Abstract
Streptococcus iniae represents a major health and economic problem in fish species worldwide. Random Tn917 mutagenesis and high-throughput screening in a hybrid striped bass (HSB) model of meningoencephalitis identified attenuated S. iniae mutants. The Tn917 insertion in one mutant disrupted an S. iniae homologue of a phosphoglucomutase (pgm) gene. Electron microscopy revealed a decrease in capsule thickness and cell wall rigidity, with ΔPGM mutant cells reaching sizes ∼3-fold larger than those of the wild type (WT). The ΔPGM mutant was cleared more rapidly in HSB blood and was more sensitive to killing by cationic antimicrobial peptides including moronecidin from HSB. In vivo, the ΔPGM mutant was severely attenuated in HSB, as intraperitoneal challenge with 1,000 times the WT lethal dose produced only 2.5% mortality. Reintroduction of an intact copy of the S. iniae pgm gene on a plasmid vector restored antimicrobial peptide resistance and virulence to the ΔPGM mutant. In analysis of the aborted infectious process, we found that ΔPGM mutant organisms initially disseminated to the blood, brain, and spleen but were eliminated by 24 h without end organ damage. Ninety to 100% of fish injected with the ΔPGM mutant and later challenged with a lethal dose of WT S. iniae survived. We conclude that the pgm gene is required for virulence in S. iniae, playing a role in normal cell wall morphology, surface capsule expression, and resistance to innate immune clearance mechanisms. An S. iniae ΔPGM mutant is able to stimulate a protective immune response and may have value as a live attenuated vaccine for aquaculture.
With increased development of intensive operations, disease has become a significant hurdle to the profitable culture of fish and shellfish. Streptococcal infections in fish, in particular those produced by the pathogen Streptococcus iniae, have increased markedly with intensification of aquaculture practices (37). S. iniae causes a fatal meningoencephalitis and is associated with large-scale mortality in a wide variety of marine and freshwater cultured fish species, as well as in wild species (5, 39, 46). More than 30 species of fish have documented susceptibility to S. iniae disease, including trout (10), yellowtail (19), tilapia (39), barramundi (6), and hybrid striped bass (HSB) (11). S. iniae is distributed globally and is estimated to cause yearly economic losses of hundreds of millions of dollars. Occasionally, S. iniae can cause serious zoonotic infections in humans who injure themselves while handling infected fish (20, 42).
Relatively little is known of the genetics of S. iniae or of the pathogenic mechanisms underlying its virulence. Here, we describe a severely attenuated mutant of S. iniae, identified through random transposon mutagenesis and direct screening for virulence in HSB. Analysis of the transposon insertion site revealed a disruption of an open reading frame (ORF) with similarity to bacterial phosphoglucomutase (pgm) genes. The enzyme phosphoglucomutase (PGM) interconverts glucose-6-phosphate and glucose-1-phosphate, and it has recently been discovered to play an important role in polysaccharide capsule production and virulence in a variety of gram-positive and gram-negative bacterial pathogens. In the present study, we characterize phenotypic changes associated with loss of PGM in S. iniae, including alterations in cell wall morphology, capsule production, and susceptibility to fish innate immune defenses. Loss of PGM is associated with markedly reduced virulence in S. iniae, and preliminary data are provided to suggest that avirulent, PGM-deficient S. iniae may serve as an effective live attenuated vaccine to protect aquacultured fish against fatal meningoencephalitis.
MATERIALS AND METHODS
Bacteria strains, culture, transformation, and DNA techniques.
Wild-type (WT) S. iniae strain K288 was isolated from the brain of a diseased HSB at Kent SeaTech. K288 was identified unambiguously as S. iniae through biochemical testing and by analysis of ribosomal 16S sequence (data not shown) and proven to be virulent in a HSB model by intraperitoneal (IP) injection of 4 × 105 CFU, a lethal dose in all fish tested. S. iniae was propagated in Todd-Hewitt broth (THB) or on Todd-Hewitt agar (THA) at 30°C unless otherwise indicated, with antibiotic selection of 2 μg/ml chloramphenicol (Cm), 5 μg/ml erythromycin (Em), and 500 μg/ml kanamycin when required. Escherichia coli was grown in Luria-Bertani (LB) medium at 37°C using 500 μg/ml of Em, 15 μg/ml of Cm, or 100 μg/ml of ampicillin for selection unless otherwise indicated. S. iniae was rendered competent for electroporetic transformation by growth in THB plus 0.6% glycine, following procedures for group B Streptococcus (12). Transformants were identified by recovery for 2 h at 30°C in SOC medium (Invitrogen Corp., Carlsbad, CA) for E. coli and THB plus 0.25 M sucrose for S. iniae, followed by plating on appropriate antibiotic selective media. For use in fish challenges, overnight cultures of S. iniae were diluted 1:20 and grown to mid-log phase (optical density at 600 nm [OD600] = 0.4) corresponding to ∼4 × 108 CFU/ml. Plasmid DNA was isolated from E. coli with QiaPrep kits (QIAGEN, Valencia, CA). For S. iniae, a 15-min incubation at 37°C in 100 U of mutanolysin (Sigma, St. Louis, MO) preceded plasmid isolation with the QiaPrep kit. Genomic DNA was isolated with the UltraClean DNA Isolation Kit (MoBio, Carlsbad, CA).
Transposon mutagenesis.
Transposon mutagenesis of strain K288 followed procedures described for Streptococcus mutans using the temperature-sensitive plasmid pTV1OK bearing transposon Tn917 (13), with slight modifications. Individual colonies of K288 transformed with pTV1OK were inoculated into THB plus kanamycin and grown to OD600 = 0.9 at the permissive temperature (30°C) for plasmid replication. Cultures were diluted 1:100 in THB plus Em and grown at a nonpermissive temperature (37°C) to an OD600 value of 0.9, then plated on THA plus Em for isolation of candidate insertion mutants. Libraries of S. iniae mutants bearing random transposon insertions into the genome were verified by Southern blot analysis (data not shown).
In vivo screening of transposon mutants for loss of virulence.
HSB (Morone chryosops × Morone saxitilis) aged <1 year and with an average weight of approximately 30 g each were used for an in vivo challenge model of S. iniae infection. HSB were challenged IP with 100 μl of WT or mutant S. iniae bacteria resuspended in phosphate-buffered saline (PBS) at known inocula and injected using a 27-gauge needle. Fish were held with aeration and flowthrough water at 24 to 27°C for 7 days after challenge and were monitored twice daily for mortalities. Brain biopsy cultures were taken from selected dead fish to confirm infection with the challenge S. iniae strain, based on the appropriate antibiotic sensitivity profile. Screening the transposon library in this manner revealed numerous attenuated mutants, including mutant TnM2 with a transposon disruption of a putative pgm gene.
Identification of the pgm gene.
The Tn917 insertion site in the attenuated mutant TnM2 was identified by direct sequencing from genomic DNA using the modified primer (fimer) 5′-GAAACATTGGTTTAGTGGGAATTTGTAC-3′ and 0.1 μl of Thermofidelase (Fidelity Systems) in a 20-μl cycle sequencing reaction mixture (Big Dye v3.0; Applied Biosystems, Inc.). Chromosome walking using a single-primer PCR technique was used to discover the entire sequence of pgm and flanking regions (17, 18). Sequence files were analyzed with Chromas (Technelysium, Tewantin, Australia), aligned with BioEdit (Ibis Therapeutics, Carlsbad, CA), and annotated with Artemis, version 5.0 (Sanger Institute, Cambridge, United Kingdom). The amino acid sequence of PGM was compared to sequences in the GenBank databases with the BlastP program (2).
Complementation and heterologous expression of S. iniae pgm.
The entire pgm gene plus the upstream putative promoter region (1865 bp) was amplified from WT S. iniae K288 genomic DNA using forward primer 5′-GAACTAGCTAGTTACTTTTGTAACTG-3′ and reverse primer 5′-CTAATTCACAAAAGTGTTGATTTCAG-3′ in a standard PCR using Platinum Supermix (Invitrogen, Carlsbad, CA) and 30 cycles of denaturation (94°C for 30 s), annealing (55°C for 30 s), and elongation (72°C for 1.5 min). The resulting product was T-A cloned into pCR2.1Topo (Invitrogen, Carlsbad, CA). The pgm gene was cut from the pCR2.1Topo intermediate with BamHI and XbaI and cloned into the corresponding sites in the E. coli-streptococcus shuttle expression vector pDC123 (7) bearing Cmr to create pSiPGM. This recombinant vector was used to transform WT S. iniae K288, S. iniae transposon mutant TnM2, and an E. coli K-12 ΔPGM mutant (1); similar transformations were performed with pDC123 to serve as a vector-only control. Transformants were identified by Cmr and confirmed by restriction enzyme and PCR analysis of plasmid preparations from transformed cells.
Phosphoglucomutase activity assays.
Cells extracts were prepared from bacteria in mid-log phase (OD600 = 0.4), collected by centrifugation, and washed twice in chilled PBS. For E. coli, cells were resuspended in 50 mM triethanolamine buffer with 5 mM MgCl2 (pH 7.2), and suspensions were frozen at −80°C. Cell lysates were prepared using sonication (three bursts of 10 s each). For S. iniae, cell pellets were frozen. Cells lysates were prepared using the Cellytic B plus reagent (Sigma Aldrich, St. Louis, MO) following the manufacturer's instructions. All cell lysates were centrifuged (30 min at 10,000 × g), and the supernatants (crude cell extracts) were stored at −80°C until use. The specific activity of PGM in cell extracts was measured as the conversion of α-glucose-1-phosphate to glucose-6-phosphate in a reaction coupled to reduction of glucose-6-phosphate by glucose-6-phosphate dehydrogenase and quantitated spectrophotometrically at 340 nm by monitoring the formation of NADPH from NADP+ at 30°C. The PGM assay solution contained final concentrations of 5 mM MgCl2, 0.4 mM NADP+, 2 U/ml glucose 6-phosphate dehydrogenase, and 50 μM α-glucose-1,6-bisphosphate. After the cell extract was added, the reaction was initiated with addition of α-glucose-1-phosphate to a concentration of 1.4 mM. Parallel phosphomannomutase assays were performed with the above reagents plus the addition of 2 U/ml each of phosphomannoisomerase and phosphoglucoisomerase and activated with a final concentration of 1.4 mM α-mannose-1-phosphate. All chemicals were obtained from Sigma Aldrich (St. Louis, MO).
Fish virulence studies.
Groups of 40 HSB (each, ∼30 g) were challenged IP with 2 × 105 to 2 × 108 CFU of WT or mutant S. iniae bacteria from log-phase (OD600 = 0.4) cultures resuspended in 100 μl of PBS. Fish were held with aeration and flowthrough water at 24 to 27°C for 14 days after challenge and were monitored twice daily for mortalities. To compare the virulence of the complemented mutant TnM2[pSiPGM] to the WT and TnM2 strains, groups of 28 fish for each treatment were injected with 4 × 105 CFU and monitored for 7 days. Three fish were selected at random from each group at 24 h postchallenge and sacrificed, and the number of CFU of S. iniae per milliliter of blood was enumerated by plating dilutions on THA.
Assays for buoyancy, surface charge, and hydrophobicity.
To measure buoyancy, sequential overlay gradients of 1 ml (each) of 70%, 60%, and 50% Percoll were prepared in 5-ml glass test tubes. One milliliter of overnight bacterial culture was placed on top of the Percoll layers, the tubes were centrifuged in a swinging bucket centrifuge for 8 min at 500 × g, and the migration of the bacteria to various Percoll interphases was recorded. To measure surface charge, bacteria were grown to stationary phase, harvested by centrifugation, and washed twice in morpholinepropanesulfonic acid (MOPS) buffer (20 mM; pH = 7.0). Cells were resuspended in MOPS to OD600 = 6.0, and cytochrome c (Sigma, St. Louis, MO) was added to a final concentration of 0.5 mg/ml. After 15-min incubation at 23°C, samples were centrifuged (13,000 × g for 5 min), and the amount of cytochrome c remaining in the supernatant was quantitated spectophotometrically at 530 nm. To measure hydrophobicity, stationary-phase cultures were washed twice in PBS and resuspended in PBS at OD600 = 1.0. A total of 300 μl of n-hexadecane was layered on top of the cell suspension, and the tubes were vortexed for 60 s. Samples were incubated for 30 min at 23°C to allow for phase separation. The aqueous phase was removed, and the OD600 value was recorded to determine the quantity of remaining bacteria.
Electron microscopy.
S. iniae were grown to late log phase (OD600 = 0.7) and collected by centrifugation. Pellets were resuspended in 0.07 M cacodylate buffer and transferred to 2.0-ml microfuge tubes. Samples were washed twice (0.07 M cacodylate buffer), resuspended in 0.2 M cacodylate plus 3.6% glutaraldehyde, incubated on ice for 1 h, and then washed three additional times. Polycationic ferritin (Sigma) was added to a final concentration of 1.0 mg/ml, and the samples were incubated for 30 min, washed twice, and stored at 4°C. Prior to electron microscopy, bacteria were washed three times in 0.1 M sodium phosphate buffer (pH 7.3), postfixed for 1 h in 0.1 M phosphate-buffered 2% osmium tetroxide, and rinsed three times in distilled water (dH2O). Dehydration was performed using ethanol at 30%, 50%, 70%, 95%, and 100% concentrations. Bacteria were immersed in two rinses of propylene oxide and incubated for 2 h in a mixture of 50% propylene oxide and 50% epoxy resin. Mollenhaure's formulation of Epon-Araldite was used to embed bacteria for thin sectioning. Sixty-nanometer sections were placed on 300-mesh copper support grids and viewed using a Zeiss EM 10 electron microscope at an acceleration voltage of 80 kV with ×16,000 and ×50,000 magnifications. Negatives were enlarged and printed to final magnifications of ×35,000 and ×109,000.
Blood survival assay.
Mid-log-phase S. iniae suspensions of ∼300 CFU in 100 μl of PBS were added to 300 μl of fresh heparinized HSB blood in 2-ml siliconized plastic tubes and incubated for 1 h at 30°C on an orbital shaker. After 1 h, 100-μl aliquots were taken from each sample in duplicate and plated on THA for enumeration of surviving bacteria. Each experiment was repeated three times, each time using the blood of a different HSB.
Antimicrobial peptide sensitivity.
Early-log-phase cultures of S. iniae (OD600 = 0.2) were diluted in fresh THB to ∼2 ×105 CFU/ml. A total of 180 μl of this bacterial suspension was added to replicate wells of a 96-well plate. Dilutions of the antimicrobial peptides (AMPs) moronecidin (2 μM) (from HSB) and CRAMP (16 μM) (from mouse) were prepared in dH2O and added to the wells in 20 μl of distilled H2O; dH2O alone was used as a negative control. To measure antimicrobial killing kinetics, 20-μl aliquots of each well were serially diluted in PBS and plated at specified time points after addition of the antimicrobial peptide for CFU determination. Each experiment was performed in triplicate.
Characterization of the S. iniae infection process.
HSB (Morone chryosops × Morone saxitilis) aged <1 year and with an average weight of ∼20 g each were used to characterize the attenuation infection process of mutant TnM2. Groups of 50 HSB were challenged by IP injection of 2.5 × 105 CFU/100 μl of PBS of log-phase WT or TnM2 S. iniae. Fifty control fish were injected with 100 μl of PBS (each). Fish were held with aeration and flowthrough water at 24 to 27°C after challenge. Three fish were sacrificed at various time points during the first 124 h of infection, and bacterial load in the brain, blood, and spleen was calculated by weighing tissue samples and homogenizing tissue samples in PBS (Tissue Tearor; Biospec Products, Bartlesville, OK). Serial dilutions of each tissue homogenate in PBS were plated in duplicate on blood agar for enumeration of CFU. All TnM2 bacteria recovered were confirmed to have maintained the Emr phenotype. Brain and spleen tissues from each of the three fish sampled at 96 h were pooled and placed in 10% buffered formalin until processing. Tissue pools were trimmed into cassettes and embedded in paraffin, and sections were cut for routine histology. Serial sections were stained with either hematoxylin and eosin (H&E) or Giemsa stain; all sections were examined in a blinded fashion and scored as the number of positive traits seen in the sections of the tissue examined. H&E-stained sections were used to score the respective spleen and brain changes, while Giemsa stain-treated sections were used to subjectively quantify the degree of bacteria associated with splenic ellipsoids and the meninges, respectively. The following criteria were used to assess the splenic response to infection: congestion, capsular hypertrophy, peritoneal inflammation, and ellipsoidal degeneration. Criteria for brain changes include meningeal inflammation and ventricle (optic lobe) inflammation.
Vaccine trial.
To test the ability of TnM2 to function as a live attenuated vaccine, groups of 40 fish (each, ∼30 g) were injected IP with 4 × 105, 4 × 106, 4 × 107, or 4 × 108 CFU of TnM2 in PBS. Forty controls were injected with PBS alone. Fish were held for 2,000 degree days (∼4 months); no mortalities were observed. Fish were then challenged by injection with the previously determined lethal dose (4 × 105 CFU) of K288, held in mixed groups for 21 days at 24 to 27°C, and monitored for mortality. Brain biopsy cultures were taken from all dead fish and cultured to confirm S. iniae meningoencephalitis as the cause of death.
Nucleotide sequence accession number.
The nucleotide sequence for pgmA identified in this study has been deposited in GenBank under accession number AY846302.
RESULTS
Sequence characteristics of S. iniae PGM.
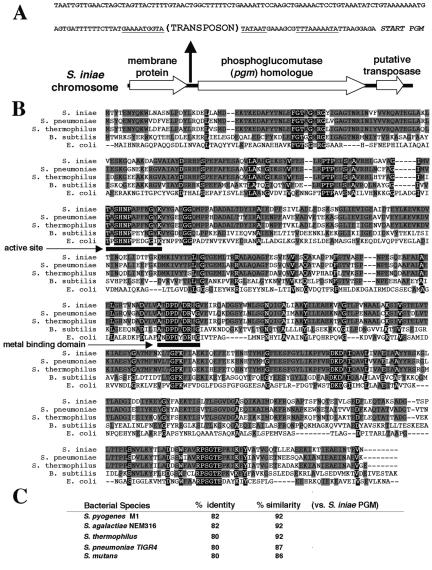
The attenuated mutant TnM2 was identified in a screen of a Tn917 chromosomal insertion library of WT S. iniae strain K288 for loss of virulence in HSB. By direct sequencing and single-primer chromosome walking, the site of the sole Tn917 insertion in TnM2 and surrounding DNA sequence was determined (Fig. 1A). Tn917 was found to have inserted in a predicted promoter region 32 bp upstream of the start ATG codon of the ORF, sharing strong sequence homology to genes encoding PGM enzymes. Herein, this ORF is designated pgmA BlastP analysis in GenBank of the deduced amino acid sequence of the candidate S. iniae PGM revealed strong sequence homology and identity with known α-PGM enzymes from other gram-positive bacteria species (Fig. 1B and C), as well as several known α-PGMs from gram-negative and vertebrate species (not shown). In particular, the functional regions characteristic of α-PGM proteins were conserved in the S. iniae homologue, including a metal binding domain and active site residues (8).
FIG. 1.
Sequence features of Streptococcus iniae PGM. (A) Chromosomal location of S. iniae PGM and site of Tn917 insertion in promoter motifs upstream of translational start site. Underlined areas indicate conserved promoter motifs. (B) Alignment of S. iniae PGM with known bacterial PGMs and indication of putative conserved functional domains. (C) Degree of homology of S. iniae PGM to other streptococcal PGMs.
PGM activity in S. iniae is linked to pgmA.
Biochemical assays were performed to confirm that the pgmA ORF encoded a functional PGM enzyme. Measured PGM activity decreased more than fivefold in mutant TnM2 compared to the WT S. iniae parent strain (Fig. 2A). Complementation of TnM2 with plasmid pSiPGM restored PGM activity to approximately WT levels. In parallel assays, we were unable to detect phosphomannomutase activity in S. iniae cell extracts and thus conclude that PGM enzyme lacks the dual activity described in a subset of known PGM enzymes. A mutant strain of E. coli (Pop458) with a disrupted native pgm gene had greatly reduced PGM activity compared to that of WT E. coli. Heterologous expression of pSiPGM in E. coli Pop458 significantly increased PGM activity (P < 0.0001) (Fig. 2B). Together, these studies demonstrate that pgmA is both necessary and sufficient for PGM enzymatic activity.
FIG. 2.
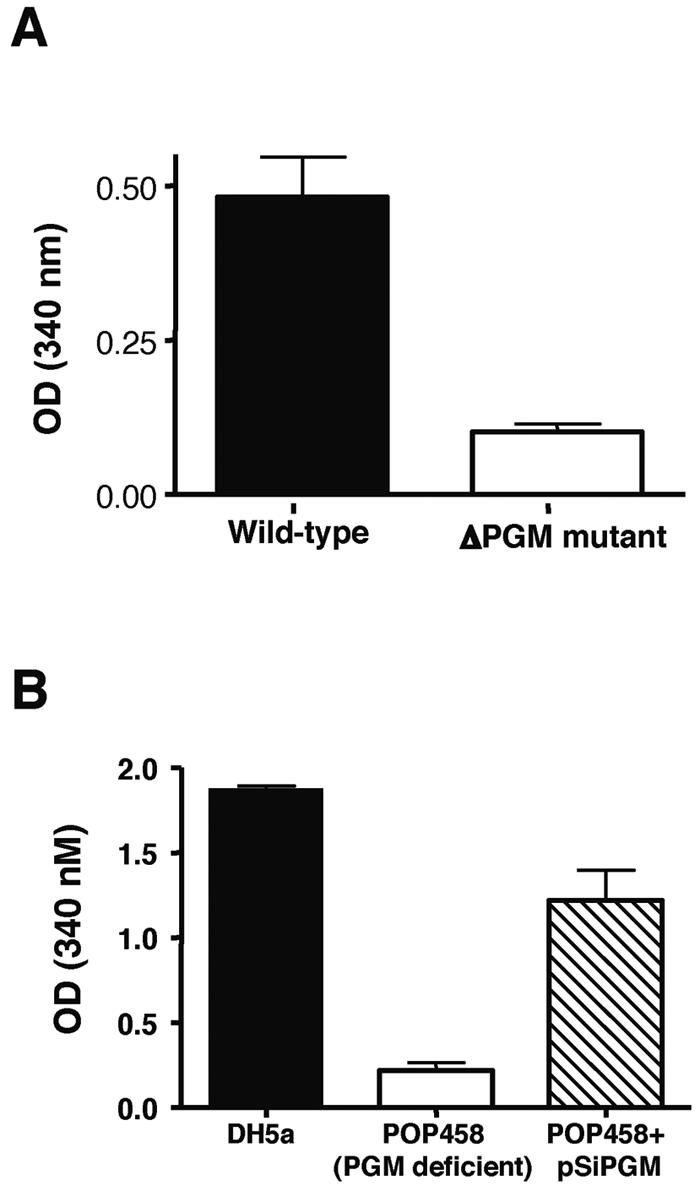
Streptococcus iniae PGM is a functional phosphoglucomutase enzyme. (A) Loss of PGM activity in S. iniae ΔPGM mutant. (B) Heterologous expression of S. iniae PGM restores phosphoglucomutase activity to E. coli PGM mutants.
S. iniae PGM expression is linked to virulence.
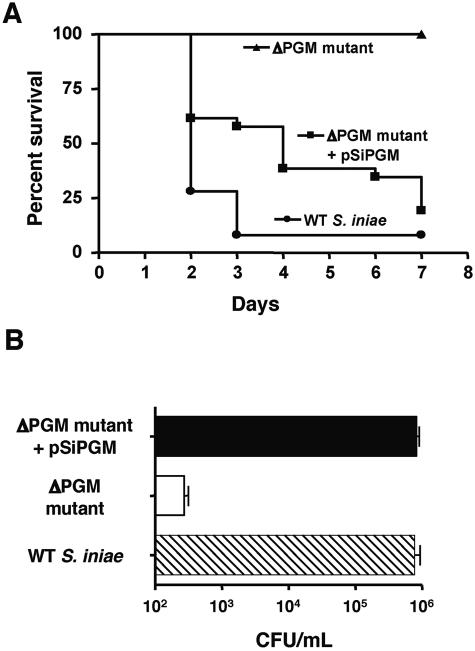
The HSB challenge model was used to evaluate the virulence potential of S. iniae mutant TnM2 with reduced PGM activity. Whereas 100% of HSB injected with 4 × 105 WT S. iniae died of meningoencephalitis, mortality was absent in fish challenged with TnM2 at the same dose; the rate of mortality was only 2.5% in fish challenged with the mutant at a 1,000-fold higher inoculum (P < 0.0001) (Table 1). In a separate experiment, survival curves were plotted for HSB after intraperitoneal challenge with either 4 × 105 CFU of WT S. iniae, mutant TnM2, or TnM2 complemented with pSiPGM. While no mortality was seen in fish infected with TnM2, reintroduction of pgmA to the mutant on a plasmid vector resulted in mortality comparable that observed with WT S. iniae (Fig. 3A). Measurement of bacterial load in the blood of infected fish at 24 h appeared to predict mortality. Bacterial levels in the blood of HSB infected with WT S. iniae or TnM2[pSiPGM] were 1,000-fold higher than those observed with TnM2 (Fig. 3B). The severe attenuation of mutant TnM2 in animal challenges identifies pgmA as being associated with one or more virulence phenotypes of S. iniae.
TABLE 1.
Attenuation of Streptococcus iniae ΔPGM mutant (TnM2) in a hybrid striped bass infection modela
| Bacterium | Dose (CFU) | No. of fish | % Mortality |
|---|---|---|---|
| WT S. iniae | 4 × 105 | 40 | 100 |
| ΔPGM mutant | 4 × 105 | 40 | 0 |
| 4 × 106 | 40 | 0 | |
| 4 × 107 | 40 | 0 | |
| 4 × 108 | 40 | 2.5 |
Fish (≈30 g each) were injected intraperitioneally with the specified bacterial dose and held at 26°C for 14 days; mortalities were identified and removed daily.
FIG. 3.
Phosphoglucomutase is a virulence factor in the pathogenesis of Streptococcus iniae meningoencephalitis. (A) Kaplan-Meier survival plot of HSB challenged with 4 × 105 CFU of wild-type S. iniae, the ΔPGM mutant, or the complemented mutant. (B) Bacterial counts in blood of fish 24 h after infection.
The S. iniae ΔPGM mutant is more sensitive to innate immune clearance mechanisms.
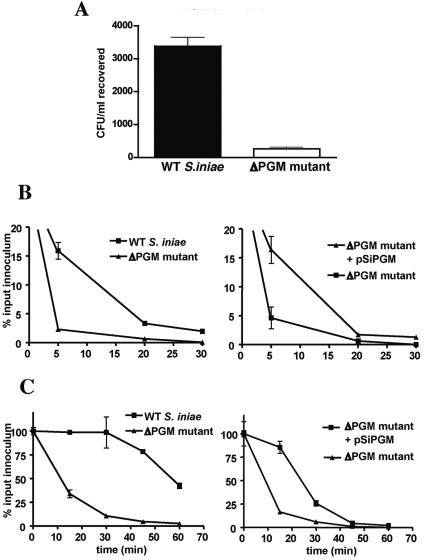
To measure the relative susceptibility of WT S. iniae and the ΔPGM mutant TnM2 to phagocytic clearance, a survival assay was performed with fresh blood isolated from HSB. Whereas the WT strain proliferated markedly (∼4-fold) in HSB blood, mutant TnM2 decreased in number (Fig. 4A). Cationic AMPs are recognized to be important components of innate defense and phagocyte killing in higher organisms (45). HSB produce the AMP moronecidin (also known as piscidin) possessing broad-spectrum antimicrobial activity (21, 40). The ΔPGM mutant TnM2 was found to be significantly more sensitive than the WT S. iniae strain to moronecidin; similar results were seen with the murine cationic AMP CRAMP. In vitro killing kinetics for each AMP showed that the rate of killing of the ΔPGM mutant was accelerated significantly compared to the WT S. iniae strain, whereas complementation of the mutant with pSiPGM produced a commensurate delay in the time course of AMP killing (Fig. 4B).
FIG. 4.
Phosphoglucomutase contributes to Streptococcus iniae bloodstream survival and resistance to AMP killing. (A) CFU recovered (mean ± standard error) of wild-type Streptococcus iniae and the isogenic ΔPGM mutant after incubation in HSB blood. Kinetics of killing of wild-type S. iniae, the ΔPGM mutant, or the complemented mutant by the HSB AMP moronecidin (B) and the murine AMP mCRAMP (C).
S. iniae PGM mutants have an altered cell phenotype.
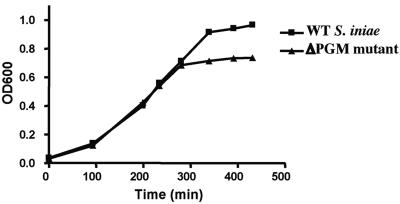
Assays were employed to identify general cell characteristics associated with loss of PGM activity in S. iniae. Logarithmic-phase growth of WT S. iniae and the ΔPGM mutant in THB were equivalent (Fig. 5). Migration through a Percoll gradient was increased in the ΔPGM mutant TnM2 compared to the WT S. iniae strain, indicative of a decrease in buoyancy (data not shown). More ΔPGM mutant cells were found to partition from the aqueous phase into n-hexadecane than did WT S. iniae cells, consistent with an overall increase in cell hydrophobicity (data not shown). The ΔPGM mutant also bound less cytochrome c than did the WT S. iniae strain (data not shown), indicating that the mutant possessed a net decrease in surface negative charge. Our transmission electron microscopic analysis of WT and ΔPGM mutant S. iniae labeled with polycationic ferritin revealed that the mutant exhibited decreased amounts of surface-associated exopolysaccharide capsule (4) and that the average cell volume of ΔPGM mutant bacteria was three to five times larger than that of the WT strain (Fig. 6).
FIG. 5.
Streptococcus iniae and the ΔPGM mutant have identical growth through the logarithmic phase in Todd-Hewitt broth as measured by optical density at 600 nm. The ΔPGM mutant does not achieve equivalent cell density at stationary phase.
FIG. 6.
Phenotypic differences observed between Streptococcus iniae and the isogenic ΔPGM mutant include increased cell size and decreased level of encapsulation, as appreciated by transmission electron microscopy.
Analysis of the aborted infectious process of ΔPGM mutant S. iniae.
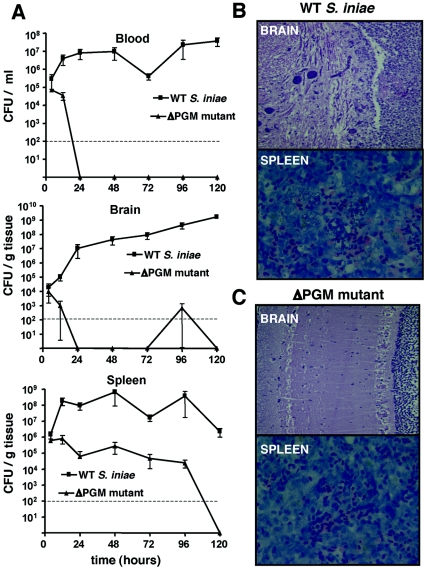
To further characterize the basis for the severely attenuated virulence of PGM-deficient S. iniae, we followed the course of infection in HSB challenged with WT and ΔPGM mutant S. iniae. At 4 h postinfection, WT bacteria were found at 105 CFU/ml in blood, >104 CFU/g in brain, and ∼106 CFU/g in spleen (Fig. 7A). Bacterial counts in WT S. iniae-infected fish increased steadily over time, surpassing 107 CFU/g in all tissues between 24 and 96 h of infection; widespread mortality was observed. Histological examination of brain revealed severe granulomatous meningeal inflammation with associated bacterial foci, and histology of the spleen showed large numbers of bacteria within degenerate splenic ellipsoids (Fig. 7B; Table 2). HSB challenged intraperitoneally with the S. iniae ΔPGM mutant similarly exhibited dissemination of the bacteria to blood, brain, and spleen such that 4-h counts of these tissues were only modestly less than those observed with WT infection (Fig. 7A). In stark contrast to the WT strain, the mutant was rapidly cleared from both blood and brain to undetectable levels within 24 h and slowly cleared from the spleen to undetectable levels by 5 days of infection (Fig. 7A). No mortalities were observed in the ΔPGM mutant-infected group of HSB. Histological examination of brain and spleen from fish infected with the ΔPGM mutant did not reveal evidence of the inflammatory damage produced in WT-infected HSB (Fig. 7B; Table 2).
FIG. 7.
Characterization of the aborted infectious process produced by the Streptococcus iniae phosphoglucomutase mutant (ΔPGM). (A) Bacterial counts in blood, spleen, and brain of HSB at different time points after intraperitoneal challenge. (B and C) Representative histological sections of brain (48 h; H&E stain, ×20 magnification) and splenic (120 h; Giemsa stain, ×40 magnification) tissues. With wild-type S. iniae infection, severe granulomatous meningeal inflammation and bacterial infiltration are seen in brain tissue (B, top), and large numbers of bacteria are associated with degenerate splenic ellipsoids (B, bottom). (C) In contrast, fish exposed to the ΔPGM mutant lacked significant histopathologic abnormalities in the meninges (top) or splenic (bottom) tissues.
TABLE 2.
Histological survey of brain and spleen of hybrid striped bass infected with wild-type Streptococcus iniae and the ΔPGM mutant
| Test and tissue studied | No. of samples infected/no. tested (h)
|
||||
|---|---|---|---|---|---|
| PBS | WT S. iniae (48) | ΔPGM (48) | WT S. iniae (96) | ΔPGM (96) | |
| Giemsa stain | |||||
| Spleen: bacteria associated with ellipsoids | 0/2 | 3/3 | 0/3 | 3/3 | 0/3 |
| Brain: bacteria associated with meninges | 0/2 | 3/3 | 0/3 | 3/3 | 0/3 |
| Brain histology | |||||
| Meningeal inflammation | 0/2 | 2/3 | 0/3 | 3/3 | 0/3 |
| Ventricle (optic lobe) inflammation | 0/2 | 0/3 | 0/3 | 2/2 | 0/3 |
| Relative severity of histological lesions (0-4) | |||||
| Spleen | 0 | 4 | 1 | 3 | 1 |
| Brain | 0 | 3 | 0 | 4 | 0 |
Efficacy of live attenuated vaccination with ΔPGM mutant against S. iniae infection.
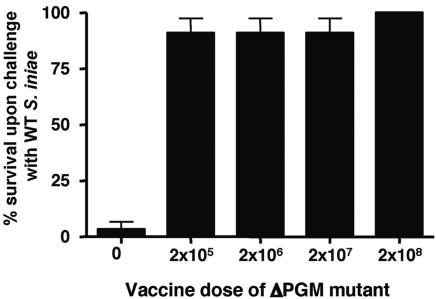
We hypothesized that the aborted infectious process of the attenuated ΔPGM mutant would elicit an immune response capable of protecting HSB against subsequent WT S. iniae infection. A vaccination trial was performed by infecting HSB with various concentrations of the S. iniae ΔPGM mutant TnM2 (or PBS as a negative control), allowing them to spontaneously clear the infection, and then housing them for a period of 2,000 degree days (∼4 months). At this point, all animals were challenged with a typically lethal dose (4 × 105 CFU) of WT S. iniae strain K288. Only a 3% survival rate was observed with control fish that were mock immunized with PBS, while all groups of fish that were previously challenged with various doses of the attenuated ΔPGM mutant experienced survival rates of at least 90% (Fig. 8). At the highest concentration of mutant used for live attenuated vaccination (2 × 108 CFU), 100% of fish survived subsequent challenge with a lethal dose of the WT S. iniae strain. This preliminary trial indicates that the ΔPGM mutant TnM2 elicits an effective immune response and may have value as a live attenuated vaccine to protect HSB against S. iniae infection.
FIG. 8.
Live attenuated vaccination with the Streptococcus iniae ΔPGM mutant protects HSB against a lethal wild-type infectious challenge. Groups of 30 HSB (each, ∼30 g) were injected IP with stated doses of ΔPGM mutant vaccine or PBS control. After 2,000 degree days, HSB from each treatment were challenged IP with a lethal dose of 2 × 105 CFU of wild-type S. iniae and survival monitored for 21 days.
DISCUSSION
Our studies have identified a mutant of S. iniae with severely attenuated virulence in a model of fish meningoencephalitis. The transposon mutation maps to the promoter of an ORF encoding the S. iniae homologue of α-PGM, sharing >80% amino acid identity to PGMs of other streptococcal species and all conserved regions for enzymatic activity. Single-gene complemenation analysis confirms that both PGM enzymatic activity and virulence attenuation are associated with the pgmA gene. The phenotype of the S. iniae ΔPGM mutant is characterized by increased cell size, decreased buoyancy, decreased encapsulation, decreased surface negative charge, more rapid clearance in blood, increased susceptibility to cationic AMPs, and a profound decrease in disease potential in vivo. To our knowledge, pgmA represents the first molecular virulence determinant identified for S. iniae pathogenicity in fish.
In bacteria, PGM catalyzes reactions at the branch point that determines whether incoming sugars will be catabolized for energy or, alternatively, utilized in the synthesis of more complex structures such as polysaccharides, usually through sugar nucleotide intermediates. In many species of bacteria, disruption of PGM activity can reduce the availability of sugar building blocks needed for key biosynthetic pathways, consequently reducing the virulence potential of the ΔPGM mutant. For example, the production of exopolysaccharide capsule in Streptococcus pneumoniae and Streptococcus thermophilus requires the presence of a functional PGM enzyme (15, 25). Cell wall polysaccharide changes associated with PGM deletion in gram-negative bacteria include altered lipopolysaccharide in Bordetella bronchiseptica (44), truncated lipo-oligosaccharide in Neisseria meningitidis (32), and shorter O-polysaccharide side chains in Stenotrophomonas maltophilia (30). Our ultrastructural examination suggests decreased but not absent expression of the recently described (4), as-yet-biochemically-uncharacterized S. iniae surface capsule in the ΔPGM mutant.
Streptococcal surface capsules are generally complex anionic polysaccharides that contribute importantly to virulence by virtue of their antiphagocytic properties (26). Streptococcal capsular polysaccharides are also frequently a principal target of protective immunity in natural infection and can represent effective vaccine antigens particularly when conjugated to protein carriers (23). Decreased buoyancy, decreased surface negative charge, and increased hydrophobicity as measured in our S. iniae ΔPGM mutant are changes consistent with diminished expression of a polyanionic surface capsule. Direct correlations between buoyancy, degree of encapsulation, and animal virulence have been demonstrated for group B Streptococcus (14), group A Streptococcus (43), and Streptococcus equi (3). Of particular interest, ΔPGM mutants of S. pneumoniae lacking surface capsule are avirulent in mice (16). In S. iniae, transposon disruption of the putative capsule operon was recently shown to be correlated with decreased buoyancy and decreased virulence in zebra fish (31). We hypothesize that diminished capsule expression is an important aspect of the increased susceptibility to phagocytic clearance and virulence attenuation of the S. iniae ΔPGM mutant.
Electron microscopy also revealed a marked increase in the cell size of the ΔPGM mutant in comparison to WT S. iniae. Precedent exists in studies of bacterial ΔPGM mutants for alterations in cell morphology in response to loss of enzyme activity. The bacteriophage C21, which is unable to infect WT E. coli strains (38), was found to efficiently bind and infect E. coli ΔPGM mutants grown in glucose media (1). This finding was theorized to reflect defects in normal cell wall polysaccharides due to the lack of production of sugar nucleotides (such as UDP-glucose) dependent upon functional PGM (1). A subsequent structural study of E. coli ΔPGM mutants measured their length to be only ∼70% that of the corresponding WT strains (27). With Lactococcus lactis, ΔPGM mutants grown on maltose (favoring catabolic processes) were observed by phase-contrast microscopy to be significantly larger than WT cells (24). Each of these phenotypes suggests significant alterations in cell wall architecture and, in the case of the gram-positive L. lactis, an increased cell size similar to that observed with our S. iniae ΔPGM mutant was documented.
AMPs are an evolutionarily conserved component of the innate defense system of higher organisms against microbial infection (45), as recently confirmed by targeted gene knockout in mammals (34). AMPs have been identified in numerous fish species (35) and represent a key component of the killing mechanism of their phagocytic cells (33). The 22-amino-acid cationic AMP moronecidin is produced in the gills, spleen, head kidney, intestine, skin, blood cells, and mast cells of HSB (21, 40). We found that loss of PGM activity in S. iniae results in markedly increased susceptibility to moronecidin antimicrobial action. We speculate that the increased cell size of the S. iniae ΔPGM mutant may reflect a decrease in cell wall rigidity that could render the bacterium more susceptible to lysis by the hypothesized pore-forming mechanism of this and other AMPs. In addition, the decreased net negative charge of the ΔPGM cell surface (as confirmed by diminished cytochrome c binding) may decrease the electrostatic attraction of the cationic AMP for its cell wall target of action, a well-described factor in determining the relative susceptibility of bacteria to AMP killing (36). Interestingly, a study of the role of PGM in B. brochiseptica determined that decreased animal virulence could be correlated with increased cationic AMP susceptibility, as corroborated experimentally with cecropin B (44).
S. iniae infection represents a serious threat to the economic viability of intensive aquaculture of several finfish species. Antibiotic treatment protocols and traditional bacterin-type vaccines to control S. iniae infection each possess significant practical limitations. Our preliminary vaccine trial indicates that ΔPGM mutant TnM2 indeed elicits an effective immune response and may have value as a live attenuated vaccine to protect HSB against S. iniae infection. This degree of attenuated infection and antigenic load was sufficient to induce an effective immune response, as live vaccinated fish demonstrated >90% survival upon exposure to a lethal dose of WT S. iniae 4 months (2,000 degree days) later. Furthermore, in our examination of the aborted infectious process of the ΔPGM mutant, we observed the mutant to disseminate to multiple organs within 4 h but to be rapidly cleared from blood and brain while persisting at diminished levels in splenic tissue for up to 5 days after infection. The use of live attenuated vaccines in aquaculture could offer benefits, including elicitation of mucosal immunity, longer presentation time of unaltered antigens, and stimulation of both cell-mediated and humoral immunity (28, 29, 41). A number of recent studies of fish have found live attenuated vaccines against other bacterial pathogens to be effective (9, 22, 28), and the development of a successful live attenuated vaccine to treat the globally important fish pathogen S. iniae would be greatly welcomed.
In summary, we have identified the PGM gene of S. iniae and linked its activity to a variety of structural and functional phenotypes that translate to dramatically decreased virulence in a model of fish meningoencephalitis. Future research avenues include unraveling the biochemistry of the S. iniae polysaccharide capsule to ascertain the role that PGM plays in its expression, identifying the PGM-dependent cell surface components of S. iniae that contribute to AMP resistance, and exploring the utility of ΔPGM mutants as live attenuated vaccines in larger scale trials for applications in aquaculture.
Acknowledgments
This work was supported by grants from the USDA CSREES National Research Initiative (V.N.) (grant number 2002-35204-11623), the California Sea Grant College program (V.N.) (grant number R/A 124), and the National Sea Grant Marine Aquaculture Initiative (to the Kent SeaTech Corporation). J.T.B. received support from UCSD Institutional NIH Training Award AI07036.
We gratefully acknowledge Mike Vicknair and Chindamony Phan for assistance with fish challenge studies, Robert Garrett for performance of electron microscopy, and Hafid Mantilla for assistance with enzymatic assays. We thank the E. coli Stock Center Database at Yale University for supplying PGM mutants.
Editor: V. J. DiRita
REFERENCES
- 1.Adhya, S., and M. Schwartz. 1971. Phosphoglucomutase mutants of Escherichia coli K-12. J. Bacteriol. 108:621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Anzai, T., J. F. Timoney, Y. Kuwamoto, Y. Fujita, R. Wada, and T. Inoue. 1999. In vivo pathogenicity and resistance to phagocytosis of Streptococcus equi strains with different levels of capsule expression. Vet. Microbiol. 67:277-286. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, A. C., F. M. Young, M. T. Horne, and A. E. Ellis. 2003. Streptococcus iniae: serological differences, presence of capsule and resistance to immune serum killing. Dis. Aquat. Org. 53:241-247. [DOI] [PubMed] [Google Scholar]
- 5.Bercovier, H., C. Ghittino, and A. Eldar. 1997. Immunization with bacterial antigens: infections with streptococci and related organisms. Dev. Biol. Stand. 90:153-160. [PubMed] [Google Scholar]
- 6.Bromage, E. S., A. Thomas, and L. Owens. 1999. Streptococcus iniae, a bacterial infection in barramundi Lates calcarifer. Dis. Aquat. Org. 36:177-181. [DOI] [PubMed] [Google Scholar]
- 7.Chaffin, D. O., and C. E. Rubens. 1998. Blue/white screening of recombinant plasmids in gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene 219:91-99. [DOI] [PubMed] [Google Scholar]
- 8.Dai, J., Y. Liu, W. Ray, Jr., and M. Konno. 1992. The crystal structure of muscle phosphoglucomutase refined at 2.7-angstrom resolution. J. Biol. Chem. 267:6322-6337. [PubMed] [Google Scholar]
- 9.Daly, J. G., S. G. Griffiths, A. K. Kew, A. R. Moore, and G. Olivier. 2001. Characterization of attenuated Renibacterium salmoninarum strains and their use as live vaccines. Dis. Aquat. Org. 44:121-126. [DOI] [PubMed] [Google Scholar]
- 10.Eldar, A., and C. Ghittino. 1999. Lactococcus garvieae and Streptococcus iniae infections in rainbow trout Oncorhynchus mykiss: similar, but different diseases. Dis. Aquat. Org. 36:227-231. [DOI] [PubMed] [Google Scholar]
- 11.Evans, J. J., C. A. Shoemaker, and P. J. Klesius. 2000. Experimental Streptococcus iniae infection of hybrid striped bass (Morone chrysops × Morone saxatilis) and tilapia (Oreochromis niloticus) by nares inoculation. Aquaculture 189:197-210. [Google Scholar]
- 12.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakansson, S., A. M. Bergholm, S. E. Holm, B. Wagner, and M. Wagner. 1988. Properties of high and low density subpopulations of group B streptococci: enhanced virulence of the low density variant. Microb. Pathog. 5:345-355. [DOI] [PubMed] [Google Scholar]
- 15.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy, G. G., A. D. Magee, C. L. Ventura, M. J. Caimano, and J. Yother. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 69:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermann, S. R., J. A. Miller, S. O'Neill, T. T. Tsao, R. M. Harding, and J. L. Dale. 2000. Single-primer amplification of flanking sequences. BioTechniques 29:1176-1178, 1180. [DOI] [PubMed] [Google Scholar]
- 18.Karlyshev, A. V., M. J. Pallen, and B. W. Wren. 2000. Single-primer PCR procedure for rapid identification of transposon insertion sites. BioTechniques 28:1078, 1080, 1082. [DOI] [PubMed] [Google Scholar]
- 19.Kitao, T. 1982. The methods for detection of Streptococcus sp., causative bacteria of streptococcal disease of cultured yellowtail (Seriola quinqueradiata)—especially their cultural, biochemical and serologic properties. Fish Pathol. 17:17-26. [Google Scholar]
- 20.Lau, S. K., P. C. Woo, H. Tse, K. W. Leung, S. S. Wong, and K. Y. Yuen. 2003. Invasive Streptococcus iniae infections outside North America. J. Clin. Microbiol. 41:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauth, X., H. Shike, J. C. Burns, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. Van Olst, V. Nizet, S. W. Taylor, C. Shimizu, and P. Bulet. 2002. Discovery and characterization of two isoforms of moronecidin, a novel antimicrobial peptide from hybrid striped bass. J. Biol. Chem. 277:5030-5039. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, M. L., R. K. Cooper, and R. L. Thune. 1997. Attenuation, persistence, and vaccine potential of an Edwardsiella ictaluri purA mutant. Infect. Immun. 65:4642-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, C. J., L. H. Lee, C. S. Lu, and A. Wu. 2001. Bacterial polysaccharides as vaccines—immunity and chemical characterization. Adv. Exp. Med. Biol. 491:453-471. [DOI] [PubMed] [Google Scholar]
- 24.Levander, F., U. Andersson, and P. Rådström. 2001. Physiological role of β-phosphoglucomutase in Lactococcus lactis. Appl. Environ. Microbiol. 67:4546-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levander, F., and P. Rådström. 2001. Requirement for phosphoglucomutase in exopolysaccharide biosynthesis in glucose- and lactose-utilizing Streptococcus thermophilus. Appl. Environ. Microbiol. 67:2734-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llull, D., R. Lopez, and E. Garcia. 2001. Genetic bases and medical relevance of capsular polysaccharide biosynthesis in pathogenic streptococci. Curr. Mol. Med. 1:475-491. [DOI] [PubMed] [Google Scholar]
- 27.Lu, M., and N. Kleckner. 1994. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J. Bacteriol. 176:5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsden, M. J., L. M. Vaughan, R. M. Fitzpatrick, T. J. Foster, and C. J. Secombes. 1998. Potency testing of a live, genetically attenuated vaccine for salmonids. Vaccine 16:1087-1094. [DOI] [PubMed] [Google Scholar]
- 29.Marsden, M. J., L. M. Vaughan, T. J. Foster, and C. J. Secombes. 1996. A live (ΔaroA) Aeromonas salmonicida vaccine for furunculosis preferentially stimulates T-cell responses relative to B-cell responses in rainbow trout (Oncorhynchus mykiss). Infect. Immun. 64:3863-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKay, G. A., D. E. Woods, K. L. MacDonald, and K. Poole. 2003. Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect. Immun. 71:3068-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. D., and M. N. Neely. 2005. Large-scale screen highlights the importance of capsule for virulence in the zoonotic pathogen Streptococcus iniae. Infect. Immun. 73:921-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monteiro, M. A., M. Fortuna-Nevin, J. Farley, and V. Pavliak. 2003. Phase-variation of the truncated lipo-oligosaccharide of Neisseria meningitidis NMB phosphoglucomutase isogenic mutant NMB-R6. Carbohydr. Res. 338:2905-2912. [DOI] [PubMed] [Google Scholar]
- 33.Neumann, N. F., J. L. Stafford, D. Barreda, A. J. Ainsworth, and M. Belosevic. 2001. Antimicrobial mechanisms of fish phagocytes and their role in host defense. Dev. Comp. Immunol. 25:807-825. [DOI] [PubMed] [Google Scholar]
- 34.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 35.Patrzykat, A., and S. E. Douglas. 2003. Gone gene fishing: how to catch novel marine antimicrobials. Trends Biotechnol. 21:362-369. [DOI] [PubMed] [Google Scholar]
- 36.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 37.Pier, G. B., and S. H. Madin. 1976. Streptococcus iniae sp. nov., a beta-hemolytic streptococcus isolated from an Amazon freshwater dolphin, Inia geoffrensis. Int. J. Syst. Bacteriol. 26:545-553. [Google Scholar]
- 38.Rapin, A. M., H. M. Kalckar, and L. Alberico. 1968. The metabolic basis for masking of receptor-sites on E. coli K-12 for C21, a lipopolysaccharide core-specific phage. Arch. Biochem. Biophys. 128:95-105. [DOI] [PubMed] [Google Scholar]
- 39.Shoemaker, C. A., P. H. Klesius, and J. J. Evans. 2001. Prevalence of Streptococcus iniae in tilapia, hybrid striped bass, and channel catfish on commercial fish farms in the United States. Am. J. Vet. Res. 62:174-177. [DOI] [PubMed] [Google Scholar]
- 40.Silphaduang, U., and E. J. Noga. 2001. Peptide antibiotics in mast cells of fish. Nature 414:268-269. [DOI] [PubMed] [Google Scholar]
- 41.Thornton, J. C., R. A. Garduno, S. G. Newman, and W. W. Kay. 1991. Surface-disorganized, attenuated mutants of Aeromonas salmonicida as furunculosis live vaccines. Microb. Pathog. 11:85-99. [DOI] [PubMed] [Google Scholar]
- 42.Weinstein, M. R., M. Litt, D. A. Kertesz, P. Wyper, D. Rose, M. Coulter, A. McGeer, R. Facklam, C. Ostach, B. M. Willey, A. Borczyk, D. E. Low, et al. 1997. Invasive infections due to a fish pathogen, Streptococcus iniae. N. Engl. J. Med. 337:589-594. [DOI] [PubMed] [Google Scholar]
- 43.Wessels, M. R., J. B. Goldberg, A. E. Moses, and T. J. DiCesare. 1994. Effects on virulence of mutations in a locus essential for hyaluronic acid capsule expression in group A streptococci. Infect. Immun. 62:433-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West, N. P., H. Jungnitz, J. T. Fitter, J. D. McArthur, C. A. Guzman, and M. J. Walker. 2000. Role of phosphoglucomutase of Bordetella bronchiseptica in lipopolysaccharide biosynthesis and virulence. Infect. Immun. 68:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 46.Zlotkin, A., H. Hershko, and A. Eldar. 1998. Possible transmission of Streptococcus iniae from wild fish to cultured marine fish. Appl. Environ. Microbiol. 64:4065-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]