Abstract
Brucella spp. are stealthy bacteria that enter host cells without major perturbation. The molecular mechanism involved is still poorly understood, although numerous studies have been published on this subject. Recently, it was reported that Brucella abortus utilizes cellular prion protein (PrPC) to enter the cells and to reach its replicative niche. The molecular mechanisms involved were not clearly defined, prompting us to analyze this process using blocking antibodies against PrPC. However, the behavior of Brucella during cellular infection under these conditions was not modified. In a next step, the behavior of Brucella in macrophages lacking the prion gene and the infection of mice knocked out for the prion gene were studied. We observed no difference from results obtained with the wild-type control. Although some contacts between PrPC and Brucella were observed on the surface of the cells by using confocal microscopy, we could not show that Brucella specifically bound recombinant PrPC. Therefore, we concluded from our results that prion protein (PrPC) was not involved in Brucella infection.
The six Brucella species are gram-negative bacteria that cause brucellosis in human and animals, a disease also known as Malta fever. Brucella spp. are facultative intracellular pathogens and infect a variety of cells including “professional” and “nonprofessional” phagocytes (14, 19, 22). Many aspects of the pathophysiology of the disease are still unclear (establishment of chronicity, for example), but it is assumed that the intracellular location is essential for bacterial multiplication and virulence (22). After penetration inside the macrophages, Brucella-containing vacuoles traffic in a complex manner (8) to become the replicative niche of the pathogen: the brucellosome (18).
Attachment to the host membrane and phagocytosis are the first steps leading to the intracellular stages, but little is known about the molecular mechanisms involved. The use of inhibitors or antibodies against putative receptors has led only to incomplete inhibition. For instance, bacterial entry into bovine macrophages was partially inhibited by the peptide RGDS, the outer membrane-peptidoglycan complex from Brucella abortus strain RB51, anti-lymphocyte function-associated antigen-1 monoclonal antibody, anti-C3 antiserum, fibronectin, purified O antigen from B. abortus lipopolysaccharide (LPS), and mannan- and heat-aggregated immunoglobulin G (5). On the other hand, recent studies have presented data suggesting that the adherence mechanism of Brucella to macrophages is mediated by cellular receptors containing sialic acid and sulfated residues, explaining the affinity for proteins of the cellular matrix (6).
Recent advances in cellular biology have attracted the attention of microbiologists to the importance of membrane cellular structures, namely, lipid rafts that exhibit many specific functions, the ability to concentrate signaling molecules in particular. An increasing number of bacteria and their products have been shown to interact with lipid rafts to promote infection (20). Brucella needs functional lipid rafts to enter macrophages (24, 36), as the disruption of lipid rafts markedly inhibits internalization and intracellular replication, indicating that the path of entry into macrophages determines the intracellular fate of the bacteria and shapes phagosome maturation (24). In particular, it was suggested that raft elements incorporated into the phagosomes containing Brucella modulate their maturation into replicative vesicles, probably by the initiation of a signaling transduction cascade (36). Some of these raft elements determining intracellular fate of the bacteria have been identified as cholesterol, gangliosides (e.g., GM1), glycosphingolipids, and glycosylphosphatidylinositol (GPI)-anchored proteins. Nevertheless, it can be supposed that bacterial phagocytosis involves many other interactions between bacterial membrane and cellular partners that are still undefined. However, because penetration needs functional rafts, it has been suggested that the cellular receptor for Brucella would be a GPI-anchored protein known to be localized inside these cholesterol-rich structures. This point of view was recently adopted by Watarai et al. (35), who concluded that the cellular prion protein (PrPC) promotes infection and could thus be one receptor for the bacteria on the membrane of macrophages.
PrPC is a 231-amino-acid GPI protein anchored on the outer leaflet of the plasma membrane of many cell types (17). It was remarkably conserved during mammalian evolution and is ubiquitously expressed in organisms, predominantly in the central nervous system. PrPC is the cellular, nonpathogenic homologue of PrPSc (for prion protein scrapie), which is suspected according to Prusiner's hypothesis (30) to be the unconventional agent responsible of transmissible spongiform encephalopathies. The two proteins share a common primary structure but differ in their tertiary structure: PrPSc exhibits a majority of β-sheets that aggregate into β-amyloid fibrils. On the contrary, PrPC exhibits mainly α-helix conformation (37). The physiological functions of PrPC are still poorly understood, and the protein has been implicated in many functions such as protection from oxidative insults, apoptosis, cellular signaling, membrane excitability and synaptic transmission, neuritogenesis, and copper(II) transport or metabolism (17, 21, 29). But how all these functions are achieved by the same protein is still enigmatic. PrPC is located in rafts of the plasmic membrane to which it is attached by its GPI anchor. This localization is compatible with a role as a membrane receptor and with cellular signaling. The use of PrPC by B. abortus (35) could be an interesting opportunity to better understand the function of this protein.
In consequence, we decided to further analyze the mechanisms involving cellular prion protein during infection of macrophages by Brucella. Surprisingly, we were not able to find any evidence of the participation of cellular prion protein during infection process, even with different species of Brucella.
MATERIALS AND METHODS
Bacteria and culture.
Wild-type (WT) strains of Brucella were American Type Culture Collection (ATCC) strains: Brucella suis 1330 S1 (ATCC 23444), B. abortus A1 544 (ATCC 23448), and B. abortus 2308/2665 (ATCC 9539). B. suis ΔmanB, B. suis WT, B. abortus 2308, constitutively expressing green fluorescent protein (GFP), were prepared as described previously for B. suis 1330 p/sog (26). B. suis virB4::Tn5 and B. suis virB5::Tn5 were described previously (19). All strains were grown overnight in tryptic soy (TS) broth (Becton Dickinson Biosciences, France) at 37°C to stationary phase; bacteria were harvested by centrifugation, washed in phosphate-buffered saline (PBS), and resuspended in serum-free RPMI 1640/Ultraglutamine medium (BioWhittaker, Tebu-bio, France) before infection of cells. The appropriate antibiotics were added at the following concentrations when needed: ampicillin, 50 μg/ml; and kanamycin, 50 μg/ml.
Macrophage-like cell lines.
The murine myelomonocytic cell line J774A.1 (ATCC TIB 67), used for infection assays, was cultured in RPMI 1640/Ultraglutamine medium supplemented with 10% heat-inactivated fetal calf serum (FCS) (Sigma-Aldrich, St. Louis, MO) at 37°C and 5% CO2.
Human THP-1 cells were obtained from ATCC. Cells were maintained at 37°C in 5% CO2 and RPMI 1640/Ultraglutamine medium supplemented with 10% (vol/vol) heat-inactivated FCS. Before infection experiments, THP-1 cells were differentiated into macrophages with 1α,25-dihydroxyvitamin D3 at a concentration of 10−7 M for 72 h.
Macrophages derived from mouse bone marrow.
Bone marrow of three mice was extracted from femurs and tibias. Myeloid precursors were pooled in serum-free RPMI 1640 medium and washed. Then, precursor cells were plated at 106 cells/well and differentiated into macrophages during 5 days in RPMI 1640 medium containing 10% (vol/vol) heat-inactivated FCS, and murine macrophage colony-stimulating factor at 50 U/ml (AbCys, France) at 37°C and 5% CO2.
Mice strains.
Control C57BL/6 mice, 8 to 14 weeks old, were purchased from Janvier (France) and kept in a specific pathogen-free animal facility. The congenic PrP-null mice of equivalent ages were described previously (15). These mice were derived from the Zurich strain described previously (4). They were iteratively backcrossed with C57BL/6 progenitors. The mice used in the present study were homozygous offspring derived from the 10th backcross. Each set of PrP−/− mice was controlled for the absence of PrPC by Western blot analysis of brain tissue.
Antibodies and recombinant prion protein.
All monoclonal antibodies against prion protein (SAF32, 12F10, and SAF61) were purchased from SPIBIO (France), except for 3F4, which was obtained from Senetek (France). Rabbit polyclonal antibody P45-66, raised against the synthetic peptide encompassing mouse PrP residues 45 to 66 (CGGNRYPPQGGTWGQPHGGGWGQ) was a gift of S. Lehmann (Institut de Génétique Humaine, Montpellier, France). All antibodies used as isotypic controls and as secondary antibodies were as follows: goat anti-mouse antibodies labeled with tetramethyl rhodamine isothiocyanate were purchased from Beckman Coulter (Immunotech, France), except for the anti-mouse secondary antibody labeled with fluoprobe 546 and purchased from FluoProbes (Interchim, France). Anti-Escherichia coli GroEL mouse monoclonal antibody 9A1/2 was obtained from Calbiochem (Tebu-bio, France). For competition experiments, we utilized anti-PrP monoclonal antibodies at a concentration of 10 μg/ml and polyclonal antibodies at 1/20 dilutions. The same concentrations of antibodies were used for the controls.
Syrian hamster recombinant prion protein (ShaPrP90-231) production was described previously (1). For labeling of recombinant PrP (rPrP) and RNase (Sigma-Aldrich) with fluorescein isothiocyanate (FITC), the two purified, unlabeled proteins were incubated with a solution of carboxyfluorescein succinimidyl ester (NH-FITC in dimethyl sulfoxide; Molecular Probes, Invitrogen, France) in an 0.5 M bicarbonate buffer for 2 h under rotation. Then, FITC-labeled proteins were purified by gel filtration chromatography on a G10 Sephadex column.
Preparation of GroEL-coated beads.
To control for GroEL recognition by the 9A1/2 antibody, latex beads (LB11-1ML; Sigma-Aldrich) were coated with recombinant E. coli GroEL (SPP-610J; Tebu-Bio, France). The latex beads, diluted to 2% in 25 mM MES (morpholineethanesulfonic acid) buffer, pH 6.1, were mixed with a solution of the recombinant protein at 1 mg/ml and incubated overnight with constant rotation. The beads were then washed twice and suspended in stock solution (0.1 M PBS, 0.1% bovine serum albumin, and 0.05% NaN3, pH 7.2). For labeling experiments, 5 μl of beads coated with GroEL was incubated with 9A1/2 monoclonal antibody at a concentration of 5 μg/ml.
Infection of macrophage cell lines.
Cells were cultured in a 24-well plate at 105 cells/well for J774 and 5.105 cells/well for THP-1 for 1 day prior to infection. Bacteria were added to the cells at a multiplicity of infection of 20 to 30 for 45 min. Extracellular bacteria were removed, and cells were washed and maintained in medium containing 30 μg/ml gentamicin (Sigma-Aldrich) to kill residual extracellular bacteria. At 90 min and 7, 24, and 48 h postinfection, cells were lysed in 0.2% Triton X-100, and dilutions of lysates were plated on TS agar. After 3 days, bacterial colonies were counted, and the number of CFU per well was determined. For phagocytosis experiments with fluorescent bacteria, the multiplicity of infection used was approximately 50.
Mouse infection protocol.
To analyze the influence of PrP on in vivo infection by B. suis, C57BL/6 control mice and mice lacking the prnP gene were infected with the B. suis 1330 strain. A total of 105 bacteria per mouse were injected intraperitoneally. At 7 and 16 days after infection, animals were euthanized, and spleens were removed. Organs were suspended in 10 ml of PBS and homogenized individually in tissue grinders. The number of CFU/spleen was determined by plating serial dilution onto TS agar plates as described.
Fluorescence microscopy.
Macrophages were cultured on Lab-Tek chambered coverslips (Nunc, Naperville, Ill.) at a concentration of 105 cells/well. We performed the infection protocol as described above, and results of infection were examined by classical fluorescence microscopy with an inverted Leica DM IRB microscope equipped with a Leica DFC350 FX digital camera. The percentage of phagocytosis was calculated by counting cells that had ingested at least one bacterium and the total number of cells per field.
Confocal microscopy.
For experiments needing confocal precision, a Leica DM IRB microscope was used. Cells were grown on glass coverslips and infected for different times with B. suis or B. abortus expressing green fluorescent protein. Then cells were fixed for 4 min with 3.7% formaldehyde in PBS and washed twice in PBS. For short infection times, i.e., 5 min and 15 min, cells were not permeabilized. On the contrary, for longer times of infection (i.e., 30 min), cells were permeabilized for 10 min with 0.05% saponin in PBS before being labeled. To label PrPC, cells were incubated for 3 h under saturating conditions with SAF61 antibody, then washed, and incubated for 1 h with the anti-mouse secondary antibody labeled with fluoprobe 546. Fusion experiments were performed by incubating cells with dextran-Texas red at 10 μg/ml as described, 16 h before the infection with GFP bacteria (24). Analysis was performed 1 h after infection.
Statistical analysis.
Statistical analysis was performed on the results of at least three independent experiments, thus taking into account the biological variability. Because normality tests (Kolmogorov-Smirnov) generally failed, we used the Mann-Whitney nonparametric test to compare the means.
RESULTS
Anti-PrP antibodies do not modify intracellular behavior of B. suis.
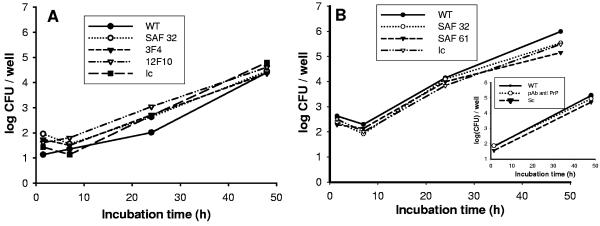
Watarai et al. (35) have published results showing that B. abortus utilizes the cellular prion protein to penetrate macrophages and to initiate multiply intracellular multiplication. However, their results do not demonstrate direct interaction between PrPC and bacteria; we therefore addressed the question as to whether such a direct interaction occurs during Brucella penetration into macrophages. To this end, we preincubated THP-1 cells, a macrophage-like human cell line expressing PrPC at its surface, with monoclonal antibodies reacting against human prion protein (i.e., SAF32, 3F4, and 12F10) before infection with B. suis. Following this treatment, flow cytometry analysis of the membrane expression revealed a reduction of approximately 20% of the surface-located PrPC labeling (data not shown); considering the concentration of antibodies used (10 μg/ml), we assume that most of surface prion proteins reacted. Infection was performed for 30 min after antibody treatment, and intracellular development of Brucella was analyzed. The results did not reveal any difference in the intracellular behavior of bacteria (neither in entry nor in multiplication), suggesting that B. suis did not interact directly with cellular PrP (Fig. 1A). To test if this result was specific to the cell line used, we repeated the experiment with the murine J774 macrophage cell line and monoclonal antibodies that target mouse cellular PrP (i.e., SAF32 and SAF 61). Again, we did not observe a significant difference in the intracellular behavior of bacteria after treatment with the different antibodies (Fig. 1B). The possibility that these two cell lines did not express PrPC at the cell surface was ruled out by Western blot and fluorescence-activated cell sorter analysis (not shown). Furthermore, it could be argued that the monoclonal antibodies used were not blocking, and the experiment was therefore performed with polyclonal antibodies. The results were identical to those described above (Fig. 1B, insert).
FIG. 1.
Antibodies against prion protein (PrPC) do not interfere with the intracellular behavior of B. suis. (A) Multiplication of B. suis in THP-1 human macrophages treated with antibodies against PrPC. WT, without pretreatment; Ic, isotype antibody as control; SAF 32, 3F4, and 12F10, monoclonal antibodies used. (B) Multiplication of B. suis in the murine J774 macrophage cell line. WT, without pretreatment; Ic, isotype control; SAF 32 and SAF 61, monoclonal antibodies used. (B, insert) Multiplication of B. suis in murine J774 cells after incubation with a purified polyclonal antibody. Sc, control serum.
To determine whether interaction with PrPC is specific to B. abortus, the experiments were repeated using B. abortus. Similar results (data not shown) were obtained with B. abortus with monoclonal antibody SAF32, suggesting interaction with PrPC is not required for successful infection of macrophage by either Brucella species.
However, in a previous publication the bacterial partner involved was identified as being GroEL in B. abortus (35). We demonstrated earlier (16) with others (10, 33) that eukaryotic Hsp60 and bacterial GroEL interact strongly with prion protein; we thus decided to search for bacterial Hsp60 on the Brucella surface.
GroEL cannot be detected on the surface of Brucella.
Since Hsp60 is able to bind PrPC, presence of an equivalent molecule on the surface of Brucella should be detectable after incubation of bacteria with rPrP. It is generally considered that recombinant PrP exhibits the same spatial structure as the genuine PrPC (see reference 1 for biochemical and biophysical analyses of our rPrP90-231 preparations). To detect this eventual interaction by fluorescence microscopy, we labeled rPrP with fluorescein, as described in Materials and Methods, and observed the binding of green rPrP on bacteria. As shown in Table 1 for all Brucella spp. tested, no significant labeling of the bacteria was observed, compared to a labeled RNase as negative control. This meant that either Brucella spp. did not express GroEL (or any other PrP receptor) at their surface or that GroEL interacts with the N terminus of the cellular prion protein (PrP1-90), absent from the recombinant molecule. We decided to control the presence of the GroEL chaperone on the B. suis external membrane directly with specific antibodies, as performed by Watarai et al. (35). Again, we were not able to detect any fluorescence that significantly exceeded the experimental background obtained with the isotypic antibody. It should be noted that rough mutants (i.e., ΔmanB) very strongly adsorbed any protein used (Table 1). A positive control was obtained by adsorbing purified GroEL on the surface of beads and revealing the protein by using the same antibody. The beads were brightly labeled (Table 1).
TABLE 1.
GroEL cannot be detected on the surface of Brucella spp.
| Labeling condition | Species or material
|
||||||
|---|---|---|---|---|---|---|---|
| B. suis | B. abortus | B. suis virB2 mutant | B. suis virB4 mutant | B. suis ΔmanB | B. suis stresseda | GroEL-labeled latex beads | |
| GroEL MAbb | − | − | − | − | + | − | + |
| Secondary Abc | − | − | − | − | + | − | − |
| PrPd | − | − | − | − | + | − | ND |
| Ribonucleasee | − | − | − | − | + | − | ND |
B. suis organisms were stressed for 3 min at 45 °C.
Anti-Escherichia coli GroEL mouse monoclonal antibody 9A1/2 was used in association with secondary anti-mouse antibody (tetramethyl rhodamine isothiocyanate conjugated).
The secondary antibody was used alone for labeling under the same conditions as described in footnote b.
Recombinant hamster PrP labeled as described in Materials and Methods.
Ribonuclease labeled as described in Materials and Methods.
In their work, Watarai et al. (35) concluded that surface expression of GroEL depends on the intact type IV secretion system VirB. We have shown that in B. suis the VirB system is expressed only inside the macrophage (2), whereas in B. abortus it is also expressed during in vitro culture (32). We reasoned that B. suis may be unable to express GroEL at its surface because of the absence of the VirB secretion system during extracellular life; we therefore repeated the experiment using B. abortus. However, we were not able to reproduce the results obtained by Watarai's group using the same B. abortus strain and the same anti-GroEL antibody (Table 1).
To increase GroEL expression, we stressed the bacteria by heat shock for 3 min at 45°C and looked for the presence of the protein at the cell surface, without success. In our hands, it was impossible to detect GroEL on the surface of Brucella spp.
Spatial proximity of Brucella and PrPC during the first steps of macrophage infection.
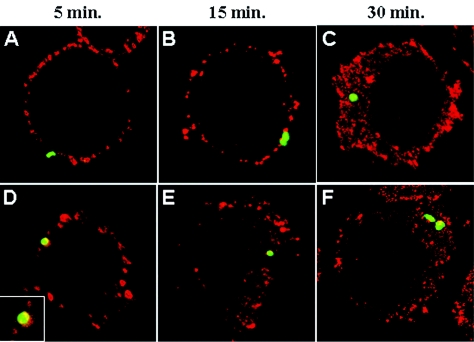
Because it was impossible to provide evidence for a direct interaction between bacteria and cellular PrP, we reasoned that an as-yet-unidentified partner could be involved. Microscopy can be used to analyze possible colocalization or close contact that does not need direct interaction. Thus, using confocal microscopy, we examined this possibility at different times after macrophage infection. Figure 2 displays the expression of PrPC (in red) in J774 cells at 5, 15, and 30 min after inoculation by B. suis GFP (in green) (Fig. 2A to C), which corresponds to the first stages, contact and penetration, of bacterial infection. It was not excluded that approximately 50% of the bacteria were in direct contact with PrPC, but due to the density of the surface-located PrPC it was difficult to decide if this proximity was fortuitous or not. On the other hand, mainly at 5 and 15 min postinfection, between 5 and 15% of B. suis and between 10 and 25% of B. abortus were surrounded partially or totally by PrPC (Fig. 2D, insert). Indeed, the results obtained with B. abortus were slightly different (Fig. 2D and E), and revealed a more engulfment-like picture of the bacteria surrounded by PrPC (Fig. 2D, insert). At 30 min postinfection, the bacteria were observed inside macrophages, but no specific interaction pattern could be evidenced (Fig. 2C and F).
FIG. 2.
Spatial proximity of Brucella and PrPC during the first steps of infection of the murine macrophage-like cell line J774. Cells cultured on coverglasses were infected as described in Materials and Methods and were analyzed by confocal microscopy. (A to C) B. suis; (D to F) B. abortus; (D, insert) magnification of the contact between B. abortus and PrPC.
The results suggested that Brucella utilized cholesterol-rich structures known as lipid rafts, where PrPC is clustered for internalization. This result was expected because it was demonstrated that Brucella enters the cells via lipid raft microdomains (24), and PrPC has been demonstrated to localize in the rafts (25). This spatial proximity appears only during the first minutes, when Brucella interacts with the cellular membrane.
Prion protein is not involved in the inhibition of phagosome-lysosome fusion.
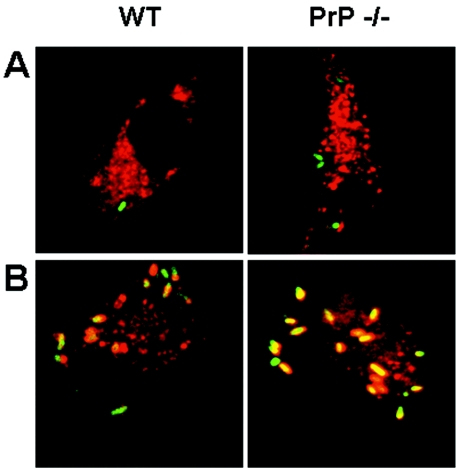
Inside the cells, Brucella avoids phagosome-lysosome fusion (18, 23, 27). Here, we evaluated the hypothesis that the expression of PrPC by the host influenced this mechanism. To verify this possibility, we used bone marrow-derived macrophages of wild-type and PrP knockout (KO) mice and confocal microscopy to observe colocalization between the GFP-labeled bacteria and a lysosome marker (dextran-Texas red), which is indicative of phagosome-lysosome fusion. As shown in Fig. 3A, B. suis was able to inhibit phagosome-lysosome fusion in both C57Bl/6 and PrP−/− macrophages.
FIG. 3.
B. suis inhibits lysosome fusion in bone marrow macrophages derived from PrP−/− mice. Bone marrow macrophages derived from WT (left panels) and PrP−/− (right panels) mice were prepared, labeled with dextran-Texas red, and infected with either WT B. suis (A) or ΔmanB mutants of B. suis (B). Analysis by confocal microscopy proceeded as described in Materials and Methods.
As a positive control for phagosome-lysosome fusion, we used the B. suis ΔmanB mutant which, due to a defect in LPS assembly, cannot inhibit the fusion of the compartment containing Brucella with lysosomes (28). Figure 3B shows that lysosomes were indeed capable to fuse with phagosomes containing B. suis ΔmanB, a PrPC-independent phenomenon in macrophages. Our results suggested that PrPC did not participate in the intracellular fate of B. suis. To test this hypothesis, we further compared intracellular growth curves of B. suis in bone-marrow derived macrophages obtained from mice either wild type or KO for the prion protein.
Infection of PrP-KO bone marrow-derived murine macrophages.
We first examined the capacity of phagocytosis of the two different types of cells: macrophages derived from wild-type mice or from mice where the gene encoding PrPC was deleted. We determined the percentage of phagocytosis, by counting the number of infected cells (cells containing one or more bacteria in their cytoplasm), using fluorescent microscopy and B. suis-GFP, and the total number of cells. We obtained a statistically equivalent percentage of phagocytosis (i.e., 52. 2% ± 2% for PrP+/+ and 48.1% ± 4% for PrP−/−) for both types of macrophages, confirming our previous observation that prion protein did not play a significant role in the penetration of Brucella in the host cells.
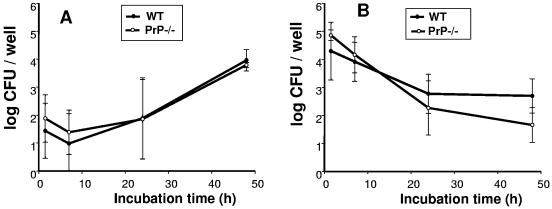
We infected the bone marrow-derived macrophages, control and knocked out for the prion protein, with B. suis, and we followed the time-dependent multiplication. Results shown in Fig. 4A represent the average of three independent experiments. No difference in intracellular behavior of B. suis was observed between the two types of cells (P > 0.5). This result confirmed that the presence or absence of PrPC expression determined neither the entry nor the multiplication of B. suis inside macrophages.
FIG. 4.
Intracellular multiplication of Brucella spp. is not affected by the absence of PrPC in macrophages. Bone marrow macrophages from PrP−/− mice and from congenic C57BL/6 controls were differentiated and infected as described in Material and Methods. The figure shows the mean of three independent experiments. (A) Intracellular multiplication of B. suis. (B) Intracellular multiplication of B. abortus. WT, macrophages derived from control C57BL/6 mice; PrP−/−, macrophages derived from PrP knockout mice.
With the aim of comparing our results to those obtained by Watarai's group, the same infection experiment was performed in parallel with the ATCC B. abortus strain. The results presented in Fig. 4B were slightly different from those obtained with B. suis. In contrast to B. suis, we observed any multiplication of B. abortus at 24 h postinfection, but similar results were obtained earlier by others (7). We did not evidence significant differences between macrophages expressing PrPC or not. Figure 4B shows also the average of three independent experiments. Comparison of the values was performed using the Mann and Whitney test for each time point, and no significant difference was evidenced. Probability that the difference of the mean observed was due to random variability was greater than 33% (P > 0.33).
In vivo infection of normal and PrP-KO mice by B. suis.
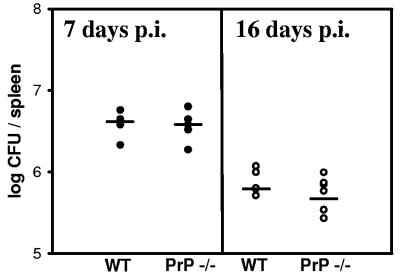
The next step, after in vitro experiments, was the infections of mice, normal C57BL/6 and congenic animals devoid of PrPC. Two sets of mice were used and experiments were performed independently; the first experiment was stopped at 7 days postinfection and the second was stopped at 16 days postinfection. The number of bacteria per spleen in wild-type or PrP-KO mice was compared. As shown in Fig. 5, no difference was observed in the spleen colonization of the two groups of animals.
FIG. 5.
B. suis infects PrP−/− mice with the same efficiency as congenic wild-type C57BL/6 mice. Mice were infected intraperitoneally with B. suis and sacrificed at day 7 or day 16; bacteria in the spleen were numbered as described in Materials and Methods.
DISCUSSION
The publication by Watarai et al. (35) described the participation of prion protein (PrPC) in the penetration and intracellular development of B. abortus, but our multiple attempts to confirm these results were unsuccessful. It was thus necessary to thoroughly compare the results to try to explain the origins of these discrepancies.
The above-cited publication (35) does not demonstrate direct contact between B. abortus and PrPC. Our first attempt was to complete this work, by addressing the question whether there was a direct interaction between Brucella and PrPC. This aim was achieved by using competing antibodies directed against cellular prion protein. Taking into account the very similar behavior of the different Brucella species, we started these experiments with B. suis, performing a pretreatment of human macrophages (THP-1 cell line) with three different anti-PrP antibodies prior to infection. If direct interaction was involved, we expected reduced Brucella penetration and/or multiplication due to the inaccessibility of the cellular prion protein of the host. In our hands, treatment with monoclonal antibodies had no effect on B. suis invasion, as we obtained similar results for bacterial intracellular behavior with or without antibody incubations. Similarly, no effect was observed with B. abortus, or a murine macrophage cell line and antibodies recognizing murine PrPC, therefore excluding an interaction with PrPC that is specific to either the host species or the Brucella species. Another possible explanation was that the monoclonal antibodies targeted exclusively specific epitopes not involved in the interaction with bacteria. To overcome this potential problem, the same experiments were performed using a polyclonal serum against murine prion protein on J774 cells, but we were still unable to evidence any differences in the bacterial behavior with or without anti-PrP treatment. Based on these results, we concluded that direct interaction between the cellular prion protein and Brucella was not required for entry or intracellular multiplication of bacteria in different macrophagic cell lines. As it is generally admitted that a negative result cannot be used to draw a definitive conclusion, we decided to further pursue this investigation.
According to Watarai's hypothesis that host PrP binds to bacterial GroEL, in line with our previous results (16) also obtained by others (10, 33), a direct interaction between bacterial chaperone and prion protein could be postulated. We considered that our negative result could be explained by the absence of GroEL on the surface of B. suis. We also know that GroEL is an essential gene in B. suis, as attempts to knock out this gene were unsuccessful (S. Köhler and J. P. Liautard, unpublished results), and it was thus impossible to generate a simple control using bacterial genetics. The intracellular (i.e., cytoplasmic) localization of GroEL is well known, but in several pathogenic bacteria surface expression of GroEL has been described, often correlating with a role of the chaperone in adherence (11, 13, 31). Our attempts to label Brucella GroEL using PrP-FITC or specific antibodies (Table 1) were unsuccessful. These results were rather qualitative and cannot be interpreted from a statistical point of view; furthermore, we cannot exclude the presence of small amounts of Hsp60 that are below the detection level. The possibility that our experiments failed because of a nonreactive antibody was ruled out by a positive control with GroEL-coated beads.
Hsp chaperones are considered to be major antigens in numerous pathogens and their expression is up-regulated during various bacterial infections in response to stress stimuli (38). It is well known that the interaction of the bacteria with host cell is a stress (3), which could result in membrane localization of GroEL. To address this possibility, a thermal stress was induced, but GroEL remained undetectable at the surface of the pathogen. We cannot completely eliminate, however, the possibility that different types of stress may result in different localizations of this stress protein.
According to Watarai et al. (35), GroEL, the putative bacterial receptor of PrPC, was transported to the surface of the bacteria by the T4SS (VirB) system. It was shown previously that B. suis does not express virB prior to phagocytosis and induces its synthesis only inside the host cell (2). It is thus quite unlikely that this cascade is involved in the internalization of B. suis into the macrophage. On the other hand, most of the other species of Brucella, including B. abortus, express VirB before interaction with the host cell (32). It is therefore not excluded that B. suis enters the macrophage by a mechanism other than the one used by the other species. Nevertheless, this possibility appears quite unlikely due to the close similarity of the species of the genus Brucella (34), and we have no specific reason to suppose that B. abortus behaves differently from B. suis during the internalization process. Furthermore, the importance of the VirB system during phagocytosis was reported only by Watarai's group. The attempts by others to demonstrate an impact of VirB on the HeLa cell penetration was unsuccessful (9), and this secretion system is generally thought to be involved in modifying the intracellular trafficking of Brucella (14).
In Watarai's paper, microscopy results show that a “tail” labeled by an antibody against PrPC was in close contact with B. abortus during phagocytosis. We therefore decided to address whether bacteria colocalized with PrPC at the earliest times of infection (5, 15, and 30 min postinfection), using confocal microscopy. Spatial proximity of GFP-Brucella and PrPC was observed by confocal microscopy. However, this result cannot be interpreted as a direct interaction for two reasons. First, the density of PrPC on the surface of the macrophage-like cell line is so high that it is difficult, when the bacteria come in contact with the membrane, not to be close to PrPC. Second, it was demonstrated that Brucella needs intact cholesterol rafts to penetrate inside the cells (24), and the rafts are the structures where PrPC is localized (25). Thus, we propose that Brucella and PrPC are picked up in a fortuitous manner on the same cargo during phagocytosis.
In our studies, we could not find any interaction between PrP molecules and Brucella of different species, and we then analyzed the possible role of PrP in the intracellular trafficking of Brucella. We studied one of the main mechanisms that ensure bacterial survival inside the macrophage: the inhibition of fusion of phagosomes containing Brucella with lysosomes. This phenomenon is well known and takes place with different Brucella species (7, 9, 12, 23, 27). The LPS O antigen seems to be involved in the early step of this phenomenon, since manB mutants that have a defect in this antigen's expression were unable to inhibit the fusion (23, 28). B. suis manB was therefore used as a positive control for fusion. To determine whether PrPC was involved in phagosome-lysosome fusion, we used macrophages derived from the bone marrow of C57BL/6 mice and congenic KO for cellular prion protein. Our results demonstrated that inhibition of phagosome-lysosome fusion by wild-type B. suis was independent of PrPC in macrophages. The fusion of B. suis ΔmanB-containing phagosomes was confirmed. This result was in clear contradiction to those published by Watarai et al. (35), as Watarai et al. observed that about 70% of the phagosomes containing Brucella were labeled with Lamp-1, a marker of the fusion with lysosomes or late endosomes. We used a method that directly monitored the fusion of lysosomes with a Brucella-containing vacuole, which is not the case for the criterion of the acquisition of Lamp-1, although it is widely accepted that this marker originates mainly from lysosomes and late endosomes. Taking into account this observation, we concluded that cellular prion protein (PrPC) of the host was not involved in directing traffic towards formation of the replicative compartment for Brucella.
The seminal experiment was focused on the analysis of intracellular multiplication of Brucella inside macrophages devoid of PrPC. The rate of phagocytosis was not significantly different for macrophages with or without surface PrPC. The number of intracellular bacteria was determined at different times postinoculation to evaluate the capacity of bacterial penetration and multiplication. Neither entry nor bacterial replication was significantly affected by the absence of PrPC, confirming that B. suis infection of macrophages was independent of the expression of cellular prion protein by the host cells. This result was independent of the Brucella species used, because essentially the same result was obtained with B. suis and B. abortus. Preparations of bone marrow-derived macrophages differed slightly between the two laboratories; terminal differentiation was obtained by culture in L-cell-conditioned medium (35), whereas we used purified (commercial) mouse macrophage colony-stimulating factor. To our knowledge, no major difference was reported between these two methods. Another explanation could be that the strains of mice used by the two groups were of different origins. We obtained congenic mice by 10 backcrosses with the strain C57BL/6, and we controlled deletion of the gene and absence of PrPC molecules from the brains of the animals, but we cannot completely exclude that an unknown important gene was also conserved during this procedure and lost in mice used by Watarai's group. Such a hypothesis may explain the discrepancy, but it rules out a major function for PrPC during infection by Brucella.
The same interpretation is also valid for mouse infection experiments which yielded comparable infection kinetics. Although we did not study B. abortus during infection of PrP KO mice, we have no indication that the two species behave differently.
Altogether, all the experiments performed and the results obtained allowed us to conclude that there was no evidence for the participation of prion protein in the infection by Brucella.
Acknowledgments
This work was supported by a grant from French GIS-Prion and a grant from the ATC-Prion of INSERM.
Editor: J. B. Bliska
REFERENCES
- 1.Alvarez-Martinez, M. T., J. Torrent, R. Lange, J. M. Verdier, C. Balny, and J. P. Liautard. 2003. Optimized overproduction, purification, characterization and high-pressure sensitivity of the prion protein in the native (PrP(C)-like) or amyloid (PrP(Sc)-like) conformation. Biochim. Biophys. Acta 1645:228-240. [DOI] [PubMed] [Google Scholar]
- 2.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmeier, N. A., and F. Heffron. 1990. Induction of Salmonella stress proteins upon infection of macrophages. Science 248:730-732. [DOI] [PubMed] [Google Scholar]
- 4.Bueler, H., A. Aguzzi, A. Sailer, R. A. Greiner, P. Autenried, M. Aguet, and C. Weissmann. 1993. Mice devoid of PrP are resistant to scrapie. Cell 73:1339-1347. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, G. A., L. G. Adams, and B. A. Sowa. 1994. Mechanisms of binding of Brucella abortus to mononuclear phagocytes from cows naturally resistant or susceptible to brucellosis. Vet. Immunol. Immunopathol. 41:295-306. [DOI] [PubMed] [Google Scholar]
- 6.Castaneda-Roldan, E. I., F. Avelino-Flores, M. Dall'Agnol, E. Freer, L. Cedillo, J. Dornand, and J. A. Giron. 2004. Adherence of Brucella to human epithelial cells and macrophages is mediated by sialic acid residues. Cell. Microbiol. 6:435-445. [DOI] [PubMed] [Google Scholar]
- 7.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizarro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celli, J., and J. P. Gorvel. 2004. Organelle robbery: Brucella interactions with the endoplasmic reticulum. Curr. Opin. Microbiol. 7:93-97. [DOI] [PubMed] [Google Scholar]
- 9.Delrue, R. M., M. Martinez-Lorenzo, P. Lestrate, I. Danese, V. Bielarz, P. Mertens, X. De Bolle, A. Tibor, J. P. Gorvel, and J. J. Letesson. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 3:487-497. [DOI] [PubMed] [Google Scholar]
- 10.Edenhofer, F., R. Rieger, M. Famulok, W. Wendler, S. Weiss, and E. L. Winnacker. 1996. Prion protein PrPc interacts with molecular chaperones of the Hsp60 family. J. Virol. 70:4724-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ensgraber, M., and M. Loos. 1992. A 66-kilodalton heat shock protein of Salmonella typhimurium is responsible for binding of the bacterium to intestinal mucus. Infect. Immun. 60:3072-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenchick, P. J., R. J. Markham, and A. H. Cochrane. 1985. Inhibition of phagosome-lysosome fusion in macrophages by soluble extracts of virulent Brucella abortus. Am. J. Vet. Res. 46:332-335. [PubMed] [Google Scholar]
- 13.Garduño, R. A., E. Garduño, and P. S. Hoffman. 1998. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66:4602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorvel, J. P., and E. Moreno. 2002. Brucella intracellular life: from invasion to intracellular replication. Vet. Microbiol. 90:281-297. [DOI] [PubMed] [Google Scholar]
- 15.Gregoire, S., C. Logre, P. Metharom, E. Loing, J. Chomilier, M. B. Rosset, P. Aucouturier, and C. Carnaud. 2004. Identification of two immunogenic domains of the prion protein—PrP—which activate class II-restricted T cells and elicit antibody responses against the native molecule. J. Leukoc. Biol. 76:125-134. [DOI] [PubMed] [Google Scholar]
- 16.Guerin, M., S. Bettache, A. Aumelas, L. Chiche, and J. Liautard. 2001. Use of surface plasmon resonance to analyse recombinant prion protein interaction with molecular chaperones. Int. J. Bio-Chromatogr. 6:121-133. [Google Scholar]
- 17.Harris, D. A. 1999. Cellular biology of prion diseases. Clin. Microbiol. Rev. 12:429-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler, S., S. Michaux-Charachon, F. Porte, M. Ramuz, and J. P. Liautard. 2003. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 11:215-219. [DOI] [PubMed] [Google Scholar]
- 19.Kohler, S., F. Porte, V. Jubier-Maurin, S. Ouahrani-Bettache, J. Teyssier, and J. P. Liautard. 2002. The intramacrophagic environment of Brucella suis and bacterial response. Vet. Microbiol. 90:299-309. [DOI] [PubMed] [Google Scholar]
- 20.Lafont, F., L. Abrami, and F. G. van der Goot. 2004. Bacterial subversion of lipid rafts. Curr. Opin. Microbiol. 7:4-10. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann, S. 2002. Metal ions and prion diseases. Curr. Opin. Chem. Biol. 6:187-192. [DOI] [PubMed] [Google Scholar]
- 22.Liautard, J. P., A. Gross, J. Dornand, and S. Kohler. 1996. Interactions between professional phagocytes and Brucella spp. Microbiologia 12:197-206. [PubMed] [Google Scholar]
- 23.Naroeni, A., N. Jouy, S. Ouahrani-Bettache, J. P. Liautard, and F. Porte. 2001. Brucella suis-impaired specific recognition of phagosomes by lysosomes due to phagosomal membrane modifications. Infect. Immun. 69:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naroeni, A., and F. Porte. 2002. Role of cholesterol and the ganglioside GM1 in entry and short-term survival of Brucella suis in murine macrophages. Infect. Immun. 70:1640-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naslavsky, N., R. Stein, A. Yanai, G. Friedlander, and A. Taraboulos. 1997. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J. Biol. Chem. 272:6324-6331. [DOI] [PubMed] [Google Scholar]
- 26.Ouahrani-Bettache, S., F. Porte, J. Teyssier, J. P. Liautard, and S. Kohler. 1999. pBBR1-GFP: a broad-host-range vector for prokaryotic promoter studies. BioTechniques 26:620-622. [DOI] [PubMed] [Google Scholar]
- 27.Pizarro-Cerda, J., E. Moreno, V. Sanguedolce, J. L. Mege, and J. P. Gorvel. 1998. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect. Immun. 66:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Porte, F., A. Naroeni, S. Ouahrani-Bettache, and J. P. Liautard. 2003. Role of the Brucella suis lipopolysaccharide O antigen in phagosomal genesis and in inhibition of phagosome-lysosome fusion in murine macrophages. Infect. Immun. 71:1481-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prado, M. A., J. Alves-Silva, A. C. Magalhaes, V. F. Prado, R. Linden, V. R. Martins, and R. R. Brentani. 2004. PrPc on the road: trafficking of the cellular prion protein. J. Neurochem. 88:769-781. [DOI] [PubMed] [Google Scholar]
- 30.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA 95:13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao, S. P., K. Ogata, S. L. Morris, and A. Catanzaro. 1994. Identification of a 68 kd surface antigen of Mycobacterium avium that binds to human macrophages. J. Lab. Clin. Med. 123:526-535. [PubMed] [Google Scholar]
- 32.Rouot, B., M. T. Alvarez-Martinez, C. Marius, P. Menanteau, L. Guilloteau, R. A. Boigegrain, R. Zumbihl, D. O'Callaghan, N. Domke, and C. Baron. 2003. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 71:1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stockel, J., and F. U. Hartl. 2001. Chaperonin-mediated de novo generation of prion protein aggregates. J. Mol. Biol. 313:861-872. [DOI] [PubMed] [Google Scholar]
- 34.Vizcaino, N., A. Cloeckaert, J. Verger, M. Grayon, and L. Fernandez-Lago. 2000. DNA polymorphism in the genus Brucella. Microbes Infect. 2:1089-1100. [DOI] [PubMed] [Google Scholar]
- 35.Watarai, M., S. Kim, J. Erdenebaatar, S. Makino, M. Horiuchi, T. Shirahata, S. Sakaguchi, and S. Katamine. 2003. Cellular prion protein promotes Brucella infection into macrophages. J. Exp. Med. 198:5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watarai, M., S. Makino, M. Michikawa, K. Yanagisawa, S. Murakami, and T. Shirahata. 2002. Macrophage plasma membrane cholesterol contributes to Brucella abortus infection of mice. Infect. Immun. 70:4818-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuthrich, K., and R. Riek. 2001. Three-dimensional structures of prion proteins. Adv. Protein Chem. 57:55-82. [DOI] [PubMed] [Google Scholar]
- 38.Zugel, U., and S. H. Kaufmann. 1999. Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin. Microbiol. Rev. 12:19-39. [DOI] [PMC free article] [PubMed] [Google Scholar]