonstrated that there was significant protection when mice were vaccinated with four different clone arrays.
The coding sequences that were shared by the four protective clone arrays were determined, and a detailed list of the 26 proteins that could potentially protect mice from M. avium subsp. paratuberculosis challenge was prepared. Among the shared proteins, the mycobacterial cell membrane-associated protease, FtsH, has been shown to induce antibody production in M. tuberculosis patients (1). The mycobacterial major membrane protein (MMP) has been reported to induce both cellular and humoral immune responses in M. leprae-infected individuals (42, 52), and a DNA vaccine study showed that MMP protects mice from leprosy challenge (36). In addition, the MMP has recently been shown to play a role in invasion of macrophages by M. avium subsp. paratuberculosis (6). Proline-rich antigens which induce both T- and B-cell responses in M. tuberculosis and M. leprae have been described (28, 49, 50). Furthermore, a DNA vaccine encoding the M. tuberculosis 36-kDa proline-rich antigen protected mice from challenge with live bacteria (49). The PE family of antigens is named after their proline- and glutamic acid-rich N-terminal ends. These antigens are highly antigenic, are located on the cell wall and cell membrane, and are involved in macrophage infection and cell-to-cell interactions (9, 12, 21). The macrophage cell entry protein, Mce, is a virulence factor involved in macrophage entry and survival (2, 16, 25). In addition, the polyketide synthase family of genes produces lipid-like molecules with a wide variety of functions, including iron acquisition and virulence (4, 46). The genes mentioned above were well represented in the protective clone array pools and likely contributed to the reduced tissue colonization observed in mice challenged with M. avium subsp. paratuberculosis.
Immunoblot analysis confirmed that mice generated humoral immune responses to vaccination and challenge. Reactivity to 15-kDa and 65-kDa M. avium subsp. paratuberculosis proteins was consistently observed in all groups of challenged mice. While the size and identity of the 15-kDa protein is speculative, it is quite possible that the 65-kDa protein is a homologue of the M. tuberculosis heat shock protein, Hsp65 (35). More importantly, sera from mice vaccinated with DNA from nonprotective clone pools recognized 35-kDa, 50-kDa, and 90-kDa proteins, while sera from protective clone groups did not recognize, or weakly recognized, the same proteins. The identities of these proteins are not known, and whether the differences in immune responses were a direct result of protection from challenge remains to be determined.
In summary, the present study demonstrated that expression library immunization of mice is an effective method of vaccination. This is the first study in which DNA vaccination as a method of protection against challenge with M. avium subsp. paratuberculosis was evaluated. In the present study we were able to demonstrate protection by reduced colonization in tissues of mice after vaccination with gene pools that contained protective sequences.
Supplementary Material
Acknowledgments
We thank Dong Wang at the Agricultural Experiment Station Consulting Service, Department of Statistics, Iowa State University, Ames, for performing the statistical analysis on the DNA vaccine data. We especially thank David Alt and the NADC Genomics Facility staff, who provided excellent assistance with clone pool sequencing. Finally, we gratefully acknowledge the other members of the Johne's Disease Research Project at the NADC for their assistance during the mouse vaccination study. The lab members include Trudy Bosworth, Janis Hansen, Tonia McNunn, and Bart Olthoff.
Editor: J. L. Flynn
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Amara, R. R., S. Shanti, and V. Satchidanandam. 1998. Characterization of novel immunodominant antigens of Mycobacterium tuberculosis. Microbiology 144:1197-1203. [DOI] [PubMed] [Google Scholar]
- 2.Arruda, S., G. Bomfim, R. Knights, T. Huima-Byron, and L. W. Riley. 1993. Cloning of an Mycobacterium tuberculosis DNA fragment associated with entry and survival inside cells. Science 261:1454-1457. [DOI] [PubMed] [Google Scholar]
- 3.Babiuk, L. A., S. L. Babiuk, B. I. Loehr, and S. van den Hurk. 2000. Nucleic acid vaccines: research tool or commercial reality. Vet. Immunol. Immunopathol. 76:1-23. [DOI] [PubMed] [Google Scholar]
- 4.Bannantine, J. P., and J. R. Stabel. 2001. Identification of two Mycobacterium avium subspecies paratuberculosis gene products differentially recognized by sera from rabbits immunized with live mycobacteria but not heat-killed mycobacteria. J. Med. Microbiol. 50:795-804. [DOI] [PubMed] [Google Scholar]
- 5.Bannantine, J. P., and J. R. Stabel. 2000. HspX is present within Mycobacterium paratuberculosis-infected macrophages and is recognized by sera from some infected cattle. Vet. Microbiol. 76:343-358. [DOI] [PubMed] [Google Scholar]
- 6.Bannantine, J. P., J. F. Huntley, E. Miltner, J. R. Stabel, and L. E. Bermudez. 2003. The Mycobacterium avium subsp. paratuberculosis 35kDa protein plays a role in invasion of bovine epithelial cells. Microbiology 149:2061-2069. [DOI] [PubMed] [Google Scholar]
- 7.Bannantine, J. P., Q. Zhang, L. Li, and V. Kapur. 2003. Genomic homogeneity between Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis belies their divergent growth rates. BMC Microbiol. 3:10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannantine, J. P., J. K. Hansen, M. L. Paustian, A. Amonsin, L. L. Li, J. R. Stabel, and V. Kapur. 2004. Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banu, S., N. Honore, B. Saint-Joanis, D. Philpott, M. C. Prevost, and S. T. Cole. 2002. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 44:9-19. [DOI] [PubMed] [Google Scholar]
- 10.Barry, M. A., and S. A. Johnston. 1997. Biological features of genetic immunization. Vaccine 15:788-791. [DOI] [PubMed] [Google Scholar]
- 11.Barry, M. A., W. C. Lai, and S. A. Johnston. 1995. Protection against mycoplasma infection using expression-library immunization. Nature 377:632-635. [DOI] [PubMed] [Google Scholar]
- 12.Brennan, M. J., G. Delogu, Y. Chen, S. Bardarov, J. Kriakov, M. Alavi, and W. R. Jacobs. 2001. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 69:7326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El-Zaatari, and M. Tizard. 2000. Characterization of IS900 loci on Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146:2185-2197. [DOI] [PubMed] [Google Scholar]
- 14.Chiodini, R. J. 1993. Abolish Mycobacterium paratuberculosis strain 18. J. Clin. Microbiol. 31:1956-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiodini, R. J., H. J. Van Kruiningen, and R. S. Merkal. 1984. Ruminant paratuberculosis (Johne's disease): the current status and future prospects. Cornell Vet. 74:218-262. [PubMed] [Google Scholar]
- 16.Chitale, S., S. Ehrt, I. Kawamura, T. Fujimura, N. Shimono, N. Anand, S. Lu, L. Cohen-Gould, L. W. Riley. 2001. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 3:247-254. [DOI] [PubMed] [Google Scholar]
- 17.Cobb, A. J., and R. Frothingham. 1999. The GroES antigens of Mycobacterium avium and Mycobacterium paratuberculosis. Vet. Microbiol. 67:31-35. [DOI] [PubMed] [Google Scholar]
- 18.Cocito, C., P. Gilot, M. Coene, M. de Kesel, P. Poupart, and P. Vannuffel. 1994. Paratuberculosis. Clin. Microbiol. Rev. 7:328-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins, M. T. 1996. Clinical approach to control of bovine paratuberculosis. J. Am. Vet. Med. Assoc. 204:208-210. [PubMed] [Google Scholar]
- 20.Colston, A., I. McConnell, and R. Bujdoso. 1994. Cloning and expression in Escherichia coli of DNA encoding a 60 kDa stress protein of Mycobacterium paratuberculosis, the causative agent of Johne's disease. Microbiology 140:3329-3336. [DOI] [PubMed] [Google Scholar]
- 21.Delogu, G., and M. J. Brennan. 2001. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect. Immun. 69:5606-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellingson, J. L., C. Bolin, and J. R. Stabel. 1998. Identification of a gene unique to Mycobacterium avium subsp. paratuberculosis and application to diagnosis of paratuberculosis. Mol. Cell. Probes 12:133-142. [DOI] [PubMed] [Google Scholar]
- 23.Fridriksdottir, V., E. Gunnarsson, S. Sigurdarson, and K. B. Gudmundsdottir. 2000. Paratuberculosis in Iceland: epidemiology and control measures. Vet. Microbiol. 77:263-267. [DOI] [PubMed] [Google Scholar]
- 24.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou, J. Y., J. E. Graham, and J. E. Clark-Curtiss. 2002. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect. Immun. 70:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huygen, K. 2003. On the use of DNA vaccines for the prophylaxis of mycobacterial diseases. Infect. Immun. 71:1613-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalis, C. H., J. W. Hesselink, H. W. Barkema, and M. T. Collins. 2001. Use of long-term vaccination with a killed vaccine to prevent fecal shedding of Mycobacterium avium subsp. paratuberculosis in dairy herds. Am. J. Vet. Res. 62:270-274. [DOI] [PubMed] [Google Scholar]
- 28.Klatser, P. R., M. Y. De Wit, A. H. Kolk, and R. A. Hartskeerl. 1991. Characterization of murine B-cell epitopes on the Mycobacterium leprae proline-rich antigen by use of synthetic peptides. Infect. Immun. 59:433-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohler, H., H. Gyra, K. Zimmer, K. G. Drager, B. Burkert, B. Lemser, D. Hausleithner, K. Cubler, W. Klawonn, and R. G. Hess. 2001. Immune reactions in cattle after immunization with a Mycobacterium paratuberculosis vaccine and implications for the diagnosis of M. paratuberculosis and M. bovis infections. J. Vet. Med. Ser. B 48:185-195. [DOI] [PubMed] [Google Scholar]
- 30.Körmendy, B. 1994. The effect of vaccination on the prevalence of paratuberculosis in large dairy herds. Vet. Microbiol. 41:117-125. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, P., R. R. Amara, V. K. Challu, V. K. Chadda, and V. Satchidanandam. 2003. The Apa protein of Mycobacterium tuberculosis stimulates gamma interferon-secreting CD4+ and CD8+ T cells from purified protein derivative-positive individuals and affords protection in a guinea pig model. Infect. Immun. 71:1929-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen, A. B., A. I. Moyle, and E. M. Himes. 1978. Experimental vaccination of cattle against paratuberculosis (Johne's disease) with killed bacterial vaccines: a controlled field study. Am. J. Vet. Res. 39:65-69. [PubMed] [Google Scholar]
- 33.Larsen, A. B., R. S. Merkal, and R. C. Cutlip. 1975. Age of cattle as related to resistance to infection with Mycobacterium paratuberculosis. Am. J. Vet. Res. 36:255-257. [PubMed] [Google Scholar]
- 34.Lima, K. M., S. A. Santos, V. M. Lima, A. A. Coelho-Castelo, J. M. Rodrigues, and C. L. Silva. 2003. Single dose of a vaccine based on DNA encoding mycobacterial hsp65 protein plus TDM-loaded PLGA microspheres protects mice against a virulent strain of Mycobacterium tuberculosis. Gene Ther. 10:678-685. [DOI] [PubMed] [Google Scholar]
- 35.Lowrie, D. B., R. E. Tascon, V. L. Bonato, V. M. Lima, L. H. Faccioli, E. Stavropoulos, M. J. Colston, R. G. Hewinson, K. Moelling, and C. L. Silva. 1999. Therapy of tuberculosis in mice by DNA vaccination. Nature 400:269-271. [DOI] [PubMed] [Google Scholar]
- 36.Martin, E., P. W. Roche, J. A. Triccas, and W. J. Britton. 2001. DNA encoding a single mycobacterial antigen protects against leprosy infection. Vaccine 19:1391-1396. [DOI] [PubMed] [Google Scholar]
- 37.Mullerad, J., A. H. Hovav, R. Nahary, Y. Fishman, and H. Bercovier. 2003. Immunogenicity of a 16.7 kDa Mycobacterium paratuberculosis antigen. Microb. Pathog. 34:81-90. [DOI] [PubMed] [Google Scholar]
- 38.Mullerad, J., A. H. Hovav, Y. Fishman, R. G. Barletta, and H. Bercovier. Antigenicity of Mycobacterium paratuberculosis superoxide dismutase in mice. FEMS Immunol. Med. Microbiol. 34:81-88. [DOI] [PubMed]
- 39.Mullerad, J., I. Michal, Y. Fishman, A. H. Hovav, R. G. Barletta, and H. Bercovier. 2002. The immunogenicity of Mycobacterium paratuberculosis 85B antigen. Med. Microbiol. Immunol. 190:179-187. [DOI] [PubMed] [Google Scholar]
- 40.Muskens, J., F. van Zijderveld, A. Eger, and D. Bakker. 2002. Evaluation of the long-term immune response in cattle after vaccination against paratuberculosis in two Dutch dairy herds. Vet. Microbiol. 86:269-278. [DOI] [PubMed] [Google Scholar]
- 41.Naser, S. A., J. Felix, H. Liping, C. Romero, N. Naser, A. Walsh, and W. Safranek. 1999. Occurrence of the IS900 gene in Mycobacterium avium complex derived from HIV patients. Mol. Cell. Probes 13:367-372. [DOI] [PubMed] [Google Scholar]
- 42.Ohyama, H., S. Matsushitab, F. Nishimuraa, N. Katoa, K. Hatanoc, S. Takashibaa, and Y. Murayama. 2001. T cell responses to major membrane protein II (MMP II) of Mycobacterium leprae are restricted by HLA-DR molecules in patients with leprosy. Vaccine 42:475-482. [DOI] [PubMed] [Google Scholar]
- 43.Olsen, I., and A. K. Storset. 2001. Innate IFN-gamma production in cattle in response to MPP14, a secreted protein from Mycobacterium avium subsp. paratuberculosis. Scand. J. Immunol. 54:306-313. [DOI] [PubMed] [Google Scholar]
- 44.Patterson, C. J., M. LaVenture, S. S. Hurley, and J. P. Davis. 1988. Accidental self-inoculation with Mycobacterium paratuberculosis bacterin (Johne's bacterin) by veterinarians in Wisconsin. J. Am. Vet. Med. Assoc. 192:1197-1199. [PubMed] [Google Scholar]
- 45.Plum, G., M. Brenden, J. E. Clark-Curtiss, and G. Pulverer. 1997. Cloning, sequencing, and expression of the mig gene of Mycobacterium avium, which codes for a secreted macrophage-induced protein. Infect. Immun. 65:4548-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rousseau, C., T. D. Sirakova, V. S. Dubey, Y. Bordat, P. E. Kolattukudy, B. Gicquel, and M. Jackson. 2003. Virulence attenuation of two Mas-like polyketide synthase mutants of Mycobacterium tuberculosis. Microbiology 149:1837-1847. [DOI] [PubMed] [Google Scholar]
- 47.Skinner, M. A., A. J. Ramsay, G. S. Buchan, D. L. Keen, C. Ranasinghe, L. Slobbe, D. M. Collins, G. W. de Lisle, and B. M. Buddle. 2003. A DNA prime-live boost strategy in mice can augment IFN-γ responses to mycobacterial antigens but does not increase the protective efficacy of two attenuated strains of Mycobacterium bovis against bovine tuberculosis. Immunology 108:548-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stabel, J. R. 2000. Johne's disease and milk: do consumers need to worry? J. Dairy Sci. 83:1659-1663. [DOI] [PubMed] [Google Scholar]
- 49.Tascon, R. E., M. J. Colston, S. Ragno, E. Stavropoulos, D. Gregory, and D. B. Lowrie. 1996. Vaccination against tuberculosis by DNA injection. Nat. Med. 28:888-892. [DOI] [PubMed] [Google Scholar]
- 50.Thole, J. E., L. F. Stabel, M. E. Suykerbuyk, M. Y. De Wit, P. R. Klatser, A. H. Kolk, and R. A. Hartskeerl. 1990. A major immunogenic 36,000-molecular-weight antigen from Mycobacterium leprae contains an immunoreactive region of proline-rich repeats. Infect. Immun. 58:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tizard, M., T. Bull, D. Millar, T. Doran, H. Martin, N. Sumar, J. Ford, and J. Hermon-Taylor. 1998. A low G+C content genetic island in Mycobacterium avium subsp. paratuberculosis and M. avium subsp. silvaticum with homologous genes in Mycobacterium tuberculosis. Microbiology 144:3413-3423. [DOI] [PubMed] [Google Scholar]
- 52.Triccas, J. A., P. W. Roche, N. Winter, C. G. Feng, C. R. Butlin, and W. J. Britton. 1996. A 35-kilodalton protein is a major target of the human immune response to Mycobacterium leprae. Infect. Immun. 64:5171-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uzonna, J. E., P. Chilton, R. H. Whitlock, P. L. Habecker, P. Scott, and R. W. Sweeney. 2003. Efficacy of commercial and field-strain Mycobacterium paratuberculosis vaccinations with recombinant IL-12 in a bovine experimental infection model. Vaccine 21:3101-3109. [DOI] [PubMed] [Google Scholar]
- 54.Van Schaik, G., C. H. Kalis, G. Benedictus, A. A. Dijkuizen, and R. B. Huirne. 1996. Cost-benefit analysis of vaccination against paratuberculosis in dairy cattle. Vet. Rec. 139:624-627. [PubMed] [Google Scholar]
- 55.Velaz-Faircloth, M., A. J. Cobb, A. L. Horstman, S. C. Henry, and R. Frothingham. 1999. Protection against Mycobacterium avium by DNA vaccines expressing mycobacterial antigens as fusion proteins with green fluorescent protein. Infect. Immun. 67:4243-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wentink, G. H., J. H. Bongers, A. A. Zeeuwen, and F. H. Jaartsveld. 1994. Incidence of paratuberculosis after vaccination against M. paratuberculosis in two infected dairy herds. J. Vet. Med. 41:517-522. [DOI] [PubMed] [Google Scholar]
- 57.Wolff, J. A., J. J. Ludtke, G. Acsadi, P. Williams, and A. Jani. 1992. Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle. Hum. Mol. Genet. 1:363-369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.