Plant growth and development are regulated by intrinsic growth regulators, as well as by both beneficial and detrimental environmental cues. Hormonal, environmental, or pathogenic signals are mostly perceived by membrane-localized receptors that transduce those signals inside plant cells to activate programs directing growth, development, and defense responses. In the fully sequenced Arabidopsis genome (1), there are more than 600 receptor-like kinase (RLK) genes, about 200 of which belong to a family called leucine-rich repeat (LRR) receptor-like kinases (LRR-RLKs; ref. 2). The main characteristics of LRR-RLKs are their extracellular domains which are composed of tandem repeats of a well conserved leucine-rich motif and intracellular protein kinase domains with Ser/Thr specificity. LRRs often participate in protein–protein interactions and, therefore, are believed to function in ligand binding (3). Despite the large numbers of LRR-RLKs in plants, fewer than 10 of them have known biological functions. For those about which some function is known, the LRR-RLKs play a role in diverse processes such as hormone perception, meristem signaling, and pathogen responses (2). Even fewer LRR-RLKs have known ligands. In this issue of PNAS, SR160, a tomato LRR-RLK, has been identified as the receptor for systemin, a peptide involved in systemic wounding signaling (4).
The LRR-RLKs for which a ligand has been defined include the plant steroid receptor BRI1 (see below), and CLV1, FLS2, and PSK receptors, for which the ligands are peptides. CLV1 and CLV3 were identified as genes controlling apical meristem proliferation based on their similar mutant phenotypes (5, 6). Whereas CLV1 encodes an LRR-RLK (6), CLV3 encodes a small and secreted protein, which is proposed to act as a peptide ligand for CLV1 (5). Indeed, CLV3 is found in an active CLV1 complex in vivo, and binds to yeast cells expressing CLV1 (7). FLS2 was identified in a genetic screen for mutants insensitive to flagellin, a bacterial peptide elicitor that induces defense responses in plants (8). Flagellin binds to membrane proteins from wild-type plants but not from mutant plants harboring mutations in FLS2 genes, suggesting that flagellin is the ligand for FLS2 (9). PSK is a five-amino acid peptide that plays a key role in cell de-differentiation (10). By using ligand-based affinity chromatography, a PSK receptor was purified from carrot cells; the corresponding gene encodes an LRR-RLK with 21 LRRs and a 36-aa island between the 17th and 18th LRRs (10). Consistent with the idea of a ligand-receptor pair, overexpression of the receptor gene increased cell de-differentiation, accompanied by increased PSK binding, whereas antisense suppression of the receptor gene expression inhibited the growth of culture cells (10).
Specific signals are generated from the damaged sites and are transduced to other undamaged parts of the plant.
What makes the identification of the systemin receptor especially notable is that, although systemin was characterized almost a decade ago, its receptor has not yet been identified. Plants respond to wounding or herbivore attack by expressing a set of defense-related genes. In tomato plants, wounding induces the expression of more than 20 genes, including those encoding insect deterrents such as proteinase inhibitors and polyphenol oxidase (11). Many of the defense responses happen in a systemic manner; for example, wound-induced proteinase-inhibitor gene expression occurs not only in the wounded leaf but also in undamaged leaves distal from the damage sites (12). It has been proposed that specific signals are generated from the damaged sites and are transduced to other undamaged parts of the plant to activate defense gene expression (12). Systemin was purified from tomato plants in 1991 as such a signaling molecule (13). Systemin is an 18-aa peptide that is derived from a 200-aa precursor called prosystemin, which is released from the wound site, triggering systemic wounding responses (11). Direct evidence for systemin as the wound signal came from transgenic studies, in which constitutive expression of prosystemin resulted in constitutive expression of defense response genes without wounding, whereas antisense suppression of prosystemin expression inhibited systemic wounding responses (11). In grafting experiments, root stock from plants that constitutively express prosystemin caused wild-type scion plants to express defense genes without wounding, demonstrating that systemin can transmit the wounding signal (11). It is not very clear, however, how systemin transduces the wounding signal over long distances.
The systemin receptor was identified by biochemical methods. Radiolabeled systemin was found to bind to a putative receptor in cell membrane fractions with high affinity, and a photoaffinity-labeled systemin cross-linked with a 160-kDa membrane protein called SR160 (14, 15). In the studies presented in this issue, Scheer and Ryan (4) used both photoaffinity- and radiolabeled systemin to purify a large amount of the receptor protein. Subsequent protein sequencing helped to identify a full-length cDNA of SR160. The predicted SR160 protein displays all of the characteristics of an LRR-RLK, including a putative signal sequence, a three-heptad leucine zipper, 25 LRRs that are interrupted by an island domain, a single transmembrane domain, and a protein kinase domain (4).
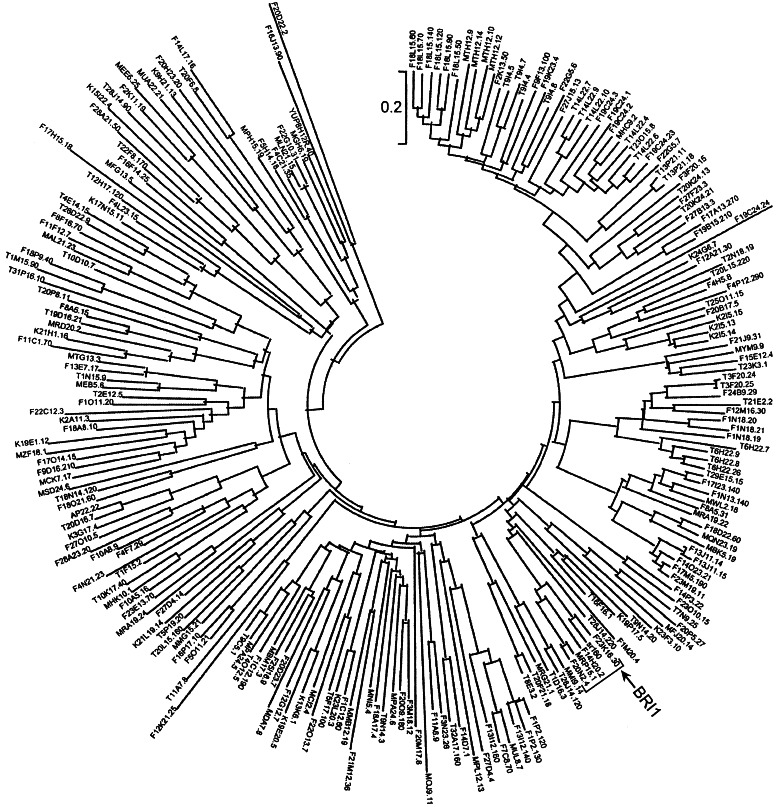
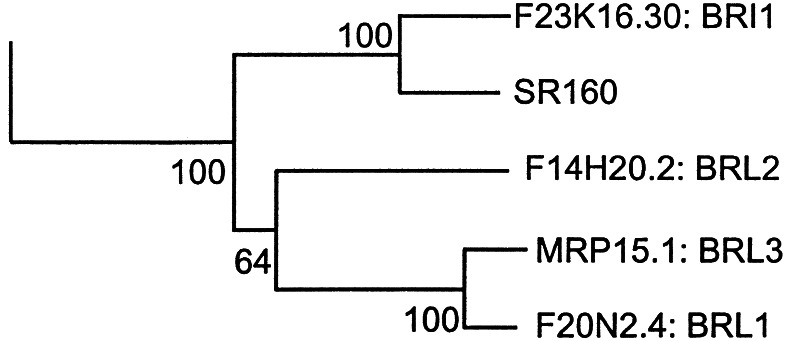
Surprisingly, of the 222 LRR-RLKs, SR160 is most closely related to BRI1 (ref. 16; Fig. 1), the receptor for plant steroids, brassinosteroids (BRs), but is less related to other LRR-RLKs with known peptide ligands. Both BRI1 and SR160 contain 25 LRRs and a unique island domain between the 21st and 22nd LRRs. Of note, three Glys in BRI1's island domain, mutations of which disrupt BR binding and signaling, are conserved in SR160. It is striking that SR160 transmembrane and kinase domains are 83% and 90% identical to BRI1 (ref. 4; Fig. 2).
Figure 1.
Arabidopsis LRR-RLKs and tomato SR160. Protein kinase domains (222) defined by Pfam PF00069.hmm from LRR protein kinases (222) in Arabidopsis genome were aligned with tomato SR160 protein kinase domain by HMMER (27). A neighbor-joining tree was created for the protein kinase domain alignments by MEGA (28). The tree was tested by bootstrapping with 1,000 replications. The branch that includes brassinosteroid receptor BRI1, tomato systemin receptor SR160, and three BRI1-like proteins is indicated by an arrow and a bracket and shown in more detail in Fig. 2.
Figure 2.
Arabidopsis LRR-RLKs and tomato SR160. SR160 is most closely linked to BRI1 and is tightly grouped with another three Arabidopsis receptor kinases with a bootstrapping value of 100.
Unlike systemin, which is present only in Solanaceous species, BRs are ubiquitously present in all plant species and play essential roles throughout plant growth and development. BRs are involved in many processes such as stem elongation, leaf development, pollen tube growth, xylem differentiation, senescence, and photomorphogenesis (17). Loss-of-function mutations in both the extracellular and the intracellular kinase domains of BRI1 lead to BR-insensitive dwarf phenotypes identical to those of BR-deficient mutants (16), providing genetic evidence that BRI1 is a BR receptor. Consistent with this idea, the BRI1 extracellular domain confers BR-responsiveness to a heterologous kinase domain from Xa21, a rice LRR-RLK for disease resistance (18). BRs were found to bind to membrane fractions from wild-type plants but not from bri1 plants harboring mutations in the island domain (19). In addition, BRs bind to a BRI1 complex immunoprecipitated from plants, and such binding triggers autophosphorylation of BRI's kinase (19). Therefore, both genetic and biochemical evidence demonstrate that BRI1 is a critical component of the BR receptor.
Considering the distinct functions of the corresponding ligands, it is intriguing that SR160 and BRI1 are highly related to each other. One possibility is that SR160 was recently derived from a tomato BRI1 and has a distinct function other than BR signaling. There are three BRI1-like proteins, BRL1–3, in Arabidopsis (Fig. 2), two of which (BRL1 and BRL3) bind brassinolide, the most active BR (Y.Y., A. Cano, and J.C., unpublished results). Although BRL2 is also closely related to BRI1, it does not bind BRs. This result suggests that small changes in these homologous proteins can alter ligand and signaling specificities. In fact, the most divergent regions between BRI1 and SR160 are in the extracellular domain (4), which could provide the basis for recognition of different ligands. Consistent with this observation, BRs do not compete for systemin binding to SR160, suggesting that these two ligands don't bind to the same site (4). On the other hand, because SR160 is even more closely related to BRI1 than BRI1 is to its homologs in Arabidopsis (Fig. 2), it is still possible that SR160 serves as a BR receptor in tomato. One scenario is that systemin and BRs bind to different sites on the receptor. In animal systems, the steroid hormone progesterone and peptide hormone oxytocin bind to the same G protein-coupled receptor and trigger opposite biological responses (20). However, a relationship between BRs and systemin seems unlikely. The identification of loss-of-function mutants of tomato SR160 gene would distinguish these two possibilities and also would be important confirmation that SR160 is the systemin receptor.
In either case, the high homology between the BR receptor BRI1 and the systemin receptor SR160 provides a unique opportunity to address the specificities of LRR-RLKs at the levels of both ligand recognition and downstream signal transduction. Because systemin and BRs are small peptide and organic compounds, respectively, it would be very interesting to determine how these two different ligands interact with very similar receptors. In addition, signaling mechanisms downstream of BRI1 and SR160 also seem to be divergent. Sytemin signaling through SR160 activates a second regulator, JA, which in turn activates defense-gene expression (11, 21, 22). In contrast, many BR-activated genes are involved in cell wall modifications (23, 24), consistent with their key role in cell elongation. BR signaling through BRI1 likely inhibits a negative kinase BIN2 (25), allowing accumulation of BES1 and BZR1 in the nucleus to activate target gene expression (24, 26). The identification of more components involved in membrane to nuclear signaling for both BRs and systemin pathways and delineation of detailed mechanisms of their actions will answer how specificity of the signaling pathway is achieved.
Footnotes
See companion article on page 9585.
References
- 1.Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2. Shiu, S. H. & Bleecker, A. B. (2001) Sci. STKE2001, http://stke.sciencemag.org/cgi/content/full/OC_sigtrans;2001/113/re22. [DOI] [PubMed]
- 3.Kobe B, Deisenhofter J. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 4.Scheer J M, Ryan C A., Jr Proc Natl Acad Sci USA. 2002;99:9585–9590. doi: 10.1073/pnas.132266499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fletcher J C, Brand U, Running M P, Simon R, Meyerowitz E M. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 6.Clark S E, Williams R W, Meyerowitz E M. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 7.Trotochaud A E, Jeong S, Clark S E. Science. 2000;289:613–617. doi: 10.1126/science.289.5479.613. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Gomez L, Boller T. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 9.Gomez-Gomez L, Bauer Z, Boller T. Plant Cell. 2001;13:1155–1163. [PMC free article] [PubMed] [Google Scholar]
- 10.Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. Science. 2002;296:1470–1472. doi: 10.1126/science.1069607. [DOI] [PubMed] [Google Scholar]
- 11.Ryan C A. Biochim Biophys Acta. 2000;1477:112–121. doi: 10.1016/s0167-4838(99)00269-1. [DOI] [PubMed] [Google Scholar]
- 12.Green T R, Ryan C A. Science. 1972;175:776–777. doi: 10.1126/science.175.4023.776. [DOI] [PubMed] [Google Scholar]
- 13.Pearce G, Strydom D, Johnson S, Ryan C A. Science. 1991;253:895–897. doi: 10.1126/science.253.5022.895. [DOI] [PubMed] [Google Scholar]
- 14.Meindl T, Boller T, Felix G. Plant Cell. 1998;10:1561–1570. doi: 10.1105/tpc.10.9.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheer J M, Ryan C A. Plant Cell. 1999;11:1525–1536. doi: 10.1105/tpc.11.8.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Chory J. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Chory J. J Exp Botany. 1999;50:332–340. [Google Scholar]
- 18.He Z, Wang Z Y, Li J, Zhu Q, Lamb C, Ronald P, Chory J. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z Y, Seto H, Fujioka S, Yoshida S, Chory J. Nature (London) 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 20.Grazzini E, Guillon G, Mouillac B, Zingg H H. Nature (London) 1998;392:509–512. doi: 10.1038/33176. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Li C, Lee G I, Howe G A. Proc Natl Acad Sci USA. 2002;99:6416–6421. doi: 10.1073/pnas.072072599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan C A, Moura D S. Proc Natl Acad Sci USA. 2002;99:6519–6520. doi: 10.1073/pnas.112196499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clouse S D. Plant J. 1996;10:1–8. doi: 10.1046/j.1365-313x.1996.10010001.x. [DOI] [PubMed] [Google Scholar]
- 24.Yin Y, Wang Z-Y, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Nam K H. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z Y, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 27.Sonnhammer E L, Eddy S R, Durbin R. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Tamura K, Jakobsen I B, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]