Abstract
We investigated secondary immunity against coccidioidomycosis by using gene expression microarrays. Surprisingly, a high percentage of B-cell-related genes were associated with protective immunity. A functional confirmation of the importance of B cells against coccidioidomycosis was achieved by demonstrating that vaccination was not fully protective in B-cell-deficient MuMT mice.
Coccidioidomycosis is a pulmonary fungal disease caused by dimorphic fungi in the genus Coccidioides (8, 13). The cumulative interpretation of studies performed in humans and mice indicates that cell-mediated immune responses are essential in primary host defenses against Coccidioides and that recovery is associated with antigen-specific delayed-type hypersensitivity and the production of T helper 1 (Th1)-associated cytokines, such as gamma interferon (IFN-γ) and interleukin 2 (1, 2, 7-10, 19, 20). Conversely, disease progression is associated with low delayed-type hypersensitivity levels to Coccidioides antigens but high levels of anti-Coccidioides antibodies.
The optimum method to induce protective immunity in mice is vaccination with formalin-killed spherules (FKS) (6, 11, 12, 18). We showed that vaccination of susceptible BALB/c mice with FKS results in both reduction in fungal tissue burden and increased survival (10, 15). In secondary responses, IFN-γ levels and antibody levels increased, indicating that protective mechanisms could utilize both T cells and B cells. This study further dissected these secondary immune mechanisms using Affymetrix gene expression microarray analysis.
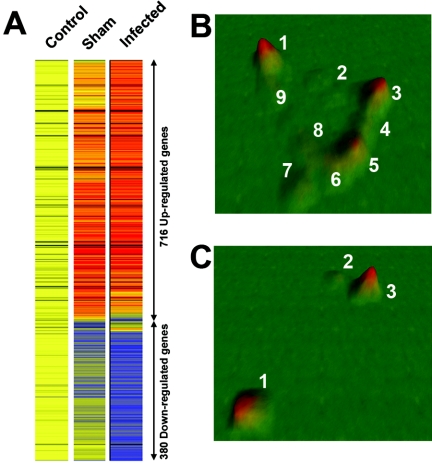
Female BALB/c mice were immunized with either FKS (by using spherules from Coccidioides posadasii strain Silveira; the FKS-immune group) or saline (the sham-immune group) and intranasally challenged with 26 arthroconidia (10). In two independent experiments, time-matched lungs were removed at day 12 postinfection and processed for RNA isolation by using TRIZOL (Invitrogen) and RNeasy kits (QIAGEN, Valencia, CA). Nonimmunized, noninfected BALB/c mice (control) lung tissue was also processed. Biotin-labeled amplified antisense RNA samples were generated and hybridized to individual Affymetrix murine MG-U74Av2 GeneChip microarrays (Affymetrix, Santa Clara, CA). Expression analysis was performed using GeneSpring 5.1 (Agilent, Redwood City, CA) and statistical analyses, including Pearson correlated measurements of gene expression similarity, were performed with P values of <0.05 considered significant. Following normalization, initial data filtering, performed to identify probe sets with signal intensities of >25 that were present in at least two of the six conditions, generated a list of 6,836 genes. We then performed filtering (n-fold change) using all possible condition-to-condition comparisons and identified 1,096 genes that were changed at least twofold in expression between any two conditions. Hierarchical cluster analysis separated these genes into two categories: those that exhibited increased gene expression (716 genes) and those that exhibited decreased gene expression (380 genes) relative to control samples (Fig. 1A).
FIG. 1.
(A) Initial hierarchical clustering of the 1,096 genes identified as >2-fold changed in expression in infected samples relative to mock-infected and control samples segregated this population into two groups: 716 up-regulated genes (red) and 380 down-regulated genes (blue). Up-regulated (B) and down-regulated (C) groups were further analyzed using VxInsight software to generate topographical terrain maps corresponding to subclusters of genes exhibiting similar expression.
Subcluster analysis was performed using VxInsight (Sandia National Labs, Albuquerque, NM) (5). This analysis creates a two-dimensional plot of genes with similar expression patterns between the conditions on the x-y plane. A third dimension represents an accumulation of genes revealing a topographical terrain of gene cluster “mountains.” The 716 up-regulated genes segregated into nine peaks (Fig. 1B), and the 380 down-regulated genes segregated into three peaks (Fig. 1C). Individual lists representing the gene populations in each of the nine peaks were imported into GeneSpring for expression pattern visualization. Peaks 1, 2, 3, and 4 (528 genes) exhibited increased gene expression in sham-immune mice relative to FKS-immune mice, and thus these genes could be associated with a failing host response following intranasal challenge. In contrast, the mean expression patterns of the genes in peaks 5, 6, 7, 8, and 9 (188 genes) exhibited increased expression in the FKS-immune mice and thus could be associated with protected host responses. Analyses of the lists revealed a marked increase in genes controlling immunoglobulin production or B-cell markers with 95 of the 188 genes (49.47%) in peaks 5 to 9, compared to 23 of the 528 genes (4.35%) in peaks 1 to 4 (P < 0.001 by chi-square analysis). While IFN-γ gene expression did not segregate in these analyses, there was an increase in interferon-regulated genes relative to control mice (Table 1). In addition, interleukin-4 and other Th2-associated gene inductions were seen in each of the up-regulated peaks. Thus, while there was no discernible Th1-Th2 cytokine bias in the FKS-protected response, both cytokine pathways were increased throughout the peaks.
TABLE 1.
Th1- and Th2-associated gene expression occurs in all peaks of the up-regulated genes
| Peak | GenBank accession no. | Description |
|---|---|---|
| 1 | M29697 | Interleukin-7 receptor mRNA |
| M33266 | Interferon-inducible protein mRNA | |
| AA144469 | Interferon-inducible protein 1-8U gene (human) | |
| Y09864 | mRNA for alpha/beta interferon receptor, IFNaR2b | |
| M31419 | 204-interferon-activatable-protein mRNA | |
| M31418 | 202-interferon-activatable-protein mRNA | |
| 2 | U22339 | Interleukin-15 receptor precursor mRNA |
| 3 | M27960 | Interleukin-4 receptor mRNA |
| M21065 | Interferon regulatory factor 1 mRNA | |
| AB019505 | Interleukin-18 binding protein gene | |
| X56602 | Interferon-induced 15-kDa protein gene | |
| 4 | U69599 | Gamma interferon receptor second chain (ifngr2) gene |
| 5 | U70430 | Interleukin-4-induced gene-1 partially spliced (Fig1ps) precursor RNA |
| M34815 | Mouse monokine induced by gamma interferon (MIG) mRNA | |
| U43428 | Inducible nitric oxide synthase mRNA | |
| 6 | U06924 | Signal transducer and activator of transcription (Stat1) mRNA |
| AF065947 | RANTES | |
| 7 | U04379 | Protein tyrosine kinase ZAP-70 mRNA |
| M12056 | Protein tyrosine kinase gene (lck gene) | |
| 8 | M32489 | Interferon consensus sequence binding protein mRNA |
| U56819 | MCP-1 receptor mRNA | |
| 9 | M21038 | Interferon-induced mutant Mx1 protein pseudo gene mRNA |
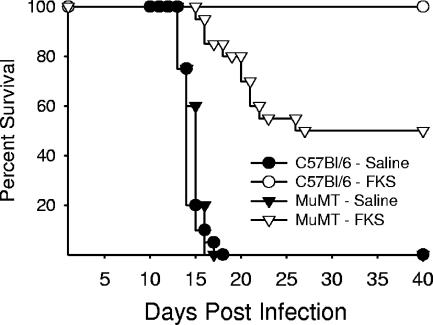
We sought to confirm that B-cell-associated genes were strongly up-regulated in FKS-immunized BALB/c mice by testing the efficacy of FKS to protect MuMT mice which are deficient in mature B cells but have relatively normal T-cell function (16). As described above, FKS-immune and sham-immune MuMT mice and C57BL/6 (wild-type controls) were intranasally infected with 30 arthroconidia and monitored (Fig. 2). All sham-immune mice succumbed within 20 days. In contrast, immunization of the C57BL/6 mice with FKS induced complete protective immunity as measured by 100 percent survival. FKS-immune MuMT mice exhibited only 50 percent survival over the observation period. This reduction in the level of protection, while significantly higher than that for the nonimmunized controls (P < 0.001; Mantel-Haenszel survival curve analysis), is significantly less than wild-type C57BL/6 mice with sufficient B cells (P < 0.007). The fact that FKS-immunized MuMT mice were significantly less protected than wild-type controls confirms the microarray results and establishes that B cells and/or antibody play a role in acquired protective immunity to Coccidioides.
FIG. 2.
Capacity for the FKS vaccine to induce immunity in B-cell-deficient MuMT mice. Results are presented as the combination of two independent experiments for a total of 15 mice per group. Triangles represent MuMT mice, and circles represent C57BL/6 (wild-type) mice, with open symbols representing those with FKS-immune treatment and solid symbols those with sham-immune treatment.
The results of this study appear to contrast with early studies of host defenses against Coccidioides which revealed that passive transfer of immune splenocytes conferred protection and that elimination of T cells from the population before transfer abrogated protection (3, 4, 17). While mice receiving whole-spleen-cell populations were completely protected, recipients given T-cell-depleted (B-cell-enriched) spleen cells succumbed by day 34 postinfection. The authors concluded that this was a total abrogation of protection; however, we extracted the data (3) and analyzed them to reveal a significant increase in the number of days the mice survived after passive transfer of immune B cells compared to nonimmune cells (P = 0.0097; Mantel-Haenszel analysis). While T cells are of major importance in host resistance to Coccidioides, when combined with the reanalysis of the results of Beaman et al. (3), our current results indicate that B cells or antibodies are also significant contributors to vaccine-induced protection against Coccidioides.
The ability to measure thousands of individual host responses can provide a biosignature that reflects the health status of the host. In this report, we utilized gene expression analysis, but this strategy could be adapted to directly detect changes in the proteome of the host for a more rapid assessment of the biosignature. These types of studies could also be used to establish the biosignature of protective versus nonprotective vaccine regimens. In addition, it may be possible to determine the infecting agent based upon the host biosignature response. For example, studies of gene expression profiles in dendritic cells challenged with viruses, bacteria, or fungi revealed distinct differences in their expression profiles (14). Based upon the biosignature, it may be possible to predict the outcome of infection and categorize those who would survive or those at high risk of mortality. The acquisition of a predictive biosignature would allow targeting therapy towards those who are at greatest risk.
Acknowledgments
This work was supported by a grant from the California HealthCare Foundation, the Department of Health Services of the State of California, and California State University, Bakersfield.
We thank Alexandra Smith and Jonathan Lawson for critical comments.
Editor: T. R. Kozel
REFERENCES
- 1.Ampel, N. M., and L. Christian. 1997. In vitro modulation of proliferation and cytokine production by human peripheral blood mononuclear cells from subjects with various forms of coccidioidomycosis. Infect. Immun. 65:4483-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ampel, N. M., L. A. Kramer, K. M. Kerekes, S. M. Johnson, and D. Pappagianis. 2001. Assessment of the human cellular immune response to T27K, a coccidioidal antigen preparation, by flow cytometry of whole blood. Med. Mycol. 39:315-320. [DOI] [PubMed] [Google Scholar]
- 3.Beaman, L., D. Pappagianis, and E. Benjamini. 1977. Significance of T cells in resistance to experimental murine coccidioidomycosis. Infect. Immun. 17:580-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaman, L. V., D. Pappagianis, and E. Benjamini. 1979. Mechanisms of resistance to infection with Coccidioides immitis in mice. Infect. Immun. 23:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyack, K. W., B. N. Wylie, and G. S. Davidson. 2002. Domain visualization using VxInsight for science and technology management. J. Am. Soc. Inf. Sci. Technol. 53:764-774. [Google Scholar]
- 6.Converse, J. L., M. W. Castleberry, A. R. Besemer, and E. M. Snyder. 1962. Immunization of mice against coccidioidomycosis. J. Bacteriol. 84:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corry, D. B., N. M. Ampel, L. Christian, R. M. Locksley, and J. N. Galgiani. 1996. Cytokine production by peripheral blood mononuclear cells in human coccidioidomycosis. J. Infect. Dis. 174:440-443. [DOI] [PubMed] [Google Scholar]
- 8.Cox, R. A. 1989. Coccidioidomycosis, p. 165-197. In R. A. Cox (ed.), Immunology of fungal diseases. CRC Press, Boca Raton, Fla.
- 9.Cox, R. A., and D. R. Arnold. 1979. Immunoglobulin E in coccidioidomycosis. J. Immunol. 123:194-200. [PubMed] [Google Scholar]
- 10.Cox, R. A., W. Kennell, L. Boncyk, and J. W. Murphy. 1988. Induction and expression of cell-mediated immune responses in inbred mice infected with Coccidioides immitis. Infect. Immun. 56:13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, R. A., and D. M. Magee. 2004. Coccidioidomycosis: host response and vaccine development. Clin. Microbiol. Rev. 17:804-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman, L., and C. E. Smith. 1956. Vaccination of mice against Coccidioides immitis. Am. Rev. Tuberc. Pulm. Dis. 74:245-248. [DOI] [PubMed] [Google Scholar]
- 13.Galgiani, J. N. 1993. Coccidioidomycosis. West. J. Med. 159:153-171. [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, Q., D. Liu, P. Majewski, L. C. Schulte, J. M. Korn, R. A. Young, E. S. Lander, and N. Hacohen. 2001. The plasticity of dendritic cell responses to pathogens and their components. Science 294:870-875. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, C., D. M. Magee, T. N. Quitugua, and R. A. Cox. 1999. Genetic vaccination against Coccidioides immitis: comparison of vaccine efficacy of recombinant antigen 2 and antigen 2 cDNA. Infect. Immun. 67:630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitamura, D., J. Roes, R. Kuhn, and K. Rajewsky. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350:423-426. [DOI] [PubMed] [Google Scholar]
- 17.Kong, Y. M., D. C. Savage, and H. B. Levine. 1965. Enhancement of immune responses in mice by a booster injection of Coccidioides spherules. J. Immunol. 95:1048-1056. [PubMed] [Google Scholar]
- 18.Levine, H. B., J. M. Cobb, and C. E. Smith. 1961. Immunogenicity of spherule-endospore vaccines of Coccidioides immitis for mice. J. Immunol. 87:218-227. [PubMed] [Google Scholar]
- 19.Magee, D. M., and R. A. Cox. 1996. Interleukin-12 regulation of host defenses against Coccidioides immitis. Infect. Immun. 64:3609-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magee, D. M., and R. A. Cox. 1995. Roles of gamma interferon and interleukin-4 in genetically determined resistance to Coccidioides immitis. Infect. Immun. 63:3514-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]