Abstract
From a library of approximately 20,000 transposon mutants, we have identified mutants affected in chromosomal genes involved in synthesis of the siderophore anguibactin, as well as in ferric anguibactin utilization. Genetic and sequence analyses of one such transport-defective mutant revealed that the transposon insertion occurred in an open reading frame (ORF) with homology to rmlC, a dTDP-rhamnose biosynthetic gene. This ORF resides within a cluster of four ORFs, all of which are predicted to function in the biosynthesis of this O side chain precursor. The same phenotype was seen in a mutant obtained by allelic exchange in rmlD, another ORF in this dTDP-rhamnose biosynthetic cluster. This mutation could be complemented with the wild-type rmlD gene, restoring both production of the O1 antigen side chain and ferric anguibactin transport. Presence of the O1 side chain was crucial for the resistance of Vibrio anguillarum to the bactericidal action of nonimmune serum from the fish host. Surprisingly, further analysis demonstrated that these mutations were pleiotropic, leading to a dramatic decrease in the levels of FatA, the outer membrane protein receptor for ferric anguibactin transport, and a concomitant reduction in iron transport. Thus, our results in this work demonstrate that the lipopolysaccharide O1 side chain is required for the operation of two critical virulence factors in V. anguillarum: serum resistance and anguibactin-mediated iron transport. These factors allow V. anguillarum to survive in serum and multiply in the iron-limiting milieu of the host vertebrate.
Iron is an essential element for the growth of most bacteria, yet in the bodily fluids of vertebrates it exists only in complex with the host high-affinity iron-binding proteins, making it unavailable for bacterial use. Pathogens that are capable of causing disseminating infections have evolved highly specialized iron-scavenging systems to utilize these host-bound iron sources and thereby overcome this nonspecific defense mechanism (10). Vibrio anguillarum is the etiologic agent of fish vibriosis, a fatal hemorrhagic septicemic disease of salmonids and other fishes (67). A large number of the virulent strains of V. anguillarum harbor pJM1-type plasmids, which provide these bacteria with a highly efficient iron-scavenging system that is essential for growth within the host during infection (16, 19, 20, 66). V. anguillarum strains comprise 11 defined serotypes; however, the most significant pathogens belong to serotypes O1 and O2. pJM1-like plasmids have only been found in serotype O1 strains, although some pathogenic strains of this serotype that do not carry a pJM1-like plasmid possess a chromosomally encoded high-affinity iron uptake system that is genetically dissimilar from the pJM1 system (14, 34, 59). The presence of a chromosomally encoded iron uptake system affecting virulence has also been described in the plasmidless pathogenic O2 serotype strains (34).
The pJM1-type plasmid-encoded iron uptake system consists of a low-molecular-weight peptide-like iron binding compound, the siderophore anguibactin, and its associated high-affinity ferric anguibactin uptake system (1, 15, 17). The genes encoding a majority of the proteins involved in this process reside in two large clusters on the pJM1 plasmid (63). Sequencing of this plasmid has revealed a total of 59 open reading frames (ORFs), of which 32% (19) are predicted by homology to function in siderophore-mediated iron scavenging (25). For eight of these genes, mutational analysis has confirmed their predicted roles in either anguibactin biosynthesis or ferric anguibactin transport (2-4, 21, 24, 25, 31, 55-57, 63). Regulation of the plasmid-encoded iron transport system and siderophore biosynthetic genes is controlled by the concentration of available iron and by the anguibactin siderophore itself. This regulation is mediated by four plasmid-encoded regulators, AngR, TAF, RNAα, and RNAβ (13, 44-46, 55, 63), as well as the chromosomally encoded Fur protein, which mediates transcriptional repression of the system (11, 58, 64). With the exception of the fur gene and chromosomal genes involved in the biosynthesis of anguibactin precursors (5, 12), little is known of the extent to which other chromosomally encoded genes participate in the biosynthesis or utilization of anguibactin.
Recently, a chromosomal region encoding putative O1 antigen genes of V. anguillarum has been analyzed by Southern blot hybridization, identifying potential dTDP-rhamnose biosynthetic genes and, immediately upstream, a JUMPstart sequence and, transcribed divergently to the rfb region, a gene apparently involved in lipopolysaccharide (LPS) core biosynthesis (28, 52). Downstream of these genes, there are two genes, wzm and wzt, whose products could be involved in O-antigen export (52). The insertion element IS1358 was found downstream of the export genes. Several ORFs found downstream of this region correspond to a number of rfb-related proteins, such as wbhM and wbhL, that, with the nomenclature VirB and VirA, respectively, have been shown to be involved in surface antigen biosynthesis in V. anguillarum (37, 39).
In this study, we screened transposition-generated mutants for chromosomal genes involved in anguibactin synthesis or ferric anguibactin utilization. We report here the identification of mutations occurring within the dTDP-rhamnose biosynthetic genes of the O1 antigen rfb region. Unexpectedly, this region is necessary not only for protection against the bactericidal action of nonimmune serum but also for the efficient utilization of ferric anguibactin and therefore for bacterial growth under the iron-limiting conditions of the vertebrate host.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Table 1 shows the strains and plasmids used in this study. V. anguillarum was cultured at 24°C in Trypticase soy broth supplemented with 1% NaCl (TSBS) or Trypticase soy agar supplemented with 1% NaCl (TSAS). To achieve iron limitation, strains were grown in M9 minimal medium (CM9) (16) supplemented with 0.2% Casamino Acids (Difco Laboratories), 1% NaCl, and the iron-chelating agent ethylenediamine-di-(o-hydroxyphenylacetic acid) (EDDA). Escherichia coli strains were grown at 37°C in Luria broth or on Luria agar. When required, antibiotics (Sigma) were added at the following concentrations: ampicillin, 500 μg/ml for V. anguillarum and 100 μg/ml for E. coli; kanamycin, 200 μg/ml for V. anguillarum and 50 μg/ml for E. coli; rifampin, 100 μg/ml for V. anguillarum; chloramphenicol, 30 μg/ml for V. anguillarum.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Escherichia coli | ||
| DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 ΔlacU169 relA1 (φ80lacZΔM15) | BRL |
| JM109λpir | recA1 endA1 gyrA96 thi hsdR17supE44 relA1 Δ(lac-proAB+) [F′ traD36 proAB lacIqlacZΔM15] λpir | Lab collection |
| Vibrio anguillarum | ||
| 531A | Wild type, Atlantic Ocean prototype | 63 |
| S531A-1 | Plasmidless derivative of 531A | 63 |
| 531A-1(pJHC1::K) | angB mutant | 63 |
| TW19 | Mini-Tn10K insertion in rmlC | This work |
| 531A-10 | rmlD mutant | This work |
| 531A-10(pJHC1::K) | rmlD mutant derivative of 531A-1(pJHC1::K) | This work |
| S531A-11 | rmlD mutant derivative of S531A-1 | This work |
| Plasmids | ||
| pBR325 | Cloning vector | 9 |
| pSC50 | fatB riboprobe template | Lab collection |
| pSC58 | fatA riboprobe template | Lab collection |
| pKAS23-Km | Derivative of pKAS32 containing Km gene from pUCK4k cloned into SalI site | 50 |
| pTW541 | rmlD gene cloned into pBR325 for expression from Tc promoter | This work |
Transposition experiments and mutant screening.
Transposon mutagenesis was carried out using mini-Tn10K according to de Lorenzo et al. (23), with the following modifications. Conjugation mixtures were allowed to incubate at 24°C for 12 to 18 h on TSAS medium, and the cells were resuspended in TSBS, diluted, and plated on TSAS containing 200 μg/ml kanamycin to select for the transposon and 100 μg/ml rifampin to counterselect the donor strain. After 2 days at 24°C, the resulting mutants were transferred onto the siderophore indicator medium chrome azurol S agar (CAS agar) and plates were then examined after 24 and 48 h, scoring colonies for siderophore synthesis at these times. Many of the genes necessary for anguibactin synthesis and utilization are on endogenous virulence plasmid pJHC1. In order to distinguish mutants with insertions in the plasmid from those with chromosomal insertions, restriction endonuclease analysis of purified plasmid DNA was performed.
Recombinant DNA procedures.
DNA purification, restriction endonuclease analysis, DNA ligations and transformations, PCR, and agarose gel electrophoresis were performed according to standard protocols (47). DNA transfer to V. anguillarum from E. coli strains was accomplished by conjugation as described previously (63). DNA sequencing reactions were carried out by the Oregon Health and Science University Molecular Microbiology and Immunology Research Core Facility (http://www.ohsu.edu/core) using a model 377 Applied Biosystems Inc. automated fluorescence sequencer. Comparative sequence analysis was carried out using the BLAST network service.
Construction of strains 531A-10 and S531A-11.
The rmlD mutant strain was generated by marker exchange mutagenesis. First, the entire rml gene cluster was amplified from wild-type V. anguillarum 531A by PCR using primers RFBU (5′-GCA TTC CGC TTC CAT CAT ATT TCA-3′) and RFBD (5′-TAC CGA GCC AGA TGT TGG AGT CAA-3′). The resulting PCR product was then cloned using the TOPO cloning reagents (Invitrogen) into the pCR2.1 vector, resulting in plasmid pTW500. The rmlD gene in this clone was then modified by cleaving the SalI site (located at codon 205) in rmlD and filling in the overhanging ends using the 5′-3′ polymerase activity of the Klenow fragment of DNA polymerase I. This modification resulted in a 4-bp insertion that caused a frameshift and premature termination of translation 19 codons downstream of this modification. The rml gene cluster from this new plasmid (pTW501) was then subcloned as an EcoRI fragment into the EcoRI site of the suicide vector pKAS32-Km, creating plasmid pTW502. pTW502 was then mobilized to V. anguillarum strains 531A (wild type) and S531A-1 (without the pJHC1 plasmid) by conjugation, and selection with kanamycin was used to identify plasmid cointegrants. Next, strains containing the plasmid cointegrate were grown to stationary phase in the absence of kanamycin for several generations before being plated onto TSAS medium. Clones that had undergone a second recombination event, leading to the excision of pTW502, were subsequently identified by their sensitivity to kanamycin and then screened for replacement of the wild-type rmlD allele by PCR (data not shown). The angB mutations were constructed as previously described (63). The rmlD gene was cloned on a multicopy plasmid for complementation analysis as follows. First, the rmlD gene was amplified from wild-type 531A by PCR using primers RFBDu (5′-GGC TCT CTT TCA GGA CAA CTT C-3′) and RFBDd (5′-ACA CGG TCG GCT CAA TAA TTT T-3′). The resulting PCR product was then cloned into the pCR2.1 vector and subsequently subcloned as an EcoRV-BamHI fragment into the tetracycline gene of pBR325. In the resulting construct, rmlD transcription is under the control of the tetracycline promoter of pBR325.
RNA isolation and RNase protection assays.
Total RNA was prepared from exponentially growing cells using the hot-phenol method (61). fatA-specific and fatB-specific riboprobe templates were prepared from linearized plasmid DNAs (Table 1). The ompU riboprobe was generated by PCR using the synthetic primers ompU (5′-ATG AAC AAA ACT CTG ATT GCT T-3′) and ompD (5′-TTG GTT GTA TAG CTC ACC AGC A-3′) and the Lig-N-Scribe kit (Ambion). Riboprobes were generated by in vitro transcription (Maxiscript kit; Ambion) in the presence of [α-32P]UTP. RNase protection assays were carried out using the RPAIII kit (Ambion) as described by the manufacturer and resolved by urea-polyacrylamide gel electrophoresis (PAGE) (6%).
Bacterial survival in trout normal serum (NS).
V. anguillarum strains to be tested were grown to log phase in TSBS before being diluted to a concentration of approximately 5 × 108 cells per ml in phosphate-buffered saline (PBS). The cells were then mixed with an equal volume of rainbow trout (Oncorhynchus mykiss) NS and cell viability measured using standard plate counting just before and at various times after the addition of NS. Serum was collected from healthy adult rainbow trout at the National Center for Cool and Cold Water Aquaculture, Kearneysville, West Virginia.
Radioactive iron (55Fe) uptake measurements.
Bacterial strains were grown for several generations at 24°C in CM9 made iron limiting by the addition of 0.5 μM EDDA. Exponentially growing cultures were then harvested by centrifugation, washed, and resuspended in M9 salts to an absorbance value of 2.0 at 600 nm. Cells were then diluted 1:1 into a cell-free culture supernatant containing anguibactin activity from V. anguillarum 531A grown in CM9 containing 40 μM EDDA. The anguibactin in this supernatant was loaded with 55Fe by preincubation for 6 h with 1 μCi/ml of carrier-free 55FeCl3. The kinetics of radioactive iron uptake was determined as previously described (3, 18).
Sodium dodecyl sulfate (SDS)-PAGE and immunoblot assays.
Protein separation was preformed as described by Laemmli (32). The resolving gel was 12.5% acrylamide (30:0.8 acrylamide-bisacrylamide). Total membrane and outer membrane (OM) proteins were isolated from equal amounts of cells as previously described (18). FatA and FatB protein abundance was evaluated using Western PAGE analysis as follows. OM proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (PVDF-PLUS; Osomonics Inc.). Membranes were then blocked for 1 h at room temperature in PBS-5% nonfat milk. After washing in PBS, membranes were incubated with FatA-specific (2) or FatB-specific (3) antiserum for 2 h at room temperature, followed by several washes in PBS-0.5% Tween 20 (Sigma). Membranes were then incubated for 1 h with goat anti-rabbit immunoglobulin G horseradish peroxidase-conjugated secondary antibody (Pierce) at a dilution of 1:10,000. After washing in PBS-0.5% Tween 20, secondary antibodies were detected using a chemiluminescent horseradish peroxidase substrate (Pierce) and radiographic film (Biomax MR; Kodak). The antisera used in these determinations are specific for the FatA and FatB proteins and do not cross-react with any other proteins in the membrane fraction of the cell (2, 3). LPS was extracted using the proteinase K digestion method of Hitchcock and Brown (27), resolved by SDS-PAGE (12.5% acrylamide), and visualized using a silver-staining kit (Bio-Rad Laboratories).
Experimental infections.
Infectivity assays were carried out on juvenile rainbow trout (O. mykiss) weighing approximately 5 g. Fish were anesthetized with tricaine methanesulfonate (MS222) and inoculated by intraperitoneal injection with 0.1 ml of each bacterial dilution, using a total of 40 fish for each dilution tested. The dilutions used were prepared with PBS from 12-h cultures grown at 24°C in TSBS. Test fish were maintained at 15°C in fresh water under flowthrough conditions for 1 month, and dead fish were removed daily and stored at −20°C. Necropsies were performed on each fish, and death due to infection by V. anguillarum was confirmed by microbiological analysis of kidney tissue. The 50% lethal doses (LD50s) were estimated using the method of Reed and Muench (42).
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in GenBank under accession number DQ109805.
RESULTS
Isolation of an O1-antigen-deficient transposon mutant.
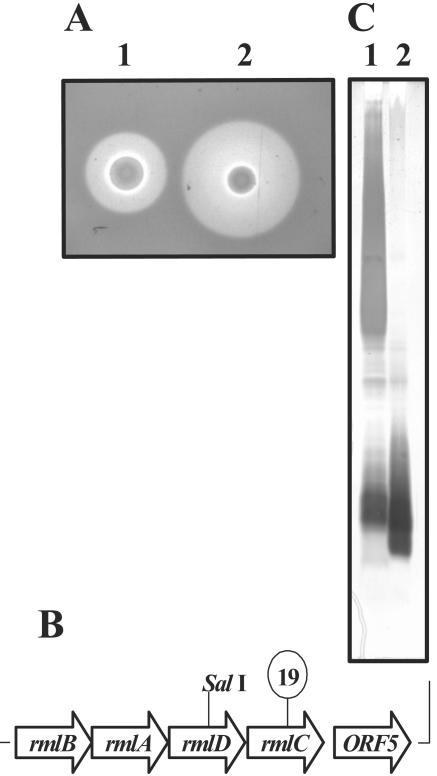
To identify chromosomally encoded genes involved in anguibactin synthesis or utilization, a bank of approximately 20,000 random mutant clones was created using the mini-transposon mini-Tn10K and screened for defects in the production of anguibactin using the siderophore indicator medium CAS agar. To differentiate between mutants with insertions in the pJM1-like virulence plasmid pJHC1 from those with chromosomal insertions, restriction endonuclease analysis of purified plasmid DNA was performed. Using this approach, several mutants were identified that formed small colonies and overproduced anguibactin when grown on CAS agar. One of these chromosomal mutants, designated TW19, is shown in Fig. 1A. The precise position of the transposon insertion in strain TW19 was determined by cloning of the complete transposon and flanking DNA as an EcoRI-EcoRV fragment from the genome of strain TW19, followed by sequencing of the DNA surrounding the site of insertion. DNA sequence analysis revealed that strain TW19 contained a transposon insertion (lollipop symbol in Fig. 1B) in an ORF that encodes a predicted protein with significant homology to the RmlC protein of E. coli K-12 and V. cholerae (35). RmlC is a dTDP-4-dehydrorhamnose 3,5-epimerase involved in the biosynthesis of dTDP-rhamnose, a common constituent of bacterial LPS O side chains. Immediately upstream, we identified three additional ORFs that also encode proteins with significant homology to enzymes involved in the biosynthesis of dTDP-rhamnose. This complete cluster of genes potentially encodes the entire pathway leading to the synthesis of dTDP-rhamnose from its precursor glucose 1-phosphate (see Table 2). The gene designations in Fig. 1B and Table 2 are based on the scheme set forth by Reeves et al. (43). Downstream of this cluster is an additional ORF (ORF5) encoding a protein which shows sequence homology to a class of ABC transporter proteins that might be needed for O-antigen export (52).
FIG. 1.
Identification of an O1-antigen-deficient mutant. (A) Detection of anguibactin using CAS agar plates (48). The identity of anguibactin as the CAS-reactive product was verified using a bioassay as previously described (55). Strains tested were as follows: 1, 531A (wild-type parent); 2, TW19 (transposon mutant). (B) Scheme of the gene cluster identified by transposon-directed cloning of mutant TW19. The point of transposon insertion in TW19 is indicated with a lollipop symbol, and the SalI restriction site modified in mutant 531A-10 is also indicated. (C) SDS-PAGE analysis of proteinase K-digested whole-cell lysates visualized by silver staining. Lanes: 1, 531A (wild-type parent); 2, TW19 (transposon mutant).
TABLE 2.
Sequence comparisons of ORFs to E. coli K-12 homologues
| Gene | Homologue (accession no.) | Proposed function | % Identitya |
|---|---|---|---|
| rmlB | RffG (NP418235) | dTDP-glucose 4,6-dehydratase | 73 (255) |
| rmlA | RffH (NP418236) | Glucose-1-phosphate thymidylyltransferase | 74 (258) |
| rmlD | RfbD (NP416544) | dTDP-6-deoxy-l-mannose dehydrogenase | 41 (289) |
| rmlC | RfbC (NP416542) | dTDP-6-deoxy-d-glucose-3,5 epimerase | 58 (184) |
The values in parentheses represent numbers of amino acid residues.
Based on the sequence analysis presented above, we hypothesized that mutant TW19 could be defective in the production of the O antigen synthesized by this strain. To test this, the LPS profile of mutant TW19 was compared to that of an isogenic wild-type strain. LPS was extracted from cells grown on CAS agar, resolved by SDS-PAGE, and visualized by silver staining. As predicted, the high-molecular-weight (HMW) O1-antigen bands seen in the wild-type strain were absent in mutant TW19 while the bands corresponding to the core remained intact (Fig. 1C), showing that the insertion mutation in strain TW19 abolishes production of the O antigen in this strain.
To further characterize this gene cluster and produce a cleaner mutation than that obtained from the transposon insertion, which could have polar effects, we decided to generate a site-directed mutation in another gene within this rhamnose biosynthetic pathway. Therefore, an unmarked rmlD mutation was constructed in wild-type V. anguillarum 531A by marker exchange mutagenesis, resulting in strain 531A-10 (see Materials and Methods for details of this construction). Strain 531A-10 contains a 4-bp insertion at a SalI site (shown in Fig. 1B) within the rmlD coding sequence. This insertion causes a frameshift at codon 185 and premature termination of translation at codon 202 of 272. This strain, like TW19, formed small colonies and overproduced siderophore when grown on CAS agar (Fig. 2B). This defect was complemented by addition of the rmlD gene supplied in trans on a multicopy plasmid (pTW541), demonstrating that this phenotype was not due to a polar effect on any downstream gene(s) and that more than one gene in the cluster could be responsible for the phenotype. Like mutant TW19, the LPS profile of strain 531A-10 was devoid of HMW O-polysaccharide bands, as assessed by silver staining of LPS extracts resolved by SDS-PAGE (data not shown). To verify that the observed loss of HMW O-polysaccharide bands seen in the rmlD mutant corresponds to a loss of O1 antigen cross-reactivity, we performed Western immunoblot analysis as described in Materials and Methods. For this experiment, cells were grown under both iron-sufficient and iron-restrictive conditions in glucose minimal medium in order to evaluate the effect of iron limitation on O-antigen production. Figure 2A shows that the mutant 531A-10, independently of the iron concentration, was devoid of the HMW antigens present in the wild-type strain and that complementation with the wild-type rmlD gene restored O-antigen production.
FIG. 2.
Genetic complementation of mutant strain 531A-10. (A) Detection of the V. anguillarum O1 antigen by Western blotting of proteinase K-digested whole-cell lysates resolved by SDS-PAGE. Strains analyzed were as follows: lanes 1 and 2, 531A/pBR325 (wild-type parent/vector); lanes 3 and 4, 531A-10/pBR325 (rmlD mutant/vector); lanes 5 and 6, 531A-10/pTW541 (rmlD mutant/complementing clone). Cells in lanes 1, 3, and 5 were grown using iron-sufficient conditions (10 μg/ml ferric ammonium citrate), while cells in lanes 2, 4, and 6 were grown using iron-deficient conditions (20 μM EDDA). (B) Detection of anguibactin using CAS agar plates as described in the legend to Fig. 1. Strains tested were as follows:1, 531A(pBR325); 2, 531A-10(pBR325); 3, 531A-10(pTW541).
O1-antigen role in resistance to the bactericidal activity of salmon serum.
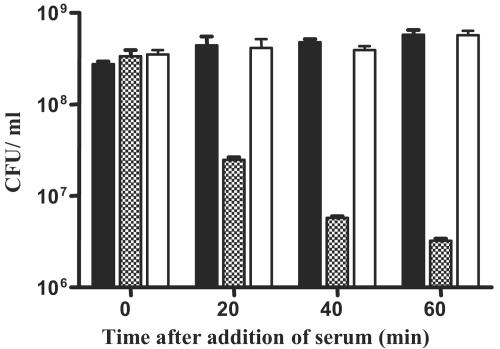
In order to determine the effect of the rml mutation on the ability of V. anguillarum to withstand the bactericidal action of nonimmune fish serum, we compared the serum sensitivity of this mutant to the isogenic wild-type and complemented mutant strains, all grown under identical conditions. This was accomplished by measuring the survival of each of these strains as a function of time after incubation in Chinook salmon NS. O1-deficient strain 531A-10 was highly sensitive to killing by NS, exhibiting a drop of approximately 2 orders of magnitude in viability over a 60-min period, while the isogenic wild-type strain showed no appreciable change in viability over the same time period (Fig. 3). Serum resistance was restored in 531A-10 by the addition of the rmlD gene supplied in trans. These results demonstrate that O1 antigen-containing LPS is necessary for protection of cells from the bactericidal effects of nonimmune salmon serum.
FIG. 3.
Sensitivity of 531A-10 to rainbow trout NS. Viability was measured by plate counting just before and at the indicated times after treatment with NS at a concentration of 50%. Strains tested were as follows: closed bars, 531A(pBR325); hatched bars, 531A-10(pBR325); open bars, 531A-10(pTW541).
Levels of the ferric anguibactin receptor FatA in mutant 531A-10.
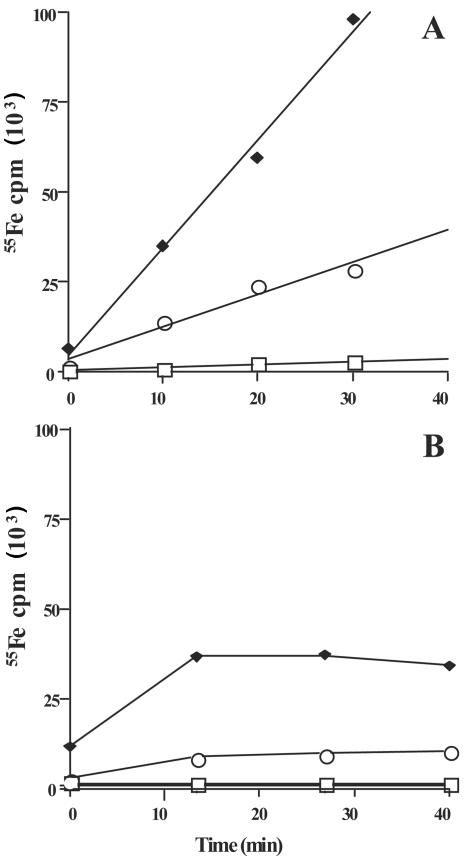
It is possible that the anguibactin overproduction phenotype seen in mutant strains TW19 and 531A-10 is the result of a defect in either the transport or utilization of ferric anguibactin. Such a mutant would be expected to experience increased iron starvation and, because of the iron regulation of siderophore production, increased anguibactin production. The increase in siderophore levels would, in turn, chelate remaining free iron, causing further iron starvation. A similar phenotype on CAS agar has been documented with ferric siderophore transport mutants of E. coli and V. anguillarum (48, 51). To test this, we first created a mutant derivative of 531A-10 that does not produce anguibactin. This was accomplished by mutating an essential anguibactin biosynthetic gene (angB) in the 531A-10 background, resulting in strain 531A-10(pJHC1::K). We then evaluated the ability of this strain to both bind and transport radioactive (55Fe) iron-loaded anguibactin added exogenously and compared it to an isogenic strain [531A(pJHC1::K)] that was O1 antigen producing and anguibactin deficient due to the same angB mutation. Figure 4A shows that mutant strain 531A-10(pJHC1::K) transports ferric anguibactin at approximately 30% of the rate of the isogenic wild-type strain and thus is transport defective. The transport of ferric anguibactin is an energy-dependent process, whereas receptor-mediated binding of ferric anguibactin is not dependent on energy (2). Therefore, ferric siderophore binding can be readily distinguished from transport by including the respiratory inhibitor KCN in the assay. Figure 4B shows that O1-antigen-deficient strain 531A-10(pJHC1::K) is defective in the binding of ferric anguibactin, exhibiting approximately 20% of the binding activity of the isogenic O1-antigen-producing parent strain. The specificity of this assay for ferric anguibactin is demonstrated by the lack of radioactive iron binding and accumulation in a strain (S531A-1) that does not contain the ferric anguibactin biosynthetic or transport machinery.
FIG. 4.
Kinetics of 55Fe-anguibactin uptake (A) and binding (B) in V. anguillarum strains grown under iron-limiting conditions as described in Materials and Methods. Strains tested were as follows: closed squares, 531A-1(pJHC1::K) (siderophore synthesis deficient); open circles, 531A-10(pJHC1::K) (siderophore synthesis deficient, O antigen deficient); open squares, S531A-1 (pJHC1-less, siderophore synthesis and utilization deficient). Transport experiments were independently replicated (three times), and the results shown are of a typical experiment.
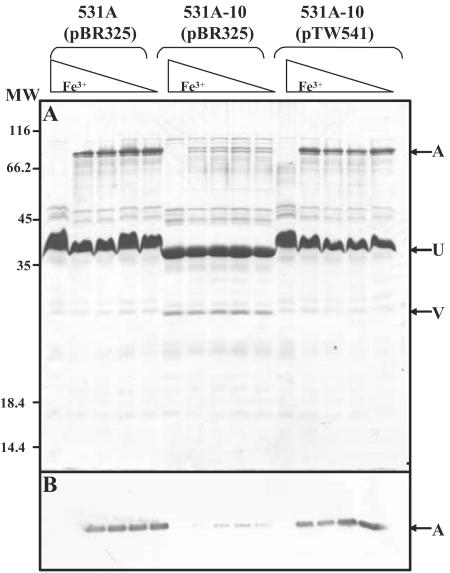
The observed defect in ferric anguibactin binding seen in this mutant suggests a decrease in ferric anguibactin receptor activity, perhaps due to a loss of the receptor protein itself. To assess this, the presence of the anguibactin receptor protein (FatA) was analyzed directly by SDS-PAGE and Western blot analysis of OM fractions purified from each of the strains under analysis. Strains were grown under both iron-sufficient and iron-deficient conditions to evaluate the effect of iron availability on FatA protein levels. The wild-type 531A strain displayed typical iron-regulated synthesis of FatA (Fig. 5), FatA production occurring only under conditions of iron limitation. In contrast, mutant strain 531A-10 (O1 deficient) displayed reduced levels of FatA in the OM fraction under both iron-limiting and iron-rich conditions. FatA levels were restored when the wild-type rmlD gene was supplied in trans in plasmid pTW541. It should be noted that LPS in the OM samples shown in this figure causes a distortion in the banding pattern in the middle of the gel due to comigration of the proteins in this area of the gel with O1 antigen-bearing LPS. This is most apparent in the retarded migration of the principal OM protein OmpU (designated U) in the samples from the wild-type and complemented mutant strains. The identity of this protein was verified in OM samples from both the wild-type (531A/pBR322) and mutant (531A-10/pBR322) strains by protein microsequencing. The N-terminal 10 amino acids of the protein purified from the wild type (slow migrating) and mutant (fast migrating) were identical to each other and to the deduced amino acid sequence of the OmpU protein (62). The 10 N-terminal amino acids of the overexpressed band marked V were identical to the previously identified OmpV protein (53).
FIG. 5.
FatA protein levels in mutant strain 531A-10. (A) Coomassie blue R-stained SDS-PAGE of OM preparations from indicated strains. (B) FatA levels measured by Western PAGE analysis of OM preparations shown in panel A and as described in Materials and Methods. Each strain was grown in CM9 supplemented with either 10 μg/ml ferric ammonium citrate (iron-sufficient conditions) or increasing concentrations of EDDA (iron-deficient conditions). EDDA concentrations in iron-restricted cultures ranged from 5 to 40 μM, increasing in twofold increments. Locations of FatA (A), OmpU (U), and OmpV (V) are denoted with arrows and were verified by protein microsequencing. MW, molecular weight (103).
Levels of the fatA transcript in mutant 531A-10.
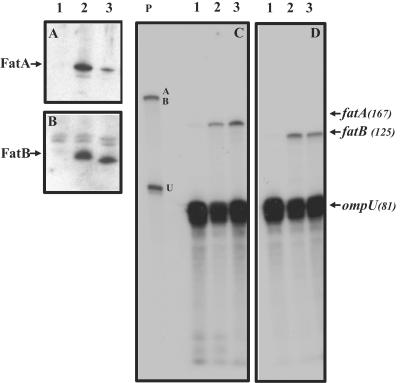
The ferric anguibactin transport genes reside in a large polycistronic message encoded on the pJM1-type plasmid pJHC1 in the order fatDCBA angRT (25, 55, 65). The decrease in FatA expression seen in strain 531A-10 could be due, at least in part, to a reduction in the synthesis of this polycistronic message or in the levels of the fatA-specific portion of this mRNA. To investigate these possibilities, we examined both fatA- and fatB-specific transcript levels in the 531A-10 mutant strain and compared them to the wild-type parent strain. The levels of both the FatA and FatB proteins were also assessed from subsamples taken from the same cultures as those used for the mRNA extractions, thereby allowing a direct comparison of the protein and mRNA levels in these strains. Since both the fatA and fatB genes are encoded in a polycistronic message, fatB-specific mRNA and FatB protein levels would be reduced in mutant 531A-10 if the observed reduction in FatA protein levels is the result of regulation of this polycistronic message. The results of this analysis are shown in Fig. 6. The levels of the fatA- and fatB-specific messages were unperturbed in the 531A-10 mutant compared to the wild-type strain (panels C and D, compare lanes 2 and 3), demonstrating that there was not reduced transcription of the fatA-specific portion of the polycistronic mRNA and thus that this was not the cause for the reduction of FatA protein levels in the mutant. Figure 6B also shows that the FatB protein levels in mutant 531A-10 are comparable to those of the wild type (compare lanes 2 and 3) while in the same samples FatA protein levels exhibited the expected decrease in mutant strain 531A-10 (panel A, compare lanes 2 and 3). These results indicate that the decrease observed in FatA protein levels in 531A-10 is the result of a process occurring after transcription of the fatA gene, most likely due to changes in the localization and/or stability of the FatA polypeptide. It is thus possible that FatA may accumulate elsewhere in the cell in mutants that lack the O1 antigen. To assess this possibility, experiments were conducted to localize FatA in 531A-10. Cells were grown in iron-depleted medium to induce FatA synthesis. Whole-cell, periplasmic, OM, total membrane, and concentrated culture supernatants were then analyzed for the presence of FatA by SDS-PAGE and Western blot analysis. FatA was detected only in the OM fraction and only in quantities as small as those shown in Fig. 5 and 6; there was no evidence suggesting FatA accumulation in any other cellular compartment or in the culture supernatant (data not shown).
FIG. 6.
Posttranscriptional regulation of FatA protein levels in 531A-10. FatA (A) and FatB (B) protein levels were assessed by Western blotting of total membrane fractions and fatA (C) and fatB (D) transcript levels assessed from total RNA using an RNase protection assay. Strains analyzed were as follows and were the same in all four panels: lanes 1 and 2, 531A (wild-type parent); lane 3, 531A-10 (rmlD mutant). Cells in lane 1 were grown using iron-sufficient conditions (10 μg/ml ferric ammonium citrate), while cells in lanes 2 and 3 were grown using iron-deficient conditions (20 μM EDDA). The size of each of the protected riboprobes is given in nucleotides. The three unprotected riboprobes are shown in lane P. The fatA and fatB full-length riboprobes resolve as a single band at approximately 220 nucleotides, while the ompU probe is 90 nucleotides in length. The ompU mRNA was used as an internal control to asses mRNA loading and quality.
Attenuation of virulence in mutant 531A-10.
Infectivity assays with rainbow trout were performed to elucidate the role of the V. anguillarum O1 antigen in virulence. An LD50 of approximately 10 bacteria was observed when fish were challenged using an intraperitoneal route of infection with wild-type strain 531A (Table 3). The LD50 of strain 531A-10 was more than 1.3 × 106 CFU, corresponding to a greater than 105-fold attenuation of virulence compared to the wild-type strain. These results demonstrate a role for the O1 antigen in virulence. However, we could not discard the possibility that this could be due to the pleiotropic effect of O1-antigen loss on the abundance of the FatA protein and subsequent diminishment of iron-scavenging activity. The observation that a strain lacking the pJHC1 plasmid (S531A-1), and therefore lacking the fatA gene, is at least 2 orders of magnitude more virulent than strain 531A-10 indicates that the low-virulence phenotype seen in mutant 531A-10 is not solely due to the loss of the FatA protein seen in this mutant strain. This is corroborated by the observation that a strain deficient in both iron transport and O1-antigen production also exhibits the low-virulence phenotype seen in mutant 531A-10 (Table 3).
TABLE 3.
Virulence assay results
| Strain | Description | LD50 (CFU/fish) |
|---|---|---|
| 531A | Wild type, Atlantic Ocean prototype | 10 |
| 531A-10 | O1-antigen-deficient derivative of 531A | >1.3 × 106a |
| S531A-1 | Plasmidless derivative of 531A | 8.4 × 104 |
| S531A-11 | Plasmidless derivative of 531A-10 | >9.0 × 106a |
No deaths occurred at this or at any lower cell number.
DISCUSSION
In this work, we have demonstrated that possession of an intact O1 side chain is essential for virulence of V. anguillarum 531A due to its effect on the maintenance of operational levels of the FatA protein and by conferring on the bacterium the ability to withstand the bactericidal action of the host nonimmune serum. The complement-mediated bactericidal effect of fish serum plays an important role in defense against bacterial pathogens. Some bacteria, however, are able to resist this nonspecific defense mechanism and thereby survive in blood and cause septicemic disease. Previous work with V. anguillarum has established a positive correlation between O-antigen size and serum resistance in strains of both the O1 and O2 serotypes (8, 59, 60). However, these studies relied on comparisons either between nonisogenic strains differing in the extent of HMW O polysaccharide or between the same strain grown under different culture conditions that affect the number of HMW O-polysaccharide forms. In the present study, a well-defined O1-antigen-deficient mutant has been constructed with which it was possible to unambiguously define the role of the O1 antigen in serum resistance.
The novel relationship between an O antigen and the levels of a ferric siderophore OM protein receptor has not been reported to occur in any other iron transport system of any bacteria. However, previous reports by others have shown a connection between LPS defects and the alteration of the function or translocation of OM proteins. For instance, both the OmpF and LamB proteins fail to trimerize in deep rough mutants of E. coli, causing a low abundance of these proteins in the OM (6, 33, 40, 49). LPS has also been shown to stimulate the assembly of OM proteins in vitro (22). Mutations causing loss of O-antigen biosynthesis in Vibrio cholerae have been shown to cause a defect in the assembly and localization of the toxin-coregulated pilus (29, 30). In Aeromonas salmonicida, it has been observed that O-antigen-deficient mutants are also defective in the assembly of the A layer (7). It is thus possible that the FatA polypeptide requires O1-antigen-bearing LPS to be correctly translocated to the OM or for stable maintenance of the protein in the OM; however, we found no evidence suggesting FatA accumulation in any other cellular compartment or in the culture supernatant. Our inability to detect FatA in these alternate locations does not rule out the possibility that FatA is mislocalized in mutant 531A-10. It is possible, for example, that FatA is not properly anchored to the OM in 531A-10 and ends up in the culture supernatant, where it is quickly degraded by extracellular protease activities, since V. anguillarum is known to produce large amounts of both secreted and cell-associated protease activities under normal culture conditions (26, 36, 38).
In the O1 side chain mutant, concomitant with the decrease in FatA levels, there was an increase in the levels of a protein that we identified as OmpV. OmpV has also been found to be overexpressed in an oxytetracycline-resistant strain of V. anguillarum (53). A homologue of this protein has also been reported in V. cholerae (41, 54). Whether there is any relationship between the increase in OmpV and the decrease in FatA is currently under study.
The observation that a strain lacking the pJM1-like plasmid pJHC1 (S531A-1), and therefore lacking the fatA gene, is still at least 2 orders of magnitude more virulent than the O1 side chain mutants indicates that the low-virulence phenotype seen in mutant 531A-10 is not solely due to the loss of the FatA protein. Underscoring this observation is the fact that an rmlD mutant constructed in the plasmidless strain resulted in an LD50 of the same order of magnitude as that of the serum-sensitive, iron transport-deficient 531A-10 strain.
There are at least 11 serotypes of V. anguillarum, although the only significant pathogens belong to the O1 and O2 types. It is noteworthy that pJM1-like virulence plasmids have only been found in serotype O1 strains of V. anguillarum; however, the opposite situation is not always true, since there are pathogenic strains of serotype O1 that do not carry the pJM1 plasmid. These plasmidless serotype O1, as well as O2, strains utilize chromosomally encoded high-affinity iron uptake systems that are genetically dissimilar from the pJM1 system (34, 59). It is tempting to speculate that the existence of the plasmid-mediated ferric anguibactin iron uptake system in serotype O1 strains is due to the requirement of the O1 side chain for an adequate level of the FatA protein and consequently for the selection and maintenance of the ferric anguibactin iron transport system in the O1 strains of V. anguillarum.
Editor: J. T. Barbieri
REFERENCES
- 1.Actis, L. A., W. Fish, J. H. Crosa, K. Kellerman, S. R. Ellenberger, F. M. Hauser, and J. Sanders-Loehr. 1986. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1). J. Bacteriol. 167:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Actis, L. A., S. A. Potter, and J. H. Crosa. 1985. Iron-regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J. Bacteriol. 161:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Actis, L. A., M. E. Tolmasky, L. M. Crosa, and J. H. Crosa. 1995. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol. Microbiol. 17:197-204. [DOI] [PubMed] [Google Scholar]
- 4.Actis, L. A., M. E. Tolmasky, D. H. Farrell, and J. H. Crosa. 1988. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J. Biol. Chem. 263:2853-2860. [PubMed] [Google Scholar]
- 5.Alice, A. F., C. S. Lopez, and J. H. Crosa. Unpublished data.
- 6.Ames, G. F., E. N. Spudich, and H. Nikaido. 1974. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J. Bacteriol. 117:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belland, R. J., and T. J. Trust. 1985. Synthesis, export, and assembly of Aeromonas salmonicida A-layer analyzed by transposon mutagenesis. J. Bacteriol. 163:877-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boesen, H. T., K. Pedersen, J. L. Larsen, C. Koch, and A. E. Ellis. 1999. Vibrio anguillarum resistance to rainbow trout (Oncorhynchus mykiss) serum: role of O-antigen structure of lipopolysaccharide. Infect. Immun. 67:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heynker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1992. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. 1977. Biotechnology 24:153-171. [PubMed] [Google Scholar]
- 10.Bullen, J. J., and E. Griffiths. 1999. Iron and infection: molecular, physiological and clinical aspects, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 11.Chai, S., T. J. Welch, and J. H. Crosa. 1998. Characterization of the interaction between Fur and the iron transport promoter of the virulence plasmid in Vibrio anguillarum. J. Biol. Chem. 273:33841-33847. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Q., L. A. Actis, M. E. Tolmasky, and J. H. Crosa. 1994. Chromosome-mediated 2,3-dihydroxybenzoic acid is a precursor in the biosynthesis of the plasmid-mediated siderophore anguibactin in Vibrio anguillarum. J. Bacteriol. 176:4226-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Q., and J. H. Crosa. 1996. Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271:18885-18891. [DOI] [PubMed] [Google Scholar]
- 14.Conchas, R. F., M. L. Lemos, J. L. Barja, and A. E. Toranzo. 1991. Distribution of plasmid- and chromosome-mediated iron uptake systems in Vibrio anguillarum strains of different origins. Appl. Environ. Microbiol. 57:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosa, J. H. 1989. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 53:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosa, J. H. 1980. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature 284:566-568. [DOI] [PubMed] [Google Scholar]
- 17.Crosa, J. H. 1984. The relationship of plasmid-mediated iron transport and bacterial virulence. Annu. Rev. Microbiol. 38:69-89. [DOI] [PubMed] [Google Scholar]
- 18.Crosa, J. H., and L. L. Hodges. 1981. Outer membrane proteins induced under conditions of iron limitation in the marine fish pathogen Vibrio anguillarum 775. Infect. Immun. 31:223-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crosa, J. H., L. L. Hodges, and M. H. Schiewe. 1980. Curing of a plasmid is correlated with an attenuation of virulence in the marine fish pathogen Vibrio anguillarum. Infect. Immun. 27:897-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosa, J. H., M. H. Schiewe, and S. Falkow. 1977. Evidence for plasmid contribution to the virulence of fish pathogen Vibrio anguillarum. Infect. Immun. 18:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Cock, H., and J. Tommassen. 1996. Lipopolysaccharides and divalent cations are involved in the formation of an assembly-competent intermediate of outer-membrane protein PhoE of E. coli. EMBO. J. 15:5567-5573. [PMC free article] [PubMed] [Google Scholar]
- 23.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Lorenzo, M., S. Poppelaars, M. Stork, M. Nagasawa, M. E. Tolmasky, and J. H. Crosa. 2004. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibrio anguillarum. J. Bacteriol. 186:7327-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Lorenzo, M., M. Stork, M. E. Tolmasky, L. A. Actis, D. Farrell, T. J. Welch, L. M. Crosa, A. M. Wertheimer, Q. Chen, P. Salinas, L. Waldbeser, and J. H. Crosa. 2003. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J. Bacteriol. 185:5822-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrell, D. H., and J. H. Crosa. 1991. Purification and characterization of a secreted protease from the pathogenic marine bacterium Vibrio anguillarum. Biochemistry 30:3432-3436. [DOI] [PubMed] [Google Scholar]
- 27.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobbs, M., and P. R. Reeves. 1994. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol. Microbiol. 12:855-856. [DOI] [PubMed] [Google Scholar]
- 29.Iredell, J. R., and P. A. Manning. 1997. Outer membrane translocation arrest of the TcpA pilin subunit in rfb mutants of Vibrio cholerae O1 strain 569B. J. Bacteriol. 179:2038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iredell, J. R., and P. A. Manning. 1997. Translocation failure in a type-4 pilin operon: rfb and tcpT mutants in Vibrio cholerae. Gene 192:71-77. [DOI] [PubMed] [Google Scholar]
- 31.Koster, W. L., L. A. Actis, L. S. Waldbeser, M. E. Tolmasky, and J. H. Crosa. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266:23829-23833. [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Laird, M. W., A. W. Kloser, and R. Misra. 1994. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. J. Bacteriol. 176:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemos, M. L., P. Salinas, A. E. Toranzo, J. L. Barja, and J. H. Crosa. 1988. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 170:1920-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Q., M. Hobbs, and P. R. Reeves. 2003. The variation of dTDP-l-rhamnose pathway genes in Vibrio cholerae. Microbiology 149:2463-2474. [DOI] [PubMed] [Google Scholar]
- 36.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1992. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milton, D. L., A. Norqvist, and H. Wolf-Watz. 1995. Sequence of a novel virulence-mediating gene, virC, from Vibrio anguillarum. Gene 164:95-100. [DOI] [PubMed] [Google Scholar]
- 38.Norqvist, A., B. Norrman, and H. Wolf-Watz. 1990. Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect. Immun. 58:3731-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norqvist, A., and H. Wolf-Watz. 1993. Characterization of a novel chromosomal virulence locus involved in expression of a major surface flagellar sheath antigen of the fish pathogen Vibrio anguillarum. Infect. Immun. 61:2434-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohlner, J., T. F. Meyer, M. B. Jalajakumari, and P. A. Manning. 1986. Nucleotide sequence of ompV, the gene for a major Vibrio cholerae outer membrane protein. Mol. Gen. Genet. 205:494-500. [DOI] [PubMed] [Google Scholar]
- 42.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 43.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 44.Salinas, P. C., and J. H. Crosa. 1995. Regulation of angR, a gene with regulatory and biosynthetic functions in the pJM1 plasmid-mediated iron uptake system of Vibrio anguillarum. Gene 160:17-23. [DOI] [PubMed] [Google Scholar]
- 45.Salinas, P. C., M. E. Tolmasky, and J. H. Crosa. 1989. Regulation of the iron uptake system in Vibrio anguillarum: evidence for a cooperative effect between two transcriptional activators. Proc. Natl. Acad. Sci. USA 86:3529-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salinas, P. C., L. S. Waldbeser, and J. H. Crosa. 1993. Regulation of the expression of bacterial iron transport genes: possible role of an antisense RNA as a repressor. Gene 123:33-38. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 49.Sen, K., and H. Nikaido. 1991. Trimerization of an in vitro synthesized OmpF porin of Escherichia coli outer membrane. J. Biol. Chem. 266:11295-11300. [PubMed] [Google Scholar]
- 50.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 51.Stork, M., M. Di Lorenzo, S. Mourino, C. R. Osorio, M. L. Lemos, and J. H. Crosa. 2004. Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect. Immun. 72:7326-7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stroeher, U. H., K. E. Jedani, and P. A. Manning. 1998. Genetic organization of the regions associated with surface polysaccharide synthesis in Vibrio cholerae O1, O139 and Vibrio anguillarum O1 and O2: a review. Gene 223:269-282. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki, S., K. Yasue, and R. Kusuda. 1998. A porin like outer membrane protein (Omp26La) appears to increase in an oxytetracycline resistant strain of marine fish pathogen Vibrio (Listonella) anguillarum. Microbes Environ. 13:197-202. [Google Scholar]
- 54.Taylor, R. K., C. Manoil, and J. J. Mekalanos. 1989. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J. Bacteriol. 171:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1988. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J. Bacteriol. 170:1913-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1995. A histidine decarboxylase gene encoded by the Vibrio anguillarum plasmid pJM1 is essential for virulence: histamine is a precursor in the biosynthesis of anguibactin. Mol. Microbiol. 15:87-95. [DOI] [PubMed] [Google Scholar]
- 57.Tolmasky, M. E., L. A. Actis, and J. H. Crosa. 1993. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect. Immun. 61:3228-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tolmasky, M. E., A. M. Wertheimer, L. A. Actis, and J. H. Crosa. 1994. Characterization of the Vibrio anguillarum fur gene: role in regulation of expression of the FatA outer membrane protein and catechols. J. Bacteriol. 176:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toranzo, A. E., J. L. Barja, S. A. Potter, R. R. Colwell, F. M. Hetrick, and J. H. Crosa. 1983. Molecular factors associated with virulence of marine vibrios isolated from striped bass in Chesapeake Bay. Infect. Immun. 39:1220-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trust, T. J., I. D. Courtice, A. G. Khouri, J. H. Crosa, and M. H. Schiewe. 1981. Serum resistance and hemagglutination ability of marine vibrios pathogenic for fish. Infect. Immun. 34:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Gabain, A., J. G. Belasco, J. L. Schottel, A. C. Chang, and S. N. Cohen. 1983. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc. Natl. Acad. Sci. USA 80:653-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, S. Y., J. Lauritz, J. Jass, and D. L. Milton. 2003. Role for the major outer-membrane protein from Vibrio anguillarum in bile resistance and biofilm formation. Microbiology 149:1061-1071. [DOI] [PubMed] [Google Scholar]
- 63.Welch, T. J., S. Chai, and J. H. Crosa. 2000. The overlapping angB and angG genes are encoded within the trans-acting factor region of the virulence plasmid in Vibrio anguillarum: essential role in siderophore biosynthesis. J. Bacteriol. 182:6762-6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wertheimer, A. M., M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1994. Structural and functional analyses of mutant Fur proteins with impaired regulatory function. J. Bacteriol. 176:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wertheimer, A. M., W. Verweij, Q. Chen, L. M. Crosa, M. Nagasawa, M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1999. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 67:6496-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolf, M. K., and J. H. Crosa. 1986. Evidence for the role of a siderophore in promoting Vibrio anguillarum infections. J. Gen. Microbiol. 132(Pt. 10):2949-2952. [DOI] [PubMed] [Google Scholar]
- 67.Woo, P. T. K., J. F. Leatherland, D. W. Bruno, and C. A. B. International. 1999. Fish diseases and disorders. CAB International, Wallingford, United Kingdom.