Comparative mapping of plant genomes has shown extensive conservation of gene content and gene order along the chromosomes, especially among the grasses, including maize, rice, sorghum, and wheat (1). This overall genomic colinearity at the recombinational map level has indicated that a few major chromosomal rearrangements have differentiated individual lineages of grasses. The comparative maps have assisted in the identification of truly orthologous loci, that is, those that are directly derived from the same ancestral gene in the last common ancestor of the investigated species. However, the first comparative sequence analyses of these orthologous regions in sorghum versus rice (2), maize versus sorghum (3), barley versus rice (4), or wheat versus barley (5, 6) have shown many small chromosomal rearrangements that involve genes, including inversions, duplications, and deletions. Hence, it appears that small rearrangements have outnumbered events that could be detected by recombinational mapping by more than 1,000-fold (7). The mechanisms, rates, and biological significance of these microrearrangements have not yet been investigated in any comparison, partly because the compared species have not been more closely related than wheat and barley, whose ancestors diverged 10–15 million years ago. In the first sequence-based investigation of the same genomic region in two different maize inbreds, Fu and Dooner (8) report in this issue of PNAS that much of the variation observed between species can also be found within the maize germplasm itself. Moreover, their results force us to rethink our general assumptions about the nature of intraspecies allelic variation.
For many years, the Dooner laboratory has used the bronze1 (bz1) locus of maize as a site for very basic studies of transposable element function and meiotic recombination (9, 10) (Fig. 1). In all of these experiments, the observed genetic phenomena could not be interpreted relative to local genome structure, because this was not known. For instance, was the frequency of intragenic recombination in bz1 influenced by the degree of structural variation between recombining bz1 alleles or in gene-adjacent regions? Were the transpositional behaviors of Ac at bz1 related to the nature and/or density of genes nearby, or interactions with adjacent heterochromatin? To place these results in a genomic context, Fu and colleagues sequenced 151 kb around bz1 in the McC inbred strains (8, 11). They found that this was an exceptionally gene-rich region, for maize, containing 13 predicted genes, with 10 of them in a contiguous span of 32 kb. This gene-rich region is interrupted by two of the long terminal repeat (LTR)–retrotransposon blocks that are seen in other areas of the maize genome (12), between genes 10 and 11 and between genes 11 and 12 (Fig. 2). In this issue of PNAS, Fu and Dooner present the sequence of the same region from a second inbred, B73, and compare these two haplotypes. Their data indicate that four of the predicted genes in the McC haplotype are missing from this region in B73, and that all of the LTR–retrotransposons are different elements representing independent insertions (Fig. 2).
Figure 1.

A rare purple kernel resulting from intragenic recombination within the bz1 locus. The linked shrunken1 (sh1) trait is also segregating on these ears, and its recessive homozygosity explains the kernels that appear somewhat shrunken. Photo courtesy of H. Dooner.
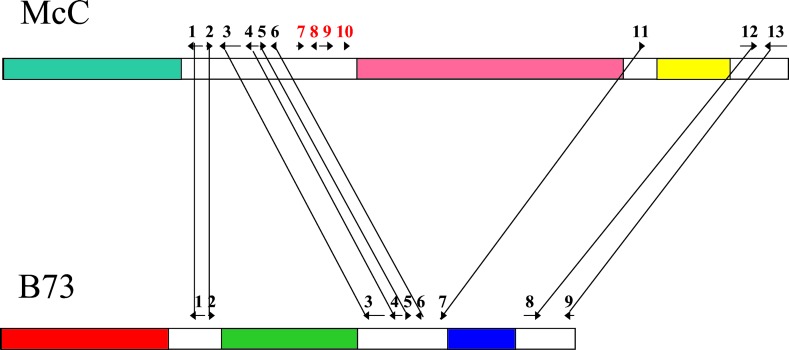
Figure 2.
A simplified schematic of the bz1 regions in maize inbreds B73 and McC. Arrows with numbers indicate the location, size, and transcriptional orientations of predicted genes in the region. The bz1 locus is gene 2 in the sequentially numbered series, and allelic loci are connected by lines. The filled regions represent LTR–retrotransposon blocks. The different colors of the fills indicate that each of these blocks is unique in its LTR–retrotransposon composition.
Previous studies of LTR–retrotransposon insertion frequencies have suggested that most elements are currently quiescent in maize and other grasses, with some significant exceptions (13–15). Because the termini of LTR–retrotransposons are usually identical at the time of insertion, divergence between the two LTRs within an element has been used to predict the approximate timing of insertion. These results have shown that most maize LTR–retrotransposons inserted within the last 2–6 million years (16). Because these elements make up over 60% of the maize genome (17, 18), this rapid amplification of LTR–retrotransposons over the last few million years has been responsible for more than a doubling of the size of the maize nuclear genome in that time span. For the bz1 haplotypes compared by Fu and Dooner, we used this LTR-dating technique and standard molecular clock analyses (19, 20) to compare the insertion times of the LTR–retrotransposons in the region with the approximate divergence dates of the 9 comparable genes. In general, the LTR–retrotransposons appear to have inserted after the divergence of the genes in these haplotypes (data not shown), in agreement with the presence of different elements in each compared region. Hence, as Fu and Dooner conclude, the LTR–retrotransposons were independently active in different maize lineages that have been carried through to the domesticated germplasm. The presence of multiple truncated LTR–retrotransposons in the bz1 region indicates a high rate of rearrangement, especially deletion, after element insertion, as has also been noted in Arabidopsis (21).
One of the recurrent observations with all transposable elements, including LTR–retrotransposons, is the existence of particular insertion/retention biases (15). Most of the abundant LTR–retrotransposons in maize insert into other LTR–retrotransposons, particularly into their LTRs (12, 15). It is interesting that the LTR–retrotransposon blocks found in both B73 and McC, although containing different elements, are found in the same location in two of four cases. Given that there are 9 shared between-gene regions in B73 and McC (Fig. 2), the presence of independent LTR–retrotransposon blocks at two identical sites suggests a bias for insertion and/or retention between specific gene pairs. Perhaps the rate-limiting step in establishing an LTR–retrotransposon block is the first LTR–retrotransposon insertion. It is possible that these two shared LTR–retrotransposon block sites once contained one or more shared LTR–retrotransposons in a common ancestor of B73 and McC. These elements may now be undetectable because of subsequent mutations, such as the unequal homologous recombination (22) and illegitimate recombination (21) that have been shown to be responsible for a very high rate of LTR–retrotransposon decay and removal in a variety of plant species. It is this balance between transposable element amplification by transposition and removal by unequal and illegitimate recombination that is largely responsible for the size of plant genomes and for the interspersion patterns of genes with mobile DNAs.
The relationship between the structure of a genomic region and the evolved functions of genes in that region remains one of the central unanswered questions in molecular genetics. For adh1, gene function and regulation seem to be the same in maize, sorghum, and rice despite the presence of adh1 on nonsyntenic chromosomes and surrounded by different genes in maize and sorghum compared with rice. Moreover, the maize adh1 gene sits alone, surrounded by large LTR–retrotransposon blocks in maize, whereas it is clustered with other genes in rice and sorghum, without any apparent differential effect on function (3, 12, 23). Hence, plants appear to have evolved very effective insulators that protect genes from the influences of local nongenic regions. The nature(s) of these insulator or boundary elements are not known, although matrix attachment regions serve as one strong candidate, especially as they appear to be conserved in location between large and small grass genomes (24). In this regard, it is also interesting that the two pairs of LTR–retrotransposon blocks that share the same locations between B73 and McC in the bz1 region also share similar sizes, despite their independent origins. The most leftward LTR–retrotransposon block is large in each haplotype (more than 35 kb), whereas the most rightward LTR–retrotransposon blocks are both about 15 kb in size. This may be a coincidence or it may indicate that a particular size (but not specific composition) of LTR–retrotransposon cluster may be selected as an important component of chromatin and chromosome folding. Only advanced mutational or transgenic studies, involving manipulated structures of such large regions, will be able to resolve these questions.
The most intriguing result from the recent work of Fu and Dooner (8) was the observation that four adjacent genes are missing from the B73 haplotype. With the current data, it is impossible to say whether these genes were deleted from the B73 lineage or inserted in the McC lineage. Deletion may seem the simpler mechanism, but movements of single genes or small blocks of adjacent genes to new locations have been observed in maize and other grasses (ref. 7; K. Ilic and J.L.B., unpublished observations). In the case of adh1, this single gene moved to a new chromosome in a common ancestor of maize and sorghum, while apparently maintaining its regulated expression profile (3, 7, 23). Each of these deleted genes has homologues elsewhere in the B73 genome, as indicated by gel blot hybridization; this may account for the ability of maize to tolerate a four-gene deletion.
Fu and Dooner (8) argue that the presence/absence of genes may be a common but previously unexpected type of allelic variation. This argument deserves further analysis. Could this difference be something unique to the bz1 region, perhaps only in these two inbred lines? The authors used gel blot hybridization to identify a minimum of four different patterns of gene presence/absence in the bz1 region in an analysis of only 10 inbred lines. This finding suggests, but does not prove, that gene loss has occurred frequently and independently from this region during the recent evolution of maize (8). The bz1 region has been a site where x-ray-induced deletion events covering as much as several centimorgans have been selected (25), but there is no evidence that deletions of a comparable size are not equally well tolerated in most of the maize genome (26). Perhaps this phenomenon is unique to maize? Maize does have an exceptionally high level of sequence polymorphism, suggesting that one or more types of DNA rearrangement are unusually frequent within the species. Maize is also an ancient allotetraploid, generated from the apparent fusion of two closely related grass genomes about 15–20 million years ago (27). Since that time, many genes have been deleted from one of the “homoeologous” genomes or the other. Perhaps B73 and McC differ for such a tolerable deletion, which has not yet been fixed within the species. Such gene losses from previously polyploid genomes appear to be common in many eukaryotes (28, 29).
Because polyploidy approximately doubles the size of every gene family, subsequent gene losses will often be tolerated. In mutational “knock-out” studies, initial investigations in all higher eukaryotes find a small minority of genes with inactivational phenotypes that can be unambiguously determined. Much of this lack of detected phenotypes for inactivational mutations is undoubtedly caused by duplicated gene functions. However, much is also probably caused by the fact that many genes perform very subtle roles that yield only Very Slightly Deleterious Mutations (30). Maize, as a recently domesticated species, may also be rapidly losing genes that were only necessary for wild populations. For instance, any positive modifiers of shattering (seed dispersal) or tillering (multiple gametic lineages) might now perform no useful function in United States field corn. Because illegitimate recombination and other deletion processes appear to be so active in maize and other plants (21, 31), the loss of unselected sequences may be relatively rapid. For Very Slightly Deleterious Mutations, population genetic theory indicates that losses of genes that have a low effect on fitness will not be cleared by natural selection, and thus will accumulate and lead to an overall degeneration in fitness (30, 32). Intragenic recombination might recover some degenerated genes, but losses caused by deletion cannot be repaired in this way.
Differences in the presence and absence of genes are not often considered as a major component of “natural” allelic variation within a species. Restriction fragment length polymorphism probes, which are usually derived from genes or gene-adjacent sequences in maize and other plants, are occasionally mapped as presence/absence differences. Hence, this may be a common form of allelic variation in plants. If so, then these results might help explain some recurrent mysteries in plant biology.
The superior vigor and yield performance of hybrid germplasms are consistent observations, and the foundation of the hybrid seed industry, in maize. The molecular basis of this hybrid vigor, otherwise known as heterosis, is not yet clear. Two models, over-dominance and dominance, have been argued, and each have some basis for support. The dominance model, proposing that any inbred will have some inferior alleles by the luck of past segregations or mutation, appears to fit most of the current data. However, over-dominant situations where two alleles at a locus provide greater fitness than either parental allele alone are known in maize and elsewhere. If allelic variation is often associated with the deletions of genes from some genetic backgrounds, then this provides obvious support for the dominance model. In either case, including with deletions, recombination could still yield an inbred with a full gene compliment, so long as each gene is present in at least one of the parents. For instance, a B73/McC cross could be used in a recurrent crossing program to bring the four missing genes from the bz1 region into the B73 background, by means of recombination between any genes proximal to the deletion (e.g., 1 and 2 or 4 and 5, Fig. 2). If these deletions do become fixed within the germplasm of the entire species because they have no strong deleterious effects (i.e., are very slightly deleterious mutations) (30), then they could still be restored by the generation of an allopolyploid with another species that does not share this deletion. Perhaps a rapid rate of gene deletion in maize and other plants partly accounts for the propensity toward polyploidy in the plant kingdom. More than 50% of flowering plants have polyploid origins, mostly as allopolyploids, and these derivative species often have much higher vigor than either parent (33).
The dramatic variation in the bz1 region between B73 and McC inbreds indicates that additional haplotype variation needs to be evaluated across multigenic regions in maize and other plants. Studies of numerous bz1 alleles, for instance, might uncover the precise nature and order of the events that gave rise to current alleles. Haplotype variation has been observed to be very high in the regions of disease resistance genes in plants that, like the major histocompatibility loci of mammals, have undergone diversifying selection to maximize variability (34–36). That similar levels of variability might be present in regions with genes that appear to be subject to purifying selection suggests that such phenomena are intrinsic outcomes of chromatin biochemistry (e.g., recombination and repair) rather than specific to diversifying gene-family regions. Moreover, analyses of orthologous regions in different plant species have assumed that the single clones analyzed for each species are generally representative. This is always a dangerous assumption, but in this case largely caused by experimental resource limitations. Regardless, future comparative studies will need to more seriously consider the possibility that the gene content, order and orientation variations seen between two species may not be fixed within the populations that they are studying, but are often variations present within a single species' germplasm.
From the first comparative plant genome studies, to the work of Fu and Dooner (8), one general observation continues to hold true. Plant genomes are surprisingly complex, both in their commonalities and their contrasts. We are just beginning to understand the components and mechanisms of genome constancy and change in plants, and we can now begin to ask what effects these changes may have on evolved gene function and organismal biology.
Footnotes
See companion article on page 9573.
References
- 1.Gale M D, Devos K M. Science. 1998;282:656–659. doi: 10.1126/science.282.5389.656. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, SanMiguel P, Bennetzen J L. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tikhonov A P, SanMiguel P, Nakajima Y, Gorenstein N D, Bennetzen J L, Avramova Z. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubcovsky J, Ramakrishna W, SanMiguel P, Busso C S, Yan L, Shiloff B A, Bennetzen J L. Plant Physiol. 2001;125:1342–1353. doi: 10.1104/pp.125.3.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feuillet C, Keller B. Proc Natl Acad Sci USA. 1999;96:8265–8270. doi: 10.1073/pnas.96.14.8265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.SanMiguel P, Ramakrishna W, Busso C S, Bennetzen J L, Dubcovsky J. Funct Integr Genomics. 2002;2:70–80. doi: 10.1007/s10142-002-0056-4. [DOI] [PubMed] [Google Scholar]
- 7.Bennetzen J L, Ramakrishna W. Plant Mol Biol. 2002;48:821–827. doi: 10.1023/a:1014841515249. [DOI] [PubMed] [Google Scholar]
- 8.Fu H, Dooner H K. Proc Natl Acad Sci USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dooner H K, Belachew A. Genetics. 1989;122:447–457. doi: 10.1093/genetics/122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooner H K, Martinez-Ferez I M. Plant Cell. 1997;9:1633–1646. doi: 10.1105/tpc.9.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu H, Park W, Yan X, Zheng Z, Shen B, Dooner H K. Proc Natl Acad Sci USA. 2001;98:8903–8908. doi: 10.1073/pnas.141221898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 13.Johns M A, Mottinger J, Freeling M. EMBO J. 1985;4:1083–1102. doi: 10.1002/j.1460-2075.1985.tb03745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirochika H. EMBO J. 1993;12:231–240. doi: 10.1002/j.1460-2075.1993.tb05907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Bennetzen J L. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 16.SanMiguel P, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 17.SanMiguel P, Bennetzen J L. Annals Bot. 1998;82:37–44. [Google Scholar]
- 18.Meyers B C, Tingey S V, Morgante M. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiffin P, Hahn W M. J Mol Evol. 2002;54:746–753. doi: 10.1007/s00239-001-0074-1. [DOI] [PubMed] [Google Scholar]
- 20. Zhang, L., Vision, T. & Gaut, B. S. (2002) Mol. Biol. Evol., in press. [DOI] [PubMed]
- 21. Devos, K. M., Brown, J. K. M. & Bennetzen, J. L. (2002) Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 22.Vicient C M, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman A H. Plant Cell. 1999;11:1769–1784. doi: 10.1105/tpc.11.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tikhonov A P, Bennetzen J L, Avramova Z. Plant Cell. 2000;12:249–264. doi: 10.1105/tpc.12.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mottinger J P. Genetics. 1970;64:259–271. doi: 10.1093/genetics/64.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopinath D M, Burnham C R. Genetics. 1956;41:382–395. doi: 10.1093/genetics/41.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaut B S, Doebley J F. Proc Natl Acad Sci USA. 1997;88:2060–2064. [Google Scholar]
- 28.Wolfe K H, Shields D. Nature (London) 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 29.Ku H-M, Vision T, Liu J, Tanksley S D. Proc Natl Acad Sci USA. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondrashov A S. J Theor Biol. 1995;175:583–594. doi: 10.1006/jtbi.1995.0167. [DOI] [PubMed] [Google Scholar]
- 31.Kirik A, Salomon S, Puchta H. EMBO J. 2000;19:5562–5566. doi: 10.1093/emboj/19.20.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch M, Conery J, Burger R. Evolution (Lawrence, Kans) 1995;49:1067–1080. doi: 10.1111/j.1558-5646.1995.tb04434.x. [DOI] [PubMed] [Google Scholar]
- 33.Wendel J F. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 34.Michelmore R W, Meyers B C. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 35.Collins N, Drake J, Ayliffe M, Sun Q, Ellis J, Hulbert S, Pryor T. Plant Cell. 1999;11:1365–1376. doi: 10.1105/tpc.11.7.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu X, Nei M. Mol Biol Evol. 1999;16:147–156. doi: 10.1093/oxfordjournals.molbev.a026097. [DOI] [PubMed] [Google Scholar]