Abstract
Flagellar structures have been shown to participate in virulence in a variety of intestinal pathogens. Here, we have identified two potential flagellar genes of Listeria monocytogenes: lmo0713, encoding a protein similar to the flagellar basal body component FliF, and lmo0716, encoding a protein similar to FliI, the cognate ATPase energizing the flagellar export apparatus. Expression of fliF and fliI appears to be downregulated at 37°C, like that of flaA, encoding flagellin. By constructing two chromosomal deletion mutants, we show that inactivation of either fliF or fliI (i) abolishes bacterial motility and flagella production, (ii) impairs adhesion and entry into nonphagocytic epithelial cells, and (iii) also reduces uptake by bone marrow-derived macrophages. However, the ΔfliF and ΔfliI mutations have only a minor impact on bacterial virulence in the mouse model, indicating that the flagellar secretion apparatus itself is not essential for survival in this animal model. Finally, among 100 human clinical isolates of L. monocytogenes tested, we found 20 strains still motile at 37°C. Notably, all these strains adhered less efficiently than strain EGD-e to Caco-2 cells at 37°C but showed no defect of intracellular multiplication. These data suggest that expression of the flagella at 37°C might hinder optimal adhesion to epithelial cells but has no impact on intracytosolic survival of L. monocytogenes.
Listeria monocytogenes is a motile, facultative intracellular bacterium that causes food-borne infections in humans and animals, with symptoms of septicemia, meningitis, and meningo-encephalitis. This gram-positive bacterium is also widely distributed in the environment and is able to grow over a wide range of temperatures, pHs, and osmotic pressures (43). In animal models, bacteria have been shown to cross the gastrointestinal barrier and possibly penetrate the intestinal epithelial cells overlaying Peyer's patches (34, 35). The organism then disseminates to the brain and to the spleen, liver, and other lymphatic systems (26). The virulence of L. monocytogenes is due to its capacity to invade and multiply within host cells, including macrophages and hepatocytes, as well as epithelial, endothelial, and neuronal cells. Each step of the infectious process is dependent on the production of virulence factors, including invasion proteins (InlA and InlB), listeriolysin O, phospholipases, and ActA, which are controlled by the pleiotropic transcriptional activator PrfA (11). The initial step of the internalization of L. monocytogenes into nonphagocytic cells involves an adhesion mediated in particular by internalin (InlA) and InlB surface proteins. InlA promotes entry of L. monocytogenes into human epithelial cells via E-cadherin as a receptor (30). InlB is involved in entry of the bacterium into a broad range of cell lines including hepatocytes and nonepithelial cells. InlB interacts with the receptor for the complement factor C1q, hepatocyte growth factor (c-Met), and glycosamine glycans (4). Recent findings suggest that other molecules are also necessary for adhesion or internalization, as for example Ami, an amidase, and FbpA, a fibronectin binding protein (11). After entry, the bacterium lyses the internalization vacuole and escapes into the host cell cytoplasm, where bacterial replication takes place. In the cytosol, L. monocytogenes uses an actin-based motility process to propel itself in the cytosol and invades neighboring cells.
It has been shown in several bacterial species that components of the flagella could act as adhesins and mediate binding to the host's mucosal surfaces (1, 46). In vitro studies suggest that one role of the flagella could be to increase host invasion (28) or survival within macrophages (45). The contribution of flagella to virulence has been demonstrated in many bacteria such as Campylobacter jejuni (15), Legionella pneumophila (9), Clostridium difficile (41), Helicobacter pylori (12), Aeromonas caviae (36), Salmonella enterica serovar Typhi (28), Vibrio cholerae (38), and Vibrio anguillarum (32). The flagellum comprises a long filament that acts as a propeller anchored to the cell envelope by a flexible hook and basal body. The flagellar “axial” proteins that polymerize to the structure are exported by the type III export apparatus to the growing flagellum (29, 47). Export of these structural components is strongly facilitated by substrate-specific ATPase (FliI). FliI is peripherally associated with the inner membrane and is assumed to couple ATP hydrolysis to substrate translocation through the membrane-export machinery. Studies in Bacillus thuringiensis have shown that a mutant strain inactivated for the flhA gene, encoding a protein of the flagellar export apparatus, was defective in both swimming and swarming motility because it lacked flagella. Strikingly, the mutant was also defective in the secretion of virulence factors (13), suggesting that some virulence factors might be translocated via the flagellar apparatus. In fact, 26% of the extracellular bacterial proteome lacks typical signal peptides and can escape from the cytoplasm by cell lysis, the holin systems, or via the flagellar export machinery and other unidentified export systems (see reference 42 for a review).
The flagellar filament of L. monocytogenes is composed of one major subunit, the flaA-encoded flagellin, that is produced and assembled at the cell surface when L. monocytogenes grows between 4°C and 30°C, whereas its production is markedly reduced at 37°C. In the present work we have identified and inactivated two potential flagellar genes of L. monocytogenes: lmo0713, encoding a protein similar to the flagellar basal body component FliF, and lmo0716, encoding a protein similar to the ATPase, FliI, energizing the flagellum component export. The two mutants were nonmotile on soft agar and did not produce flagella. We have also analyzed a series of 100 clinical isolates for their motility at 37°C or 30°C and tested host cell adhesion of selected samples. Our data demonstrate that FliF and FliI are essential for flagella assembly in L. monocytogenes. We show that the flagella are involved in the entry of L. monocytogenes in tissue culture cells but are not crucial for virulence in the mouse model. The role of the flagella in bacterial pathogenicity is discussed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Brain heart infusion (BHI) and Luria-Bertani (Difco Laboratories) broth and agar were used to grow Listeria and Escherichia coli strains, respectively. We used the reference strains EGD-e for L. monocytogenes (serovar 1/2a) and CLIP11262 for Listeria innocua, for which sequencing was recently completed (14). E. coli DH5α and TOP10 (Invitrogen Corp.) strains were used as recipients for plasmid construction and propagation. Strains harboring antibiotic resistance were grown with the following antibiotic concentrations: ampicillin, 100 μg/ml (E. coli); erythromycin (Em), 150 μg/ml (E. coli) and 5 μg/ml (L. monocytogenes); kanamycin (Km), 50 μg/ml.
Genetic manipulations.
Chromosomal DNA, plasmid isolation, restriction reactions, and PCR amplification were performed according to standard protocols (39). Restriction enzymes and ligase were purchased from New England Biolabs. DNA was amplified with the Taq DNA polymerase DyNAzyme II from Finnzymes, according to the manufacturer's recommendations, in a Bio-Rad iCycler thermal cycler. Oligonucleotides were synthesized by Eurogentec. Nucleotide sequencing was carried out with a BigDye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer) on an ABI Prism 310 sequencer (Applied Biosystems).
Plasmid pUC19 (Ampr) was used to clone PCR-amplified sequence and construct deletions. Deletions were then transferred to the shuttle vector pAUL-A (Emr) (5). Wild-type L. monocytogenes EGD-e was transformed by electroporation (33).
Construction of the ΔfliF and ΔfliI mutants of L. monocytogenes EGD-e.
Two mutants were constructed by allelic replacement of the open reading frame of lmo0713 (fliF) or lmo0716 (fliI) by the promotorless aphA3 cassette (conferring Km resistance). A 740-bp EcoRI-SmaI-digested double-stranded DNA fragment (fragment A) upstream of the fliF gene and a 770-bp SmaI-XbaI DNA fragment (fragment B) downstream of the fliF gene were amplified by PCR using L. monocytogenes EGD-e genomic DNA and the following primer pairs: for fragment A, 5′-CCGGAATTCCAAGCGGCTCTGCGTTAAAT-3′ and 5′-TCCCCCGGGCCGACTAGTCTAACCGCTAC-3′; for fragment B, 5′-TCCCCCGGGAAGGCAGGAATGACAATGGC-3′ and 5′-TGCTCTAGATCGTTCTAAACCGCGGGTAA-3′. After digestion, the two fragments A and B were successively cloned into the pUC19 vector between the EcoRI and the XbaI sites. The pUC19-AB plasmid was digested by SmaI, and the 900-kb aphA3 cassette flanked by two SmaI restriction sites was inserted in pUC19-AB. The 2.4-kb EcoRI-XbaI A-aphA3-B fragment was then cloned into the thermosensitive shuttle vector pAUL-A. Plasmid pAUL-A-aphA3-B was then electroporated into L. monocytogenes EGD-e (33), and the transformants were selected for Emr at 30°C. The L. monocytogenes ΔfliF chromosomal mutant was obtained by allelic replacement in a two-step procedure: first, culture of a transformant at 37°C allowed selection of the integration of the vector into the chromosome by homologous recombination in the A or B region; then at replication-permissive temperature (30°C) excision events suppressing the plasmid and the fliF gene allowed detection of a ΔfliF mutant strain.
The construction of the ΔfliI mutant was carried out by a similar procedure, by using the following primers for amplifications of fragments A and B: for fragment A, 5′-CGGAATTCGGGAACAGGATGTCCTTATC-3′ and 5′-TCCCCCGGGCGGTTTATCGCCTCCTAGAA-3′; for fragment B, 5′-TCCCCCGGGAAGGAGTGCCGTGGATGAAT-3′ and 5′-GCTCTAGATGGTTTATTCTCCCTTACCC-3′. The constructions were confirmed by PCR sequence analysis of chromosomal DNA from the mutants.
RNA extraction and real-time RT-PCR assays.
L. monocytogenes was grown in BHI broth, and bacteria in exponential phase (optical density at 600 nm [OD600] from 0.4 to 0.6) were collected by centrifugation. RNA extraction proceeded as described previously (3). DNA contamination was removed by digestion with DNase I (Roche Diagnostic), and the RNA preparation was purified using an RNeasy kit (QIAGEN S.A.). Reverse transcription (RT)-PCR experiments were carried out with 1 μg of RNA and 2.5 pmol of specific reverse primer for gyrA and the studied gene (flaA, fliF, or fliI) in a volume of 8 μl. After denaturation at 65°C for 10 min, 12 μl of the mixture containing 2 μl of each deoxynucleoside triphosphate (25 mM), 4 μl of 4× buffer, 2 μl of dithiothreitol, 1 μl of RNasin (Promega), and 1.5 μl of Superscript II (Invitrogen) was added. Samples were incubated for 60 min at 42°C, heated at 75°C for 15 min, and then chilled on ice. Samples were diluted with 40 μl of H2O and then stored at −20°C.
The following pairs of primers were designed (using Primer Express software) in order to amplify mRNAs: flaA forward primer, 5′-ACTCATCCATGGGTATTGACTTGTT-3′; flaA reverse primer, 5′-AATTGACGCATACGTTGCAAGA-3′; fliF forward primer, 5′-AAAGCTGAACCAGCAAGCAAAG-3′; fliF reverse primer, 5′-GCGATGACGGCGAAGAGTAA-3′; fliI forward primer, 5′-ACTGCGCAAAAGCGTGATT-3′; fliI reverse primer, 5′-CGGCGCGTAGTTGCATAA-3′; gyrA forward primer, 5′-AAATGCGGACATCATTCCTAGACT-3′; and gyrA reverse primer, 5′-TTTAACCCGTCACGAACATCAG-3′.
Real-time PCRs were carried out using an ABI Prism 7700 sequence detection system. The conditions were identical for all reactions. The 25-μl mix consisted of 4 μl of template, 12.5 μl of SYBR Green Jump Start Taq ready mix (Sigma), and 5 pmol of each primer. The reactions were carried out in sealed tubes. Results were normalized to the amount of gyrA mRNA. The gyrA gene was chosen because its expression remains constant under the different experimental conditions used here (37). Each assay was performed at least in triplicate.
For classical reverse transcription PCR, amplification of flaA cDNA was carried out with the forward primer 5′-GCATGACTCAAGCGCAAGAA-3′ and the previously described reverse primer.
Motility assay.
Motility was assayed on soft agar plates. BHI broth overnight cultures were point inoculated in 0.4% BHI agar plates. Swimming bacteria are able to move within the agar. Plates were incubated for 24 to 48 h at 25, 30, or 37°C.
Electron microscopy.
Bacteria from 0.3% BHI agar plates were fixed with a 3% paraformaldehyde solution on an electron microscopy grid. Grids were washed three times, and then negative contrast was obtained by a 10-min treatment with phosphotungstic acid (2%). Bacteria were observed with a JEON 100 CX II electron transmission microscope.
SDS-polyacrylamide gel electrophoresis and Western blot analysis.
Total extract from bacteria in exponential (OD600 of 0.4) or stationary phase (OD600 of 1.5) was prepared. Bacterial cultures were recovered by centrifugation, washed in phosphate-buffered saline, and resuspended in a 1-ml suspension in order to be broken in a Fast Prep apparatus (30-s pulse on setting 6.5). Lysates were mixed with sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis buffer (130 mM Tris-HCl, pH 6.8, 1% SDS, 7% 2-β-mercaptoethanol, 7% sucrose, 0.01% bromophenol blue) and loaded on a 12% polyacrylamide gel (Protean II; Bio-Rad). Electrophoresis, transfer onto nitrocellulose, and immunodetection were carried out according to standard procedures. FlaA protein was visualized using a rabbit anti-Listeria FlaA antibody (H-AB antiserum; Eurobio, France) and an anti-rabbit peroxidase-conjugated secondary antibody.
An E. coli BL21 strain expressing a six-His-tagged L. monocytogenes flaA gene in a pET28 vector was constructed and used as a positive control in the Western blotting.
Culture of cell lines.
The human colon carcinoma cell line Caco-2 (ATCC HTB37) and the human hepatocellular carcinoma cell line HepG-2 (ATCC BH8065; obtained from S. Dramsi and P. Cossart, Institut Pasteur, Paris, France) were propagated in Dulbecco modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum at 37°C with 5% CO2. Cells were seeded at approximately 2 × 105 cells per well in 12-well tissue culture plates. Monolayers were used 24 h (Caco-2) or 48 h (HepG-2) after seeding. Cells were infected (following the experiment) with bacteria grown at 30°C or 37°C overnight at a multiplicity of 100 bacteria per cell.
Bone marrow-derived macrophages.
Bone marrow-derived macrophages from BALB/c mice were obtained and cultured as described previously (7) and then infected at 1 bacterium per 10 macrophages with bacteria grown at 30°C (or 37°C) overnight.
Invasion and adhesion assay. (i) Invasion assay.
Cell monolayers were incubated for 30 min after infection with 1 ml of bacterial suspension in DMEM to allow bacteria to enter. Then after three washes with RPMI medium, cells were either lysed with cold distilled water (time zero of the kinetic) or reincubated with fresh DMEM containing 10 μg/ml gentamicin to kill extracellular bacteria. The kinetic was followed for 5 or 6 h. At each point, cells were washed three times before lysis with distilled water (incubation for 20 min at 4°C with 1 ml of water before recovery of the cells lysate), and then lysates were diluted and spread on BHI agar plates to determine the number of viable intracellular bacteria. Each point was carried out in triplicate and each experiment was repeated at least twice.
(ii) Adhesion assay.
Cell line monolayers were incubated with bacteria from 15 min up to 60 min and then washed three times with RPMI medium before lysis of the cells with cold distilled water. Lysates were diluted and spread on BHI agar plates to number the amount of adherent or intracellular bacteria.
Virulence in mice. (i) LD50.
Pathogen-free Swiss female mice (Janvier), 6 to 8 weeks old, were used. The bacterial inoculum was diluted in 0.15 M NaCl and inoculated intravenously (0.5 ml per mouse) in the lateral tail vein. Groups of five mice were challenged intravenously (i.v.) with various doses of bacteria, and mortality was followed for 10 days. The virulence of the strains was estimated by the 50% lethal dose (LD50) using the Probit method.
(ii) Oral infection.
Pathogen-free BALB/c female mice (Janvier), 6 to 8 weeks old, were infected intragastrically with 1.5 × 1010 bacteria grown at 30°C overnight (10 mice per group). Mice were sacrificed 24 or 72 h after infection. Intestinal tissues were removed and washed three times with phosphate-buffered saline and then treated with gentamicin (10 μg/ml). Mesentery and spleen were also removed, and each organ was homogenized in 0.5 M NaCl. Serial dilutions (10-fold) of the lysates in 0.5 M NaCl were plated on BHI agar to determine the number of CFU per organ.
RESULTS
Temperature-regulated expression of flaA, fliF, and fliI in L. monocytogenes.
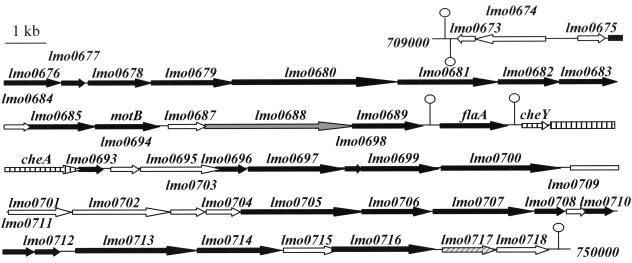
At 25°C, L. monocytogenes is actively motile, with four to six peritrichous cells (40), and produces abundant flagellin (flaA encoded). In contrast, at 37°C, it is virtually nonmotile and produces little or no detectable flagellin (22). Here, we have identified two potential flagellar genes of L. monocytogenes: lmo0713, encoding a protein of 551 amino acids having 23% identity (and 45% similarity) with FliF of B. subtilis, the first structure to be assembled during flagellar biogenesis (23); and lmo0716, encoding a protein of 434 amino acids, having 43% amino acid identity (and 62% similarity) with FliI, an ATPase of B. subtilis that drives flagellar protein export. The two genes are located in the same portion of the L. monocytogenes chromosome (Fig. 1) that comprises 26 other genes encoding putative flagellar machinery proteins (Table 1). This locus of 38 kb is composed of three clusters flanked by predicted transcription terminators. In contrast to the other genes, flaA seems to be transcribed independently (Fig. 1).
FIG. 1.
Genetic organization of the flagellar genes of L. monocytogenes EGD-e (between 709 and 750 kb). Black arrow, flagellar genes; vertically striped arrow, signalization pathways; diagonally striped arrow, other functions; gray arrow, similar to unknown protein; white arrow, no similarity; open circle with line, transcription terminator.
TABLE 1.
Flagellar genes of L. monocytogenes EGD-e clustered between 709 and 750 kba
| Gene | Description (no. of base pairs) |
|---|---|
| lmo0676 | Similar to flagellar biosynthesic protein FliP (768) |
| lmo0677 | Similar to flagellar biosynthesic protein FliQ (273) |
| lmo0678 | Similar to flagellar biosynthesic protein FliR (762) |
| lmo0679 | Similar to flagellar biosynthesic protein FlhB (1047) |
| lmo0680 | Similar to flagellar associated protein FlhA (2076) |
| lmo0681 | Similar to flagellar biosynthetic protein FlhF (1224) |
| lmo0682 | Similar to flagellar hook-basal body protein FlgG (780) |
| lmo0685 | Similar to motility protein (flagellar motor rotation) MotA (852) |
| motB | Similar to motility protein (flagellar motor rotation) MotB (828) |
| flaA | Flagellin protein (864) |
| lmo0693 | Similar to flagellar motor switch protein FliY C-terminal part (300) |
| lmo0696 | Similar to flagellar hook assembly protein (423) |
| lmo0697 | Similar to flagellar hook protein FlgE (1236) |
| lmo0698 | Weakly similar to flagellar switch protein FliY (237) |
| lmo0699 | Similar to flagellar switch protein FliM (993) |
| lmo0700 | Similar to flagellar switch protein FliY (1548) |
| lmo0705 | Similar to flagellar hook associated protein FlgK (1521) |
| lmo0706 | Similar to flagellar hook associated protein 3 FlgL (876) |
| lmo0707 | Similar to flagellar hook associated protein 2 FliD (1290) |
| lmo0708 | Similar to a hypothetical flagellar protein (387) |
| lmo0710 | Similar to flagellar basal-body rod protein FlgB (402) |
| lmo0711 | Similar to flagellar basal body rod protein FlgC (411) |
| lmo0712 | Similar to flagellar hook-basal body complex protein FliE (297) |
| lmo0713 | Similar to flagellar basal-body M-ring protein FliF (1653) |
| lmo0714 | Similar to flagellar motor switch FliG (1107) |
| lmo0716 | Similar to flagellar specific FliI ATPase alpha chain (1302) |
Schematic diagram is shown in Fig 1.
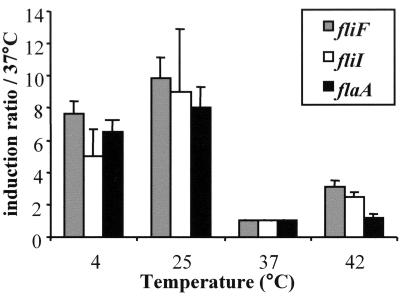
We monitored by real-time PCR the transcription of flaA, lmo0713, and lmo0716 in EGD-e grown at different temperatures (Fig. 2). All three genes appeared to be upregulated at 4°C and 25°C (sixfold and ninefold, respectively) compared to growth at 37°C. At 42°C, flagellar gene expression was not significantly different from that at 37°C. Thus, at the temperature encountered in a mammalian host, L. monocytogenes shuts down expression of flagellin as well as that of the proteins of the flagellar basal body.
FIG. 2.
Transcriptional analysis of the expression of flaA, fliF, and fliI at different temperatures. Gene expression was measured by real-time PCR from bacteria in exponential phase of growth (in BHI broth). The amount of each gene mRNA was normalized by the amount of gyr mRNA (constant under these conditions). Induction ratios were then compared to the 37°C condition. Values shown are means of results from at least three assays for different mRNA preparations for each temperature. Error bars indicate the standard deviations.
ΔfliF and ΔfliI mutants impair flagellar assembly.
Experiments were performed to determine the role of FliF and FliI in flagellar assembly and function. We constructed two chromosomal deletion mutants of EGD-e by allelic exchange (see Materials and Methods), denoted ΔfliF and ΔfliI. Each mutant corresponds to the in-frame deletion of the entire coding sequence (lmo0713 and lmo0716, respectively). We confirmed by RT-PCR that the genes immediately downstream of fliF were transcribed by monitoring transcription of fliG and fliI (data not shown).
We first evaluated the growth of the two mutants in BHI medium at 4°C, 25°C, 37°C, and 42°C. At each condition tested, the ΔfliF and ΔfliI mutant strains exhibited a normal colony morphology on solid medium and a growth rate identical to that of the wild-type strain EGD-e in liquid medium (data not shown).
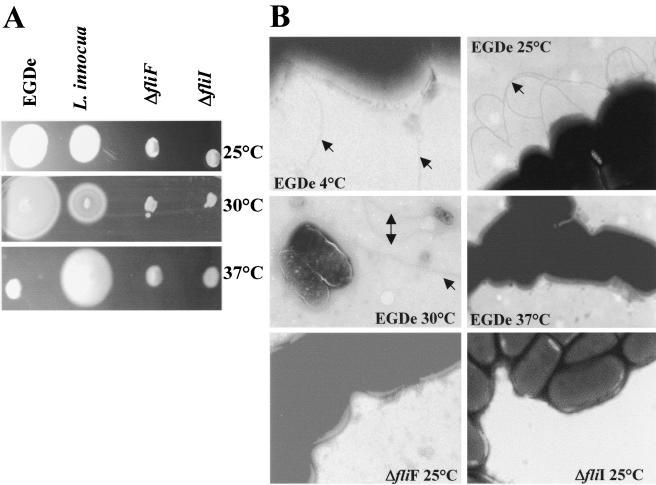
The two mutants were assayed for mobility in semisolid BHI medium, as described by Kathariou and coworkers (22). The ability to swim is thought to be a ubiquitous behavioral trait exhibited by flagellated eubacteria. Wild-type EGD-e swims in 0.4% agar at 25°C, 30°C (Fig. 3A), and 4°C (data not shown) but is unable to spread in agar medium at 37°C (Fig. 3A). The ΔfliF and ΔfliI mutant strains were nonmotile at 37°C as well as at permissive temperatures, i.e., 25°C and 30°C (Fig. 3A). The mutant strains were also nonmotile at 4°C (data not shown). Electron microscopy experiments showed that absence of motility at 37°C was correlated with an absence of visible flagella at the bacterial surface (for EGD-e as well as for the two isogenic mutants). Flagella were visible at the surface of the wild-type strain grown at 25°C, 30°C, or 4°C (Fig. 3B). In contrast, flagella were not visible at the surface of any of the two mutants at 25°C (Fig. 3B), 37°C, or 4°C (not shown), demonstrating that lmo0713 and lmo0716 are involved in flagellar biogenesis of L. monocytogenes.
FIG. 3.
Motility and flagellation of L. monocytogenes EGD-e, ΔfliF, ΔfliI, and L. innocua. (A) Motility assayed on 0.4% BHI agar plates. (B) Flagellation observed with electron microscopy (negative contrast obtained with 2% phosphotungstic acid). Flagella are indicated with arrows.
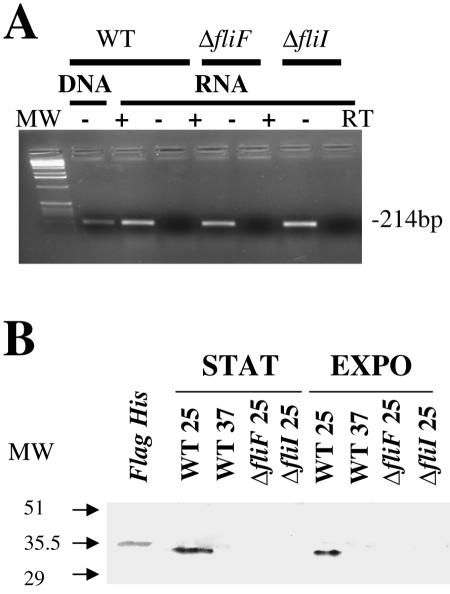
We then compared the transcription of the flaA gene in a ΔfliF or ΔfliI genetic background to that in the wild-type strain at 25°C (Fig. 4A). RT-PCR experiments showed that flaA transcription was unaffected in the ΔfliF and ΔfliI mutants. Thus, in contrast to B. subtilis, where the transcription of flagellin is dependent on the formation of the flagellar basal body (2), in L. monocytogenes the lack of motility and the absence of flagella in ΔfliF and ΔfliI mutant strains do not provoke a repression of flaA transcription. Confirming the RT-PCR data indicating that flaA was downregulated at 37°C (Fig. 2), our Western blot analyses showed that the protein FlaA was detected in the total protein extracts of EGD-e grown at 25°C but not in extracts from bacteria grown at 37°C (Fig. 4B). Notably, in B. thuringiensis a defect of flagellar filament assembly leads to an accumulation of flagellin in the cytosol (13). In contrast, in L. monocytogenes FlaA was not detected in whole-cell protein extracts of the ΔfliF and ΔfliI mutant strains grown at 25°C (Fig. 4B). The lack of detection of nonsecreted FlaA in these mutants is likely to be due to its rapid cytosolic degradation.
FIG. 4.
Expression of flagellin in EGD-e, ΔfliF, and ΔfliI strains. (A) Transcriptional analysis by RT-PCR using primers inside the flaA sequence. (B) Western blot analysis. Bacterial cultures from exponential (EXPO; OD600 of 0.4) or stationary phase (STAT; OD600 of 1.5) were collected by centrifugation, and total extract was obtained with a Fast Prep apparatus. Anti-Listeria serum H-AB purchased from Eurobio (France) was used as the primary antibody at a dilution of 1/500. An E. coli BL21 strain expressing EGD-e FlaA protein with a six-histidine tag (Flag His panel) was used as a positive control for protein detection.
Role of the flagellar apparatus in cell invasion.
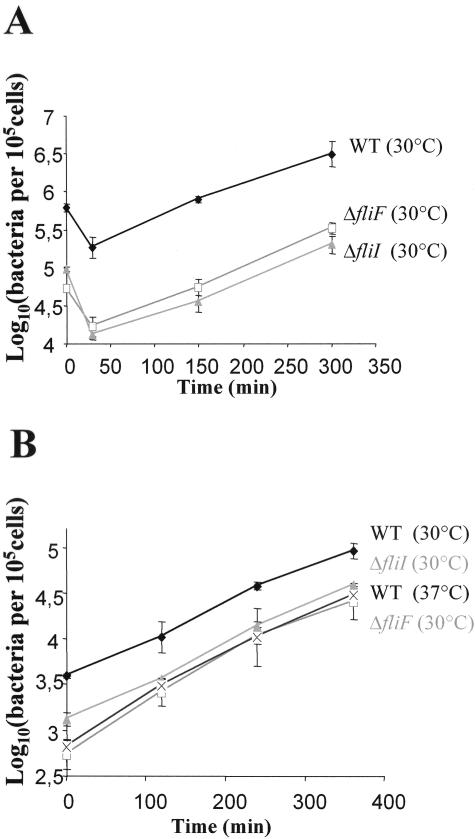
The ability of the two mutants to penetrate and replicate within cells was then studied in two different types of mammalian cells: the human epithelial Caco-2 cell line and mouse bone marrow-derived macrophages (BMM), with bacteria grown at 30°C overnight prior to infection. In both cell types, adhesion and entry of the two mutants were reduced: in Caco-2 cells, invasion of the two mutants was 10-fold lower than that of EGD-e (Fig. 5A), and in BMM, a more modest but reproducible defect of invasion was also observed. (ca. three to five times lower than EGD-e, after 30 min of infection) (Fig. 5B).
FIG. 5.
Invasion kinetics of L. monocytogenes EGD-e, ΔfliF, and ΔfliI strains in two different cell types: Caco-2 cells (A) and bone marrow-derived macrophages (B). Bacteria grown overnight at 30°C (or 37°C) were used to infect cell monolayers (100 bacteria per cell for Caco-2 cells; 0.1 bacteria per cell for macrophages). The temperatures given in the figure are the temperatures of bacterial growth before infection. All the infections were carried out at 37°C. After 30 min of infection, cells were washed and reincubated in fresh medium containing gentamicin in order to kill extracellular bacteria. After incubation (time given in abscissa) following washing, cells were lysed, and viable bacteria were counted on BHI agar plates. Values and error bars represent the means and standard deviations of the numbers of bacteria per well (three wells per assay and two different assays).
Notably, entry of EGD-e in BMM was more efficient when bacteria were grown at 30°C prior to infection than when they were grown at 37°C (Fig. 5B), confirming the idea that flagella participate in the entry of L. monocytogenes into macrophages. In contrast, in Caco-2 cells, entry of EGD-e was 10-fold higher at 37°C than at 30°C (data not shown), most likely because of the PrfA-dependent upregulation of the internalins and other adhesins (27).
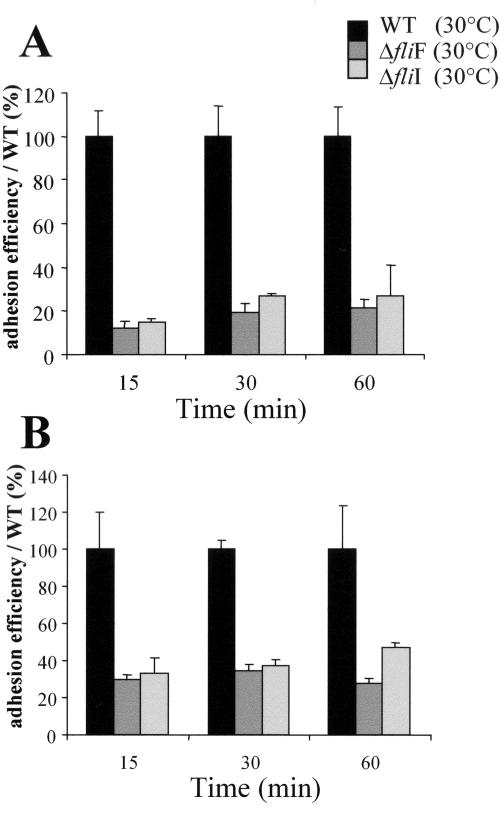
The rates of intracellular multiplication of the ΔfliF and ΔfliI mutants were similar to that of the wild-type strain in each experiment in Caco-2 cells or BMM (Fig. 5), indicating that after internalization, the intracellular multiplication was not affected in the mutants. In order to explain this defect of entry, we performed an adhesion assay between bacteria grown at 30°C and Caco-2 cells. After 15, 30, or 60 min of infection, the adhesion efficiency of both mutants was about 10 to 25% that of the wild-type strain (Fig. 6A). This adhesion defect was confirmed on another nonphagocytic cell line: the human hepatocyte cell line Hep-G2 (Fig. 6B), where the adhesion efficiency of the two mutants was 30% to 40% inferior to that of the wild-type strain. Mutant and wild-type strains grown at 37°C prior to infection showed similar adhesion and entry efficiencies in all the cell lines tested (data not shown).
FIG. 6.
Adhesion assay on Caco-2 (A) or Hep-G2 (B) cell lines. Cell monolayers were infected with 100 bacteria (grown at 30°C) per cell. Adhesion efficiency is defined as the percentage of adherent bacteria (enumerated after 15, 30, or 60 min of incubation and three washes) compared to the initial inoculum. All adhesion efficiency rates are compared to that of the wild-type strain, which is set to 100%.
Altogether, these data demonstrate that deletion of either fliF or fliI abolishes flagella production and causes a defect of bacterial adhesion to nonphagocytic cells. Expression of the flagella also favors the phagocytosis of L. monocytogenes by bone marrow macrophages. However, intracellular multiplication of L. monocytogenes is apparently not affected by the absence of flagella in any of the cell types tested.
Virulence of the ΔfliF and ΔfliI mutant strains in the mouse model.
We then evaluated the possible impact of the deletions on the in vivo survival of L. monocytogenes in the mouse model of infection. Therefore, we first determined the LD50 of the ΔfliF and ΔfliI mutant strains, after i.v. inoculation of Swiss mice. The LD50 of the two strains was estimated at 105.1 (compared to 104.6 for the wild-type strain, EGD-e), reflecting a very modest attenuation of virulence by this route of infection. We also addressed the role of the flagella in promoting initial bacterial colonization of the intestinal tract and crossing of the intestinal barrier by evaluating the virulence of the ΔfliF mutant after oral infection. Female BALB/c mice were infected after intragastric inoculation with 1.5 × 1010 bacteria per mouse. Bacterial counts were enumerated in intestinal tissue, mesentery, and spleen 1 day and 3 days after oral infection (Table 2). For both strains, a significant increase in bacterial counts was recorded in the mesentery between day 1 and day 3 (>100-fold), reflecting active bacterial multiplication in this tissue. Conversely, the counts in the intestinal tissue slightly decreased from day 1 to day 3, as a result of bacterial translocation. However, the counts recorded with the ΔfliF mutant strain (in the intestinal tissue or mesentery, 1 day and 3 days after infection) were not significantly different from those recorded with the wild-type EGD-e. In agreement with earlier observations, no bacteria were found in the spleen 1 day after infection, but counts reached 104.7 and 105.6 per spleen at day 3 for EGD-e and ΔfliF, respectively. Although not statistically significant, the higher average number of bacteria recorded in the spleens of animals infected with the ΔfliF mutant would even suggest a slight increase of virulence.
TABLE 2.
Bacterial dissemination in organs after oral inoculation of BALB/c mice
| Organ | No. of viable bacteriaa
|
|
|---|---|---|
| 1 day p.i. | 3 days p.i. | |
| Intestinal tissue | ||
| WT | 4.55 ± 0.72 | 4.29 ± 0.15 |
| ΔfliF | 4.68 ± 0.61 | 4.45 ± 0.35 |
| Mesentery | ||
| WT | 3.05 ± 0.47 | 5.71 ± 0.19 |
| ΔfliF | 2.92 ± 0.18 | 5.60 ± 0.54 |
| Spleen | ||
| WT | ND | 4.76 ± 0.49 |
| ΔfliF | ND | 5.60 ± 0.54 |
Values given are means ± standard deviations of decimal logarithm of the number of viable bacteria per organ (10 mice per assay). p.i., postinfection; ND, not done.
Altogether, these results show that the ΔfliF mutant can normally cross the intestinal barrier and efficiently multiply in the spleen of infected mice.
Study of clinical isolates for motility and adhesion properties.
A total of 100 clinical isolates, from human sporadic cases of listeriosis obtained in France in a single year (1995), were tested for motility at 30°C and 37°C on semisolid agar plates. The panel, chosen among 300 isolates, comprised three major forms of infection: (i) materno-neonatal forms of listeriosis (20%), (ii) septicemiae (50%), and (iii) meningitis or meningo-encephalitis (21%). These bacteria were collected from blood samples (67/100), cerebrospinal fluid (14/100), placenta (6/100), and ascitic fluids (6/100), and seven samples were of unknown origin.
At 30°C, 10 strains were less motile than wild-type EGD-e after 24 h. Notably, only one of them was totally nonmotile after 48 h. However, when the initial isolate (CLIP67887, serotype 1/2b, collected from a blood sample) was rechecked, it appeared to be positive in agglutination assays with antiflagellar antibodies and totally motile. Thus, the nonmotile mutant strain is likely to be a spontaneous derivative of CLIP67887. We further studied this totally nonmotile strain by (i) monitoring flaA transcription and FlaA production and (ii) evaluating its ability to adhere to and multiply within Caco-2 cells. In all the assays, strain CLIP67887 behaved like the ΔfliF and ΔfliI mutants of EGD-e: (i) the flaA gene was transcribed but the FlaA protein was not detected; (ii) strain CLIP67887 showed a significant defect of adhesion to Caco-2 cells (ca. 10-fold lower than that of EGD-e). The kinetics of invasion and intracellular multiplication of strain CLIP67887, grown at 30°C, confirmed the results of the adhesion assay. A significant decrease in bacteria number (ca. 10-fold) was recorded at time zero of bacterial entry into in Caco-2 cells. After 1 h of intracellular survival, CLIP67887 and EGD-e both started to grow at the same rate, confirming that the role of the flagella is limited to the entry process. Previous studies have shown that all the clinical isolates tested here expressed listeriolysin O, InlB, InlA, and ActA, the main virulence actors of L. monocytogenes (19, 20). Thus, the adhesion defect of CLIP67887 is most likely due to the absence of flagellation.
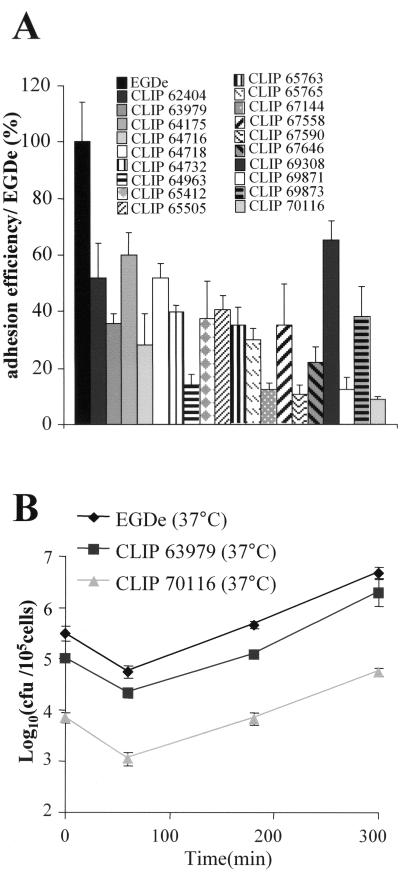
As mentioned earlier, at 37°C, the flaA gene is repressed and EGD-e is nonmotile. We found that 20% of the clinical isolates were still able to swim in semisolid agar at 37°C. This value is similar to that reported very recently by Way et al. based on 25 clinical isolates (44). We monitored adhesion to Caco-2 cells and intracellular multiplication of these strains grown at 37°C. All the strains appeared to adhere less efficiently (between 2 and 10 times less) than EGD-e to Caco-2 cells (Fig. 7). However, intracellular survival was apparently not affected in any of the isolates. These data suggest that expression of the flagella at 37°C might hinder optimal adhesion to epithelial cells but has no impact on intracytosolic multiplication.
FIG. 7.
Analysis of the clinical isolates motile at 37°C. (A) Adhesion efficiency to Caco-2 cells of the 19 clinical isolates that were motile at 37°C. The laboratory EGD-e strain is set to 100%. All strains were grown at 37°C before infection. (B) Caco-2 invasion kinetics of two isolates (CLIP63979 and CLIP70116) compared to strain EGD-e.
DISCUSSION
L. monocytogenes produces and assembles flagella when grown between 4°C and 30°C, whereas flagella production is markedly reduced at 37°C. We have identified two genes of the flagellar apparatus of L. monocytogenes: fliF, encoding the basal body protein FliF, and fliI, encoding the cognate ATPase energizing this type III-like secretion apparatus. We show that inactivation of either fliF or fliI abolishes flagella production and bacterial motility. Strikingly, when the bacteria are grown at 30°C, the two mutants show a significant reduction of entry into both nonphagocytic and phagocytic cells, compared to wild-type EGD-e. However, once inside cells, intracellular bacterial survival and multiplication are unaffected.
We have also performed a motility analysis on 100 human clinical isolates of L. monocytogenes. While only 1 strain was totally nonmotile, 20 strains appeared to be still motile at 37°C. Notably, these strains grown at 37°C showed reduced efficiencies of adhesion and entry into Caco-2 cells but had rates of intracytosolic multiplication comparable to that of EGD-e. These data suggest that expression of the flagella, as well as its down-regulation at 37°C, is not critical for the pathogenicity of L. monocytogenes in humans.
FliF and FliI are involved in flagellar assembly.
The requirement of fliF and fliI activity for flagellar filament assembly, flagellum-driven motility, and swimming behavior of L. monocytogenes was demonstrated by constructing chromosomal ΔfliF and ΔfliI mutations. The deletions abolished the ability of L. monocytogenes to swim in semisolid medium at 30°C. A detailed analysis of the morphogenetic pathway of the Salmonella organelle indicates the sequential assembly of the various elements, from the inner components of the basal body to the last addition of the flagellin monomers to form the filament (24). It is likely that the flagellar assembly process of Listeria is also sequential and requires, in the first place, a functional type III export apparatus permitting FlaA export. Transcription of flagellar genes generally occurs in a hierarchical fashion that mirrors assembly of the nascent flagellum, the genes encoding the early flagellar components being transcribed prior to the genes encoding late flagellar components such as flagellin subunits (25). In Salmonella, the fliC gene, encoding H1 flagellin, is transcribed only after the full assembly of the basal body has been completed. This regulation is mediated by the product of flgM. In the absence of the basal body, FlgM acts as an anti-sigma factor, inhibiting the action of sigma 28, the sigma factor responsible for the transcription of fliC and other late genes (18). In B. subtilis, it has been shown that flaA transcription is dependent on the formation of the flagellar basal body but that such a requirement can be bypassed by overexpression of sigD. Moreover, expression of flaA is defective in a ΔfliI mutant background (2). In contrast, in B. thuringiensis, inactivation of the gene flhA, encoding a protein of the flagellar type III export apparatus, provokes the accumulation of intracellular flagellin, indicating that flaA is still expressed (13).
Our data indicate that flagellar gene expression in L. monocytogenes differs from that in B. subtilis and B. thuringiensis, since the flaA gene is expressed but flagellin is not detected in ΔfliF or ΔfliI backgrounds. The lack of FliF or FliI prevents flagellin export, and the nonexported product is likely to be rapidly degraded in the bacterial cytoplasm. Thus, fliF and fliI are essential for the flagellar assembly in L. monocytogenes.
The flagellum is involved in cell adhesion but not essential for virulence in the mouse model.
The two mutants of EGD-e (ΔfliF and ΔfliI) constructed in this work showed a reduced capacity to adhere to epithelial cells as well as to enter into macrophages when bacteria were grown at 30°C. However, subsequent intracellular multiplication was apparently not affected, suggesting that flagella are mainly involved in L. monocytogenes-host cell contacts. The human enterocyte Caco-2 cells require the presence of an intact InlA at the surface of L. monocytogenes for an efficient E-cadherin-mediated adhesion and subsequent internalization. At 37°C, the expression of the pleiotropic regulator PrfA is upregulated (21) and participates in the induction of InlA production. It is likely that at 30°C adhesion of L. monocytogenes to eukaryotic host cells involves other surface structures such as the flagella. In order to rule out a possible impact of the fliF and fliI deletions on the expression of InlA, we verified by Western blotting that InlA was produced at the same level in the mutants and in the wild-type strain (data not shown).
Similar effects of mutations in motility-related genes on cell association have been shown for other bacteria. For example, mutations in the Yersinia enterocolitica flhDC or fliA genes, which are transcriptional regulators of the flagellar regulon required for motility, affect the effective migration of bacteria to the host cells (48). In S. enterica, the flagella assist in colonization of epithelial cells by enabling motility rather than providing an adhesin (8).
We found that expression of the flagella also favored the entry of L. monocytogenes into macrophages when bacteria were grown at 30°C. Interestingly, in bone marrow macrophages, the efficiency of entry of EGD-e grown at 30°C appeared to be better than that of EGD-e grown at 37°C (i.e., nonflagellated). Since bacterial internalization by bone marrow macrophages does not require specific internalins, components of the flagella may act as adhesins and/or mediate the binding to the macrophage surface.
The absence of flagellar apparatus in the ΔfliF and ΔfliI mutants had no significant consequence on the virulence of L. monocytogenes after either i.v. or oral inoculation in the mouse model. A very recent study, published during the course of our work, has shown that nonflagellated mutants of L. monocytogenes deleted in flaA, as well as flagellated but nonmotile mutants deleted in the chemotactic response genes cheY and cheA, had a reduced association with, and invasion of, Caco-2 cells (10). A ΔcheYA double mutant behaved like the wild-type strain during mouse infection, but the ΔflaA mutant was more virulent, suggesting that the presence of flagella might be deleterious to systemic infection (10). In agreement with these data, we also observed a modest increase of bacterial counts in the spleens of mice infected with the ΔfliF mutant compared to EGD-e 3 days after oral infection.
What is the advantage to the organism to be flagellated at 30°C and not at 37°C?
Motility and chemotaxis genes such as motA, cheR, cheA, and cheY have been found to have a temperature-dependent transcription (10, 31). We found here that the expression of fliF and fliI was also downregulated at 37°C. The three induction rates were similar, indicating that the operon encoding fliF and fliI is regulated in the same way as the flaA monocistronic transcriptional unit.
As recalled above, the PrfA regulon is upregulated at 37°C and leads to the increased expression of many genes, including the internalins. In a ΔprfA strain, expression of the motility gene motA is derepressed at 37°C. Therefore, it has been suggested that PrfA would participate in the downregulation of motility genes in L. monocytogenes at 37°C (31). It is possible that repression of the flagella at 37°C would favor optimal expression and binding of the internalins to their cell surface receptors. This hypothesis is supported by the fact that clinical isolates that are motile at 37°C show decreased adherence to Caco-2 cells (Fig. 7). A very recent study has shown that another regulatory protein, MogR, directly bound to the flaA promoter region (16). The authors provide evidence that MogR acts as a repressor of motility gene expression during intracellular infection and is required for full virulence of L. monocytogenes. It has been proposed that this temperature regulation and the presence of a repressor during infection might serve as a mechanism to evade recognition by the innate immune system. Indeed, bacterial flagellins have been shown to serve as pattern recognition molecules for Toll-like receptor 5-mediated signaling, leading to activation of innate immune responses to infection (6, 17). It has been shown that FlaA of L. monocytogenes binds to Toll-like receptor 5 expressed by intestinal epithelial cells, monocytes, and dendritic cells (17).
Flagellar motility helps bacteria to reach the most favorable environments and to successfully compete with other microorganisms. The fact that 99% of the human clinical isolates that we tested here were motile may suggest that flagella participate to some extent in L. monocytogenes infection in humans. Alternatively, this high percentage of flagellated strains may simply reflect a selective advantage in the extracellular environment. Therefore, flagellated strains of L. monocytogenes would be the strains most commonly encountered by humans.
Finally, the flagellar apparatus is involved in the secretion of virulence factor in several bacterial species. For example, the flhA mutant of B. thuringiensis is defective in the secretion of hemolysin BL and phosphatidylcholine-preferring phospholipase C, two enzymes involved in bacteria virulence. At present, a possible role of the flagellar apparatus of L. monocytogenes as a type III-like secretion system for heterologous proteins is purely speculative, and the molecular mechanisms underlying the flagella-dependent entry processes of L. monocytogenes into eukaryotic cells remain to be elucidated. At this stage, we can only conclude that the complete inactivation of the flagellar translocation apparatus provoked by the ΔfliF and ΔfliI mutants has no significant impact on virulence in the mouse model. A possible impact of the mutations on human infections cannot be ruled out.
Acknowledgments
This work was supported by CNRS, INSERM, and University Paris V. C. Raynaud was supported by a fellowship from INSERM.
Editor: V. J. DiRita
REFERENCES
- 1.Arora, S. K., B. W. Ritchings, E. C. Almira, S. Lory, and R. Ramphal. 1998. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect. Immun. 66:1000-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barilla, D., T. Caramori, and A. Galizzi. 1994. Coupling of flagellin gene transcription to flagellar assembly in Bacillus subtilis. J. Bacteriol. 176:4558-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnemain, C., C. Raynaud, H. Reglier-Poupet, I. Dubail, C. Frehel, M. A. Lety, P. Berche, and A. Charbit. 2004. Differential roles of multiple signal peptidases in the virulence of Listeria monocytogenes. Mol. Microbiol. 51:1251-1266. [DOI] [PubMed] [Google Scholar]
- 4.Braun, L., B. Ghebrehiwet, and P. Cossart. 2000. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 3:1458-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty, T., M. Leimeister-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciacci-Woolwine, F., P. F. McDermott, and S. B. Mizel. 1999. Induction of cytokine synthesis by flagella from gram-negative bacteria may be dependent on the activation or differentiation state of human monocytes. Infect. Immun. 67:5176-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae- and flagella-mediated association with an invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023-1031. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dons, L., E. Eriksson, Y. Jin, M. E. Rottenberg, K. Kristensson, C. N. Larsen, J. Bresciani, and J. E. Olsen. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 72:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dussurget, O., J. Pizarro-Cerda, and P. Cossart. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587-610. [DOI] [PubMed] [Google Scholar]
- 12.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser, P., L. Frangeul, A. Buchrieser, A. Amend, et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 15.Grant, C. C., M. E. Konkel, W. J. Cieplak, and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundling, A., L. S. Burrack, H. G. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 101:12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 19.Jacquet, C., M. Doumith, J. I. Gordon, P. M. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 20.Jacquet, C., E. Gouin, D. Jeannel, P. Cossart, and J. Rocourt. 2002. Expression of ActA, Ami, InlB, and listeriolysin O in Listeria monocytogenes of human and food origin. Appl. Environ. Microbiol. 68:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M., Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 22.Kathariou, S., R. Kanenaka, R. D. Allen, A. K. Fok, and C. Mizumoto. 1995. Repression of motility and flagellin production at 37 degrees C is stronger in Listeria monocytogenes than in the nonpathogenic species Listeria innocua. Can. J. Microbiol. 41:572-577. [DOI] [PubMed] [Google Scholar]
- 23.Kubori, T., M. Okumura, N. Kobayashi, D. Nakamura, M. Iwakura, and S. I. Aizawa. 1997. Purification and characterization of the flagellar hook-basal body complex of Bacillus subtilis. Mol. Microbiol. 24:399-410. [DOI] [PubMed] [Google Scholar]
- 24.Kubori, T., N. Shimamoto, S. Yamaguchi, K. Namba, and S. Aizawa. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433-446. [DOI] [PubMed] [Google Scholar]
- 25.Kutsukake, K., S. Iyoda, K. Ohnishi, and T. Iino. 1994. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and anti-sigma factors in Salmonella typhimurium. EMBO J. 13:4568-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lecuit, M., and P. Cossart. 2002. Genetically-modified-animal models for human infections: the Listeria paradigm. Trends Mol. Med. 8:537-542. [DOI] [PubMed] [Google Scholar]
- 27.Leimeister-Wachter, M., E. Domann, and T. Chakraborty. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174:947-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S. L., T. Ezaki, H. Miura, K. Matsui, and E. Yabuuchi. 1988. Intact motility as a Salmonella typhi invasion-related factor. Infect. Immun. 56:1967-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macnab, R. M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131-158. [DOI] [PubMed] [Google Scholar]
- 30.Mengaud, J., M. Lecuit, M. Lebrun, F. Nato, J. C. Mazie, and P. Cossart. 1996. Antibodies to the leucine-rich repeat region of internalin block entry of Listeria monocytogenes into cells expressing E-cadherin. Infect. Immun. 64:5430-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michel, E., J. Mengaud, S. Galsworthy, and P. Cossart. 1998. Characterization of a large motility gene cluster containing the cheR, motAB genes of Listeria monocytogenes and evidence that PrfA downregulates motility genes. FEMS Microbiol. Lett. 169:341-347. [DOI] [PubMed] [Google Scholar]
- 32.O'Toole, R., D. L. Milton, and H. Wolf-Watz. 1996. Chemotactic motility is required for invasion of the host by the fish pathogen Vibrio anguillarum. Mol. Microbiol. 19:625-637. [DOI] [PubMed] [Google Scholar]
- 33.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 34.Pron, B., C. Boumaila, F. Jaubert, P. Berche, G. Milon, F. Geissmann, and J. L. Gaillard. 2001. Dendritic cells are early cellular targets of Listeria monocytogenes after intestinal delivery and are involved in bacterial spread in the host. Cell Microbiol. 3:331-340. [DOI] [PubMed] [Google Scholar]
- 35.Pron, B., C. Boumaila, F. Jaubert, S. Sarnacki, J. P. Monnet, P. Berche, and J. L. Gaillard. 1998. Comprehensive study of the intestinal stage of listeriosis in a rat ligated ileal loop system. Infect. Immun. 66:747-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rabaan, A. A., I. Gryllos, J. M. Tomas, and J. G. Shaw. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to Hep-2 cells. Infect. Immun. 69:4257-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Réglier-Poupet, H., C. Frehel, I. Dubail, J. L. Beretti, P. Berche, A. Charbit, and C. Raynaud. 2003. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J. Biol. Chem. 278:49469-49477. [DOI] [PubMed] [Google Scholar]
- 38.Richardson, K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Seeliger, H. P. R. 1961. Listeriosis. Karger Verlag, Basel, Switzerland.
- 41.Tasteyre, A., M. C. Barc, A. Collignon, H. Boureau, and T. Karjalainen. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. Van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Way, S. S., L. J. Tompson, J. E. Lopes, A. M. Hajjar, T. R. Kollmann, N. E. Freitag, and C. B. Wilson. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol. 6:235-242. [DOI] [PubMed] [Google Scholar]
- 45.Weinstein, D. L., M. Carsiotis, C. R. Lissner, and A. D. O'Brien. 1984. Flagella help Salmonella typhimurium survive within murine macrophages. Infect. Immun. 46:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 47.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2002. Growth mechanism of the bacterial flagellar filament. Res. Microbiol. 153:191-197. [DOI] [PubMed] [Google Scholar]
- 48.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]