Abstract
Sera of kala-azar patients from Bihar, India, were used to identify Leishmania donovani antigens encoded by a phage expression library. Ten antigens were identified, five of which have not been described as leishmania antigens before. The antigens specifically react with sera of leishmania-infected patients but not of toxoplasma- or plasmodium-infected patients.
Leishmania donovani is the causative agent of visceral leishmaniasis (VL) in India. Like other parasites, Leishmania donovani induces vigorous immune responses which, however, do not control the infection in patients with acute disease. If not treated, VL is lethal. Notwithstanding, there are indications of immunity to VL in populations cured of VL and in individuals in disease-endemic regions who are seropositive without signs of disease. Knowledge of antigens targeted during infection may, therefore, be instrumental in the development of Leishmania donovani vaccines and immunodiagnostics (6, 8). To identify serological antigens in VL, a Leishmania donovani λTriplEx2 phage expression library consisting of 1.5 × 106 primary PFU was established from oligo(dT)-primed cDNA of mid-log-phase promastigotes of Leishmania donovani strain AG83 (3). By use of 10 antisera from VL patients (5 obtained from untreated patients and 5 obtained under or shortly after therapy) from highly endemic foci around Muzaffarpur, Bihar, India, 30 positive phage plaques were identified and subcloned to monoclonality as determined by PCR. The inserts were recloned in pTriplEx2 plasmids and sequenced by standard protocols. A comparison of the sequences with the National Center for Biotechnology Information, EMBL, and Leishmania Genome Network databases revealed that they correspond to 10 different genes each represented by 1 to 8 clones of various lengths (Table 1). Sequences for LDHSP70 (5), HASPB1 (4), P0 (10), ARP-1 (11), and α-tubulin (9) genes had been published as Leishmania donovani antigen gene sequences before. However, the sequences of our LDHSP70 and P0 inserts deviate from the database entries. One of our LDHSP70 gene sequences covers 1,002 bp, of which the 5′ 762 bp code for 254 amino acids of the known LDHSP70. The 3′ 240 bp are located 1,630 bp downstream of the hsp70 gene and correspond to no known translation product. Since these 240 bp are in-frame with the LDHSP70 gene sequence, the insert can be translated into a continuous polypeptide. All four LDHSP70 clones differ at the 3′ end and deviate from the published cDNA sequence X52314. The database entry for the Leishmania donovani P0 gene (GenBank accession number AY180912) includes a 220-bp insertion missing from our clone as well as from Leishmania chagasi and Leishmania infantum database sequences. L. infantum has two P0 genes, LiP0-A and LiP0-B, of which LiP0-B shows highest similarity to our sequence and may be its homologue. This suggests that Leishmania donovani, like L. infantum, has two P0 genes. In the case of these two antigens, it is not clear whether the published antigenic determinants are the same as ours. Five of the 10 antigens had not been identified before. LePa2 is an α subunit of the 20S proteasome, of which another subunit, LePa, had been described as a Leishmania donovani antigen (2). Our sequence was not assigned for Leishmania donovani but is identical to the sequence of the gene for L. infantum Y13059 gene sequence. This antigen, therefore, is a new 20S proteasome α subunit of Leishmania donovani. The second new antigen is the putative elongation factor 1β (EF1B). The other three new antigens are products of unique genes without identifiable homologues in other species. We tentatively named them LDAG1, LDAG2, and LDAG3 for Leishmania donovani antigens 1, 2, and 3.
TABLE 1.
Antigens identified by immunoscreening of the L. donovani AG83 cDNA expression library with sera of kala-azar patients from Bihar, India
| Antigen | Highest sequence identity (description) | GenBank accession no.a | No. of analyzed primary plaques | No. of corresponding sera in primary immunoscreens |
|---|---|---|---|---|
| LDHSP70 | 71.2-kDa L. donovani heat shock protein LDHSP70 (X52314) | AY702003 | 4 | 4 |
| HASPB1 | 42.2-kDa L. donovani hydrophilic cell surface protein HASPB1 (AJ011810) | AY702004 | 3 | 3 |
| LePa2 | 25.1-kDa L. infantum 20 S proteasome α-2 subunit LePa (Y13059) | AY702005 | 3 | 3 |
| P0 | 35-kDa L. chagasi 60 S ribosomal protein P0 (L29300) | AY702006 | 3 | 3 |
| EF1B | 25.6-kDa Leishmania major putative elongation factor 1β LmjF34.0820 | AY702007 | 1 | 1 |
| ARP-1 | 10.4-kDa L. donovani acidic ribosomal protein ARP-1(LiP, P2β; AF034539) | AY702008 | 4 | 2 |
| α-Tubulin | 49.7-kDa L. donovani α-tubulin (U09612) | AY702009 | 8 | 2 |
| LDAG1 | 33.1-kDa L. major hypothetical protein LmjF36.0540 | AY702010 | 2 | 1 |
| LDAG2 | 14.5-kDa L. major hypothetical protein LmjF30.0770 | AY702011 | 1 | 1 |
| LDAG3 | 28.8-kDa L. major hypothetical protein LmjF34.2700 | AY702012 | 1 | 1 |
The sequences of the inserts coding for the Leishmania donovani antigens were submitted to GenBank.
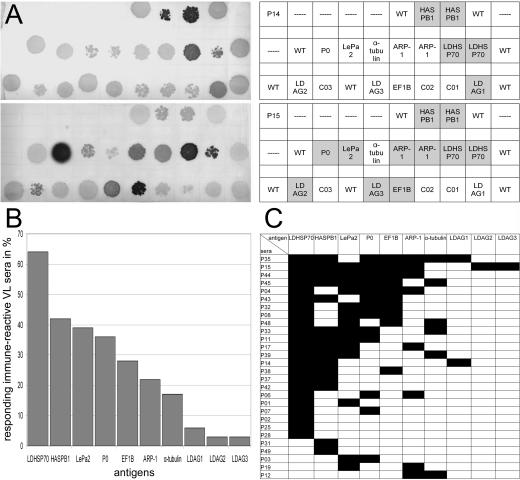
The frequencies of seropositivity for the 10 antigens in VL patients were determined by spotting the phages that code for the antigens onto a growing bacteria layer as shown in Fig. 1A, blotting the antigens onto nitrocellulose, and probing the filters with sera from 44 VL patients, 10 control sera from healthy relatives of the patients, and 11 sera from healthy donors from areas in which disease was not endemic (1 from India and 10 from Germany). Eleven of the VL patients had received chemotherapy at the time of serum preparation, and 33 had not. All sera had been tested before against the total amount of Leishmania donovani lysate separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (see the supplemental material). Thirty-six of the patient sera were positive at a dilution of 1:640. Except for one serum from an Indian donor who was the healthy relative of a patient, none of the control sera were positive. At 1:60, faint signals were detectable with sera of healthy Indian donors. The statistics of seropositivity for the 10 identified antigens are summarized in Fig. 1B and C. Sixty-four percent of the 44 tested patients and 80% of the 36 seroreactive patients were seropositive for at least one of the defined antigens. LDHSP70 was most prominent, with 64% of the 36 immune sera reacting. The frequencies of seropositivity among the seroreactive patients for the other previously known antigens HASPB1, P0, ARP-1, and α-tubulin were 42, 36, 22, and 17%, respectively. The newly identified antigens LePa2, EF1B, LDAG1, LDAG2, and LDAG3 were detected in 39, 28, 6, 3, and 3% of seroreactive patients, respectively. There is no correlation of the extent of seropositivity with the ages or sexes of the patients. The average age of all the 44 VL patients was 24 years (range, 6 to 50 years), that of the 28 patients with responses to the antigens defined herein was 21.8 years (range, 6 to 50 years), and that of the remaining nonreactive 16 patients was 27.8 years (range, 10 to 50 years). Fourteen of the 28 patients who responded to the defined antigens were male, and 12 were female. For two patients, no gender information was available.
FIG. 1.
Analysis of the frequencies of seropositivity in kala-azar patients from the Kala-Azar Medical Research Center in Muzaffarpur, Bihar, India. The recombinant λ phages for the 10 antigens identified in this study were spotted onto E. coli layers according to the scheme in panel A. After the induction of antigen expression, the antigens were blotted onto nitrocellulose and probed with the sera for antibodies with the corresponding specificities. (A) Examples of the nitrocellulose filters. Replicas of these filters were prepared for and probed with the sera of the patients and healthy controls. The upper filter shown is for patient P14, the lower for patient P15. The charts give the positions of the antigens on the filter. Serodetected antigens are highlighted. For HASPB1, LDHSP70, and ARP-1, two independently identified phage clones were employed. WT, wild-type phages included as negative control; C01 through C03, phages with undefined polyclonal cDNA inserts. (B) Frequencies of seropositivity for the indicated antigens in seroreactive patients. (C) Seroreactivity of the Indian kala-azar patient sera against the identified antigens. Black fields indicate that the patient is seropositive for the corresponding antigen. Only the results for patients who are seropositive for at least 1 of the 10 antigens are shown. Fifteen patients, the 10 healthy relatives of patients, and 11 outgroup individuals (1 from India and 10 from Germany), were seronegative for the antigens.
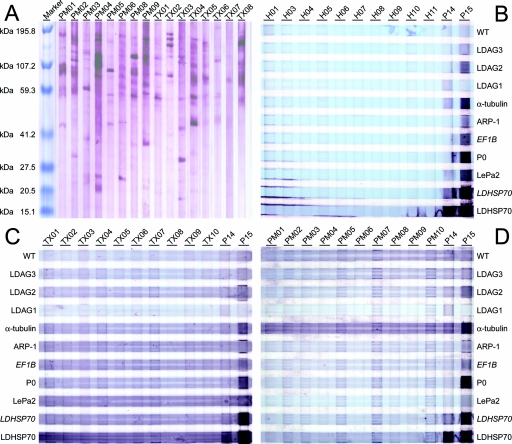
As specificity controls, eight sera each were collected from Toxoplasma gondii-, Plasmodium falciparum-, or Plasmodium vivax-infected patients in Germany who had been infected while traveling in disease-endemic areas and, most likely, had not been exposed to Leishmania donovani. Full-length expression clones coding for the identified Leishmania donovani antigens, with the exception of EF1B, were isolated from Leishmania donovani DNA and overexpressed in Escherichia coli. For EF1B, the original insert that codes for a 10-kDa fragment was used. For HSP70, in addition to the full-length clone, the 36-kDa fusion protein (see above) coded for by the corresponding phage clone was tested. The recombinant bacteria were homogenized, and the solutions were cleared of debris, blotted onto nitrocellulose, and probed with the antisera. None of the sera from healthy donors or the sera from Toxoplasma- or Plasmodium-infected patients reacted with the 10 Leishmania donovani antigens described herein (Fig. 2B, C, and D), although the sera from Toxoplasma- and Plasmodium-infected patients cross-reacted with other Leishmania donovani antigens as detected by Western blot analysis of Leishmania donovani lysates after SDS-PAGE (Fig. 2A). The elevated background seen with all patient sera correlates with the low serum dilution of 1:60 in these analyses, and hypergammaglobulinemia typically associated with parasitic infections was confirmed for the sera of this study.
FIG. 2.
Serological responses of antisera from healthy donors and patients infected with Toxoplasma or Plasmodium to the 10 identified Leishmania donovani antigens. (A) The proteins of Leishmania donovani AG83 lysate were separated by SDS-PAGE and probed with eight sera each of Plasmodium-infected patients (PM01 through PM06, PM08, and PM09) or Toxoplasma-infected patients (TX01 through TX08) by using a serum dilution of 1:60. (B through D) Lysates of wild-type E. coli (WT) or E. coli overexpressing the antigens were probed with nine control sera from healthy relatives of VL patients (H01 and H03 through H10) and one serum from a healthy donor from a nonendemic area in India (H11) (panel B), sera of Toxoplasma-infected patients (panel C) or sera of Plasmodium-infected patient (panel D), as well as two kala-azar patients (P14 and P15) in all panels B through D at dilutions of 1:60. The antigens were expressed from full-length expression clones amplified from Leishmania donovani DNA. Exceptions were EF1B and a second expression clone of LDHSP70 that were expressed from the original phage expression clone insert (both are in italics). The lysates of the bacteria that overexpress the antigens were colored with bromophenol blue and blotted as stripes onto nitrocellulose. The sera were incubated with these blots in stripes perpendicular to the antigen stripes resulting in the grid pattern in panels C and D. The light blue horizontal stripes indicate the distribution of the antigens, the dark purple stripes the Western blot stain of Nitro Blue Tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate).
The role of the strong antibody responses in leishmaniasis has been a controversial issue. As recently reported, high immunoglobulin G titers can cause antigen-independent suppression of immunity to leishmaniasis via induction of interleukin-10 (1, 7). In contrast, an earlier report demonstrated a correlation of protective cellular immune responses with antibody responses to the leishmania antigen HASBP-1 (12). Together, these reports suggest that the specificity of the serological responses might determine the outcome of the infection. Ranges and specificities of natural antileishmania responses can provide important information on the suitability of vaccine candidate antigens. As shown with this report, the serological responses in leishmaniasis are very heterogeneous (see the supplementary material). Even LDHSP70, which is rated among the most dominant antigens in leishmaniasis, induces specific antibodies in no more than 60% of the patients. Seropositivity for HASBP1, which, based on studies with mice, is considered a promising vaccine candidate (13), is found in less than 40% of the patients. Future studies will follow up the patients described herein to see whether they are protected against reinfection and whether such protection correlates with serological responses to such previously known antigens and the antigens described in this report.
Nucleotide sequence accession numbers.
Sequences for the genes encoding the newly discovered antigens have been deposited in GenBank under the accession numbers given in Table 1.
Supplementary Material
Acknowledgments
The technical assistance by Ulrike Fritz and Arthur O'Connor is gratefully acknowledged. We also thank Kaushik Roychoudhury for his help in collecting sera and parasites and Patricia Zambon for her assistance in preparing the manuscript. We thank Thomas Weitzel, Institut für Tropenmedizin, Diagnostisches Labor, Charité, for Plasmodium sera and PD Renate Bollmann, Institut für Mikrobiologie und Hygiene, Abt. Mikrobiologie-Serologie, Charité, for Toxoplasma sera.
The project was supported by the Volkswagen Foundation, Hannover, Germany (grant I/77 908), and the Deutsche Forschungsgemeinschaft, Germany (grant 446 IND 121/3/03).
Editor: J. F. Urban, Jr.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Buxbaum, L. U., and P. Scott. 2005. Interleukin 10- and Fcγ receptor-deficient mice resolve Leishmania mexicana lesions. Infect. Immun. 73:2101-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen, C. B., L. Jorgensen, A. T. Jensen, S. Gasim, M. Chen, A. Kharazmi, T. G. Theander, and K. Andresen. 2000. Molecular characterization of a Leishmania donovani cDNA clone with similarity to human 20S proteasome a-type subunit. Biochim. Biophys. Acta 1500:77-87. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh, A. K., F. K. Bhattacharyya, and D. K. Ghosh. 1985. Leishmania donovani: amastigote inhibition and mode of action of berberine. Exp. Parasitol. 60:404-413. [DOI] [PubMed] [Google Scholar]
- 4.Jensen, A. T., S. Gasim, T. Moller, A. Ismail, A. Gaafar, M. Kemp, A. M. el Hassan, A. Kharazmi, T. M. Alce, D. F. Smith, and T. G. Theander. 1999. Serodiagnosis of Leishmania donovani infections: assessment of enzyme-linked immunosorbent assays using recombinant L. donovani gene B protein (GBP) and a peptide sequence of L. donovani GBP. Trans. R. Soc. Trop. Med. Hyg. 93:157-160. [DOI] [PubMed] [Google Scholar]
- 5.MacFarlane, J., M. L. Blaxter, R. P. Bishop, M. A. Miles, and J. M. Kelly. 1990. Identification and characterisation of a Leishmania donovani antigen belonging to the 70-kDa heat-shock protein family. Eur. J. Biochem. 190:377-384. [DOI] [PubMed] [Google Scholar]
- 6.Martin-Sanchez, J., J. A. Pineda, F. Morillas-Marquez, J. A. Garcia-Garcia, C. Acedo, and J. Macias. 2004. Detection of Leishmania infantum kinetoplast DNA in peripheral blood from asymptomatic individuals at risk for parenterally transmitted infections: relationship between polymerase chain reaction results and other Leishmania infection markers. Am. J. Trop. Med. Hyg. 70:545-548. [PubMed] [Google Scholar]
- 7.Miles, S. A., S. M. Conrad, R. G. Alves, S. M. Jeronimo, and D. M. Mosser. 2005. A role for IgG immune complexes during infection with the intracellular pathogen Leishmania. J. Exp. Med. 201:747-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riera, C., R. Fisa, M. Udina, M. Gallego, and M. Portus. 2004. Detection of Leishmania infantum cryptic infection in asymptomatic blood donors living in an endemic area (Eivissa, Balearic Islands, Spain) by different diagnostic methods. Trans. R. Soc. Trop. Med. Hyg. 98:102-110. [DOI] [PubMed] [Google Scholar]
- 9.Sheppard, H. W., and D. M. Dwyer. 1986. Cloning of Leishmania donovani genes encoding antigens recognized during human visceral leishmaniasis. Mol. Biochem. Parasitol. 19:35-43. [DOI] [PubMed] [Google Scholar]
- 10.Soto, M., J. M. Requena, and C. Alonso. 1993. Isolation, characterization and analysis of the expression of the Leishmania ribosomal PO protein genes. Mol. Biochem. Parasitol. 61:265-274. [DOI] [PubMed] [Google Scholar]
- 11.Soto, M., J. M. Requena, M. Garcia, L. C. Gomez, I. Navarrete, and C. Alonso. 1993. Genomic organization and expression of two independent gene arrays coding for two antigenic acidic ribosomal proteins of Leishmania. J. Biol. Chem. 268:21835-21843. [PubMed] [Google Scholar]
- 12.Stager, S., J. Alexander, A. C. Kirby, M. Botto, N. V. Rooijen, D. F. Smith, F. Brombacher, and P. M. Kaye. 2003. Natural antibodies and complement are endogenous adjuvants for vaccine-induced CD8+ T-cell responses. Nat. Med. 9:1287-1292. [DOI] [PubMed] [Google Scholar]
- 13.Stager, S., D. F. Smith, and P. M. Kaye. 2000. Immunization with a recombinant stage-regulated surface protein from Leishmania donovani induces protection against visceral leishmaniasis. J. Immunol. 165:7064-7071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.