Abstract
Previously, we identified bovine CD18 as the receptor for leukotoxin secreted by Mannheimia (Pasteurella) haemolytica. In this study, we constructed bovine-murine CD18 chimeras to locate the leukotoxin binding domain on CD18. Leukotoxin specifically lysed transfectants expressing bovine CD18 fragment encompassing amino acids 1 to 291, indicating that leukotoxin binding domain lies within amino acids 1 to 291 of bovine CD18.
Mannheimia (Pasteurella) haemolytica A1 is the most important bacterial pathogen of bovine pneumonic pasteurellosis, which causes extensive economic losses to the cattle industry in the United States and elsewhere (3). M. haemolytica produces several virulence factors. Of these, the extracellular leukotoxin (Lkt) is widely accepted as the most important virulence factor of this organism (2, 10). Previously, we (1) and others (8, 9, 12) identified β2 integrins as the receptor for Lkt on bovine leukocytes.
β2 integrins are leukocyte-specific integrins which have a common β subunit, CD18. The β subunit CD18 associates with three distinct α subunits, CD11a, CD11b, and CD11c to give rise to three different β2 integrins: CD11a/CD18 (LFA-1), CD11b/CD18 (Mac-1), and CD11c/CD18 (CR4), respectively. Both the α subunit, CD11, and the β subunit, CD18, are type I membrane glycoproteins consisting of a short C-terminal cytoplasmic domain, a transmembrane domain, and an extracellular domain (6).
In a previous study in our laboratory, by rendering Lkt-nonsusceptible murine cells susceptible to Lkt by recombinant expression of bovine CD18, we demonstrated in a biologically relevant system that bovine CD18 is necessary and sufficient to mediate Lkt-induced cytolysis of bovine leukocytes (4). Therefore, the next logical step was to identify the Lkt binding domain on CD18, which formed the objective of this study. Although bovine and murine CD18 molecules have 80% amino acid sequence identity, Lkt does not bind to murine CD18. Alignment of amino acid sequence of bovine and murine CD18 (GenPept accession no. AAA30438 and CAA33077, respectively), by use of ClustalW and Boxshade 3.21 (Swiss Embnet servers, http://www.ch.embnet.org/software/ClustalW.html and http://www.ch.embnet.org/software/BOX_form.html), revealed that N-terminal and C-terminal regions have the most variation. Therefore, we hypothesized that the Lkt binding domain lies in the N- or C-terminal region of bovine CD18. We reasoned that chimeric CD18 made with different domains of bovine and murine CD18 should reveal the location of Lkt binding site. In this study, we used transfectant cell lines expressing bovine-murine chimeric CD18 molecules to map the Lkt binding domain on bovine CD18.
Cell lines, antibodies, and leukotoxin.
P815 is a murine mastocytoma cell line which expresses murine β2 integrin LFA-1 (CD11a/CD18). This cell line is neither lysed nor bound by Lkt. 2B2, a transfectant cell line expressing full-length bovine CD18 on the cell surface, was previously developed in our laboratory by transfecting P815 with cDNA for bovine CD18 (4). The monoclonal antibody (MAb) MM-605, previously developed in our laboratory (5), specifically binds Lkt but does not neutralize it. Fluorescein isothiocyanate (FITC)-conjugated MM-605 was used to detect Lkt binding in flow cytometry experiments. Production and affinity purification of Lkt from M. haemolytica A1 has been described by us previously (5).
Construction of bovine-murine chimeric CD18 cDNA.
Bovine CD18 cDNA cloned in pBluescript SK(−) was obtained from Marcus Kehrli (11). Murine CD18 cDNA (13) cloned in pBluescript SK(−) was acquired from the American Type Culture Collection (catalog no. 63056). The nucleotide (nt) sequences of bovine and murine CD18 (GenBank accession no. M81233 and X14951, respectively) were compared to identify unique restriction sites present in both sequences at identical positions. The restriction sites BsrGI (at nt 875) and BglII (at nt 1583) were identified. Cleavage at these sites divided CD18 cDNA into three fragments containing nucleotides encoding amino acid residues 1 to 291 (fragment A), residues 292 to 527 (fragment B), and residues 528 to 770 (fragment C) (Fig. 1). We used two restriction sites in the pBluescript SK(−) vector, viz., SacI at nt 759 and KpnI at nt 657 for releasing the A and C fragments from the vector. The acquired murine CD18 cDNA cloned in pBluescript SK(−) was in the reverse orientation in the vector compared to that of the bovine CD18 cDNA clone. The murine CD18 cDNA was brought to the same orientation as that of the bovine CD18 cDNA so that fragments A, B, and C originating from restriction digestion could be exchanged with the bovine counterparts. The murine CD18 cDNA clone in pBluescript SK(−) with the same orientation as that of bovine CD18 cDNA was chosen to be spliced for making chimeras.
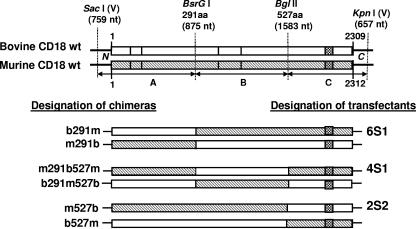
FIG. 1.
Construction of bovine-murine CD18 chimeric cDNA. The wild-type (wt) bovine and murine CD18 cDNAs were cut by using two unique restriction sites present in both sequences (BsrGI [at nt 875], and BglII [at nt 1583]), generating three fragments, A, B, and C. One set of three chimeric CD18 cDNAs was constructed by deleting fragment A, B, or C, from murine CD18 and inserting the corresponding fragment of bovine CD18; these chimeras were designated b291m, m291b527m, and m527b, respectively, following the nomenclature adopted by Huang et al. (7). The designation b291m indicates that residues 1 to 291 are from bovine CD18 and residues from 292 to the C terminus are from murine CD18. The designation m291b527m indicates that residues 1 to 291 are from murine CD18, residues 292 to 527 are from bovine CD18 and residue 528 to the C terminus are from murine CD18. The designation m527b indicates that residues 1 to 527 are from murine CD18 and residues from 528 to the C terminus are from bovine CD18. The chimeras b291m, m291b527m, and m527b were transfected into the murine mastocytoma cell line P815, and the resultant stable transfectant cell lines were designated 6S1, 4S1, and 2S2, respectively. Another set of three chimeras was constructed by a method opposite to the method described above, i.e., by deleting a fragment from bovine CD18 and inserting the corresponding fragment of murine CD18 (m291b, b291m527b, and b527m). Transfectant cell lines expressing this set of chimeras were not developed because of the reasons presented in the text. aa, amino acid.
We constructed six chimeras by exchanging fragments A, B, and C of bovine CD18 cDNA with fragments A, B, and C of murine CD18 cDNA (Fig. 1). All the chimeras were generated by using the common restriction sites found in bovine and murine CD18 cDNA and by exchanging the fragments produced by double digestion with chosen restriction enzymes. First, the fragment A of murine and bovine CD18 was exchanged by cutting with enzymes SacI (vector) and BsrGI (at nt 875). The vector and the insert DNA fragments obtained by restriction digestion were gel purified and ligated after exchanging the counterparts, viz., the vector DNA fragment from bovine was ligated with murine insert and vice versa. The chimeras were designated b291m and m291b, following the nomenclature adopted by Huang et al. (7). The designation b291m indicates that residues 1 to 291 are from the bovine CD18 and residues from 291 to the C terminus are from the murine CD18. The designation m291b indicates that residues 1 to 291 are from the murine CD18 and the residues from 291 to the C terminus are from the bovine CD18. The chimeras generated are shown in Fig. 1. Next, region B of murine and bovine were exchanged by cutting with enzymes BsrGI (at nt 875) and BglII (at nt 1583), and the chimeras m291b527m and b291m527b were generated (designation follows the same nomenclature as that for b291m and m291b). The C regions of murine and bovine were exchanged by replacing the BglII-KpnI fragment, yielding chimeras m527b and b527m. The three chimeric cDNAs in which a fragment of murine cDNA was replaced with the corresponding fragment from bovine cDNA (b291m, m291b527m, and m527b) were selected for development of transfectant cell lines. We chose these three chimeric cDNAs based on the following reasoning. If the insertion of a fragment of bovine CD18 into murine CD18 enables the chimeric CD18 to bind Lkt and mediate Lkt-induced cytolysis, it will indicate that the bovine CD18 segment that was inserted is necessary and sufficient to bind Lkt and mediate Lkt-induced cytolysis. On the other hand, if the deletion of a fragment of bovine CD18 and its replacement with the corresponding murine CD18 fragment disables the chimeric CD18 from binding Lkt and mediating Lkt-induced cytolysis, it will indicate only that the bovine CD18 segment that was deleted is necessary but may not be sufficient to bind Lkt and mediate Lkt-induced cytolysis, since additional amino acid sequences from the rest of the bovine CD18 may also be necessary.
Stable expression of bovine-murine chimeras in P815 cells.
P815 cells were transfected for stable expression of bovine-murine CD18 chimeras by use of Superfect transfection agent (QIAGEN, Valencia, CA) according to the manufacturer's instructions. One stable transfectant cell line per chimera was selected for analysis of Lkt binding and lysis: 6S1 for b291m, 4S1 for m291b527m, and 2S2 for m527b.
Lkt binds to the transfectant cell line expressing bovine CD18 fragment encompassing amino acids 1 to 291.
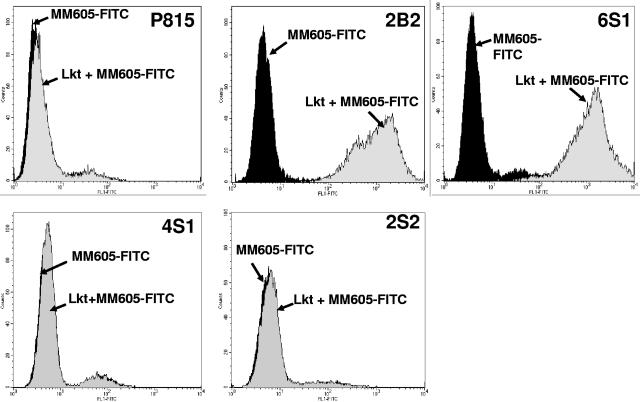
The bovine-murine chimeric transfectant cell lines 6S1, 4S1, 2S2, and the transfectant cell line 2B2 expressing the full-length bovine CD18, and the parent cells, P815, were tested for Lkt binding by flow cytometric analysis, according to procedures previously published by us (1, 4). The cells were incubated with Lkt, followed by FITC-conjugated anti-Lkt MAb MM-605. As expected, the Lkt bound to 2B2 but not to the parent cells (Fig. 2). Of the three transfectant cell lines, Lkt bound to 6S1 but not to 4S1 and 2S2, indicating that the Lkt binding domain lies within amino acids 1 to 291 of bovine CD18.
FIG. 2.
Lkt binds to the transfectant cell line expressing bovine CD18 fragment encompassing amino acids 1 to 291. The bovine-murine chimeric CD18 transfectant cell lines 6S1 (b291m), 4S1 (m291b527m), and 2S2 (m527b) were tested for Lkt binding by flow cytometry. The parent cell line, P815, and the previously developed transfectant cell line, 2B2, expressing the full-length bovine CD18, were used as the negative and positive controls, respectively. The cells were treated with Lkt, and the binding of Lkt to the cells was detected by treating the cells with FITC-conjugated anti-Lkt MAb MM-605. Results of one representative experiment out of three are shown.
Lkt lyses the transfectant cell line expressing bovine CD18 fragment encompassing amino acids 1 to 291.
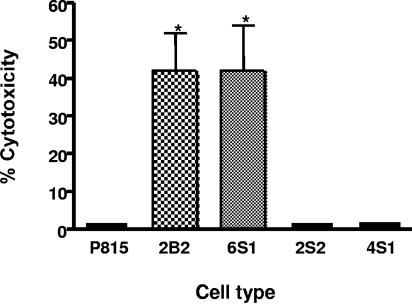
The susceptibility of the transfectant cell lines to Lkt-induced cytolysis was determined by a cytotoxicity assay previously described by us (4, 5). This assay measures the ability of the endoplasmic reticulum-resident enzymes in viable cells to convert a tetrazolium dye {MTT [3-(4,5-dimethylthiazoyl-2-yl)-2,5-diphenyl tetrazolium bromide; Sigma]} into a purple formazan precipitate, which is later dissolved in acid isopropanol. The optical density (OD) of the end product, representing the intensity of the purple color which developed, is directly proportional to the viability of the cells. Briefly, the cells were suspended in colorless RPMI 1640 medium at a concentration of 107/ml and seeded into 96-well round-bottomed microtiter plates in duplicates (50 μl per well). Fifty microliters of Lkt (culture supernatant at a 1:4 dilution) in colorless RPMI 1640 medium was added into each well, and the plates were incubated at 37°C for 1 h. The cells were centrifuged at 600 × g for 5 min following incubation, and the supernatant fluid was discarded. The cells were resuspended in 100 μl of colorless RPMI 1640 medium, and 20 μl of 0.5% MTT was added to each well. Following incubation at 37°C for 2 h, the cells were centrifuged at 600 × g for 5 min and the supernatant fluid was discarded. The remaining formazan precipitate was thoroughly dissolved in 100 μl of acid isopropanol, and the OD of the samples was measured by use of an enzyme-linked immunosorbent assay reader. The percent cytotoxicity was calculated as follows: percent cytotoxicity = [1 − (OD of toxin-treated cells/OD of untreated cells)] × 100. Lkt-induced cytolysis of the transfectant cell lines expressing the bovine-murine chimeric CD18 (6S1, 4S1, and 2S2), the full-length bovine CD18 (2B2) and the parent cells, P815, was tested by this assay. As shown in Fig. 3, the Lkt lysed 2B2 (as expected) and the transfectant cell line 6S1 but not 4S1 and 2S2, confirming the results of the Lkt binding assay, that the Lkt binding domain lies within amino acids 1 to 291 of bovine CD18. Thus, we have shown convincingly that the Lkt binding domain lies in the N-terminal region of bovine CD18 encompassed by amino acids 1 to 291.
FIG. 3.
Lkt lyses the transfectant cell line expressing bovine CD18 fragment encompassing amino acids 1 to 291. The bovine-murine chimeric CD18 transfectant cell lines 6S1 (b291m), 4S1 (m291b527m), and 2S2 (m527b) were subjected to an MTT dye reduction cytotoxicity assay with Lkt (culture supernatant at 1:4 dilution). The parent cell line, P815, and the previously developed transfectant cell line, 2B2, expressing the full-length bovine CD18, were used as the negative and positive controls, respectively. The percent cytotoxicity was calculated as follows: percent cytotoxicity = [1 − (OD of toxin-treated cells/OD of untreated cells)] × 100. Results shown are the means of two independent experiments. The error bars indicate standard deviations of the means. Asterisks indicate that the values for 2B2 and 6S1 are statistically significantly greater than the value for P815, the negative control cell line (P < 0.05). Statistical analysis also revealed that the values for 2B2 and 6S1 are not significantly different from each other.
Statistical analysis.
Statistical analyses were performed by using Prism 4.0 software (GraphPad Software Inc., San Diego, CA). Analyses were performed by one-way analysis of variance and Newman-Keul's posttest for multiple comparisons.
Acknowledgments
This work was supported by National Research Initiative Competitive Grant 2002-35204-12212 from the United States Department of Agriculture.
Editor: A. D. O'Brien
REFERENCES
- 1.Ambagala, T. C., A. P. N. Ambagala, and S. Srikumaran. 1999. The leukotoxin of Pasteurella haemolytica binds to β2 integrins on bovine leukocytes. FEMS Microbiol. Lett. 179:161-167. [DOI] [PubMed] [Google Scholar]
- 2.Berggren, K. A., C. S. Baluyut, R. R. Simonson, W. J. Bemrick, and S. K. Maheswaran. 1981. Cytotoxic effects of Pasteurella haemolytica on bovine neutrophils. Am. J. Vet. Res. 42:1383-1388. [PubMed] [Google Scholar]
- 3.Bowland, S. L., and P. E. Shewen. 2000. Bovine respiratory disease: commercial vaccines currently available in Canada. Can. Vet. J. 41:33-48. [PMC free article] [PubMed] [Google Scholar]
- 4.Deshpande, M. S., T. C. Ambagala, A. P. N. Ambagala, M. E. Kehrli, Jr., and S. Srikumaran. 2002. Bovine CD18 is necessary and sufficient to mediate Mannheimia (Pasteurella) haemolytica leukotoxin-induced cytolysis. Infect. Immun. 70:5058-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentry, M. J., and S. Srikumaran. 1991. Neutralizing monoclonal antibodies to Pasteurella haemolytica leukotoxin affinity-purify the toxin from crude culture supernatants. Microb. Pathog. 10:411-417. [DOI] [PubMed] [Google Scholar]
- 6.Harris, E. S., T. M. McIntyre, S. M. Prescott, and G. A. Zimmerman. 2000. The leukocyte integrins. J. Biol. Chem. 275:23409-23412. [DOI] [PubMed] [Google Scholar]
- 7.Huang, C., Q. Zang, J. Takagi, and T. A. Springer. 2000. Structural and functional studies with antibodies to the integrin β2 subunit. J. Biol. Chem. 275:21514-21524. [DOI] [PubMed] [Google Scholar]
- 8.Jeyaseelan, S., S. L. Hsuan, M. S. Kannan, B. Walcheck, J. F. Wang, M. E. Kehrli, E. T. Lally, G. C. Sieck, and S. K. Maheswaran. 2000. Lymphocyte function-associated antigen 1 is a receptor for Pasteurella haemolytica leukotoxin in bovine leukocytes. Infect. Immun. 68:72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, J., K. D. Clinkenbeard, and J. W. Ritchey. 1999. Bovine CD18 identified as a species specific receptor for Pasteurella haemolytica leukotoxin. Vet. Microbiol. 67:91-97. [DOI] [PubMed] [Google Scholar]
- 10.Shewen, P. E., and B. N. Wilkie. 1982. Cytotoxin of Pasteurella haemolytica acting on bovine leukocytes. Infect. Immun. 35:91-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuster, D. E., B. T. Bosworth, and M. E. Kehrli, Jr. 1992. Sequence of the bovine CD18-encoding cDNA: comparison with the human and murine glycoproteins. Gene 114:267-271. [DOI] [PubMed] [Google Scholar]
- 12.Wang, J. F., I. R. Kieba, J. Korostoff, T. L. Guo, N. Yamaguchi, H. Rozmiarek, P. C. Billings, B. J. Shenker, and E. T. Lally. 1998. Molecular and biochemical mechanisms of Pasteurella haemolytica leukotoxin-induced cell death. Microb. Pathog. 25:317-331. [DOI] [PubMed] [Google Scholar]
- 13.Wilson, R. W., W. E. O'Brien, and L. A. Beaudet. 1989. Nucleotide sequence of the cDNA from the mouse leukocyte adhesion protein CD18. Nucleic Acids Res. 17:5397. [DOI] [PMC free article] [PubMed] [Google Scholar]