Abstract
Pathogenic biovars of Yersinia enterocolitica maintain the well-studied plasmid-encoded Ysc type III secretion (TTS) system, which has a definitive role in virulence. Y. enterocolitica biovar 1B additionally has a distinct chromosomal locus, the Yersinia secretion apparatus pathogenicity island (YSA PI) that encodes the Ysa TTS system. The signals to which the Ysa TTS system responds and its role in virulence remain obscure. This exploratory study was conducted to define environmental cues that promote the expression of Ysa TTS genes and to define how the Ysa TTS system influences bacterium-host interactions. Using a genetic approach, a collection of Y. enterocolitica Ysa TTS mutants was generated by mutagenesis with a transposon carrying promoterless lacZYA. This approach identified genes both within and outside of the YSA PI that contribute to Ysa TTS. Expression of these genes was regulated in response to growth phase, temperature, NaCl, and pH. Additional genetic analysis demonstrated that two regulatory genes encoding components of the YsrR-YsrS (ysrS) and RcsC-YojN-RcsB (rcsB) phosphorelay systems affect the expression of YSA PI genes and each other. The collection of Ysa TTS-defective transposon mutants, along with other strains carrying defined mutations that block Ysa and Ysc TTS, was examined for changes in virulence properties by using the BALB/c mouse model of infection. This analysis revealed that the Ysa TTS system impacts the ability of Y. enterocolitica to colonize gastrointestinal tissues. These results reveal facets of how Y. enterocolitica controls the function of the Ysa TTS system and uncovers a role for the Ysa TTS during the gastrointestinal phase of infection.
Three species of pathogenic Yersinia infect humans: Y. pestis causes bubonic and pneumonic plague, while Y. enterocolitica and Y. pseudotuberculosis cause a diverse range of gastrointestinal system-associated syndromes (3, 4, 8). Most infections of humans by members of this bacterial genus are due to Y. enterocolitica. Infection by this species of bacterium is often a self-limiting disease in healthy human adults, restricted to bacterial colonization of gastrointestinal tissues. In individuals that have a limited capacity to mount a strong immune response, however, the disease is more invasive, with bacteria spreading systemically to vital organs and a mortality rate that approaches 50% (8). Clinical isolates of Y. enterocolitica are categorized based on biochemical and physiological testing which separates them into six groups designated biovars 1A, 1B, and 2 to 5. This method of classification is usually augmented by serological analysis to detect the presence of specific O antigens. Pathogenic strains belonging to each of the six biovars of Y. enterocolitica maintain a common set of factors that are important for virulence, but there is growing evidence that strains from selected biovars, such as biovar 1B, have additional factors that influence pathogenic outcomes (5, 13).
One of the common sets of virulence factors is the plasmid-borne Ysc type III secretion (TTS) system encoded by genes of plasmid pYV, which delivers Yop effector proteins into targeted host cells (23). In the mouse model of infection, the Ysc TTS system plays an important role in systemic phases of disease and is essential for Y. enterocolitica to cause mortality. The activities of several Yop effectors have been investigated, and collectively they are known to modulate host cell physiology by affecting the activities of GTPases, altering the phosphorylation states of various proteins and changing host protein stability. This, in turn, affects the host innate and adaptive immune response, thereby influencing the progression of an infection. Other well-conserved virulence factors include invasin and urease. Invasin is an outer membrane protein that promotes bacterial attachment to and invasion of mammalian cells (20). This factor is not essential for systemic infection, but it is necessary for the efficient invasion of the gastrointestinal tract by promoting bacterial uptake by M cells, an event that results in efficient colonization of the Peyer's patches during the earliest phases of infection (31). Urease contributes to virulence by protecting bacteria from the lethal assault of rapid acidification, a situation that Y. enterocolitica endures as it passes though the host gastric regions (16, 40).
Examples of biovar-restricted virulence factors have been described for members of Y. enterocolitica biovar 1B, and genes encoding these factors cluster in two distinct regions of the chromosome. One region is called the high-pathogenicity island (PI), which contains genes that encode components for the synthesis and uptake of the siderophore yersiniabactin (23). A number of studies have established that mutations in the high-pathogenicity island reduce the ability of Y. enterocolitica biovar 1B strains to cause a systemic and lethal infection in experimentally infected mice (23). More recently, another chromosomal locus called the Yersinia secretion apparatus (YSA) that has features of a PI was described (13, 17). The YSA PI contains a set of genes that encode a second contact-dependent TTS system that functions independently of the well-characterized Ysc TTS system (17, 39). The Ysa TTS system is required for the export of a set of proteins called Yersinia secreted proteins (Ysps), but there is a limited amount of information concerning the role of this TTS system in Y. enterocolitica pathogenesis. Secretion of Ysps is promoted in the laboratory by growth of bacterial cultures at 26°C in a rich medium containing relatively high concentrations of NaCl (17, 39). The factors that lead to function of the Ysa TTS system under these conditions are not fully understood. In part, these conditions appear to stimulate the transcription of genes within the YSA PI. Recent studies have established that transcription of at least some of these genes depends on the transcriptional regulators CRP, YsrR, and YsaE (32, 37). However, there has not been a systematic survey to define environmental conditions that influence the expression of these genes.
The Ysa TTS system exports at least 11 Ysp proteins; the primary amino acid sequence has been deduced for 7 of these proteins (14, 38, 39). Four of these proteins are designated YspA, -B, -C, and -D (14). No function has been ascribed to YspA. Based on homology to components of TTS systems from other bacteria, it is predicted that YspB, -C, and -D are important for the translocation of Ysp effector proteins into host cells, but this proposed function remains to be established (14). The three other characterized Ysp proteins were recently identified as YopE, YopN, and YopP (38). Each of these Yop proteins is also exported by the Ysc TTS system, indicating that there is some overlap in the group of proteins targeted by the Ysa and Ysc TTS systems. Several studies of YopP from Y. enterocolitica and of the orthologous protein YopJ from Y. pseudotuberculosis and Y. pestis established that translocation of this effector by the Ysc TTS system influences pathogenic outcomes by altering NF-κβ-mediated expression of tumor necrosis factor alpha (TNF-α) and other cytokines by targeted host cells (23). Likewise, a recent study demonstrated that the Ysa TTS system delivered YopP into cultured macrophages, which suppressed the production of the cytokine TNF-α (38). The contribution of the other proteins secreted by the Ysa TTS system to host-pathogen interactions has not been investigated in detail. However, the lethality of a Ysa TTS system mutant has been tested and was reportedly reduced 10-fold in BALB/c mice when it was administered by the oral route (17).
Here we report a broadly based exploratory study intended to provide a foundation for examining the specific role of the Ysa TTS system in the virulence of Y. enterocolitica biovar 1B. In the first part of the study, we generated a collection of mutants carrying transcriptional fusions between genes required for Ysa TTS and the reporter operon lacZYA. These reporter strains were used to systematically evaluate different laboratory conditions for effects on gene expression to define environmental cues that are important for promoting the Ysa TTS system function. In the second part of the study, we used these Ysa TTS mutants, along with other defined mutants, to examine how the Ysa TTS system influences host-pathogen interactions in susceptible BALB/c mice.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Unless otherwise noted, the E. coli strains were routinely grown at 37°C, and the Y. enterocolitica strains were grown at 26°C in Luria broth (LB medium; 1% tryptone, 0.5% yeast extract, 90 mM NaCl) or Luria agar (Difco). The medium used for the examination of protein secretion by Y. enterocolitica was Luria broth base (L medium; 1% tryptone, 0.5% yeast extract) adjusted to contain NaCl at the final concentrations indicated in the text and as described previously (39). When necessary, depletion of calcium from the medium was accomplished by addition of 20 mM sodium oxalate and 20 mM MgCl2. Dipryridyl at a concentration of 100 μm was added to deplete the media of iron (Fe2+). Where indicated below, the following buffers were included to obtain buffered media: 100 mM citrate, pH 5.0; 100 mM MES (morpholineethanesulfonic acid), pH 6.0; 100 mM MES, pH 7.0; and 100 mM MOPS (morpholinepropanesulfonic acid), pH 8.0. Phospholipase indicator agar (PLA) consisted of MacConkey agar base (Difco) supplement with 1% (vol/vol) Tween 80 and 1 mM CaCl2 (39, 41). MacConkey lactose agar (Difco) was used to phenotypically screen strains for the ability to transport and metabolize lactose (Lac+). Antibiotics were used at the following concentrations: chloramphenicol (25 μg/ml for E. coli; 12 μg/ml for Y. enterocolitica), kanamycin (50 μg/ml), nalidixic acid (20 μg/ml), and tetracycline (15 μg/ml for E. coli; 7 μg/ml for Y. enterocolitica).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| Strains | ||
| Y. enterocolitica | ||
| JB580v | Serogroup O:8, Nalr ΔyenR (R− M+) | 21 |
| GY1087 | yscR::mTn5-Km2 ysaV::pEP185.2 | 39 |
| GY4428 | ysaV::pEP185.2 | This study |
| GY4478 | pYV8081− | 39 |
| GY4531 | crp::str | 32 |
| GY4572 | ysaU::TnMod-lacZYA-RKm′ | This study |
| GY4573 | orf6 orf7::TnMod-lacZYA-RKm′ | This study |
| GY4574 | ddhB::TnMod-lacZYA-RKm′ | This study |
| GY4613 | rcsB::TnMod-lacZYA-RKm′ | This study |
| GY4639 | ysaR::TnMod-lacZYA-RKm′ | This study |
| GY4641 | ysaE::TnMod-lacZYA-RKm′ | This study |
| GY4664 | ysaU::TnMod-lacZYA-RKm′ | This study |
| GY4665 | ysaC::TnMod-lacZYA-RKm′ | This study |
| GY4668 | orf6 orf7::TnMod-lacZYA-RKm′ | This study |
| GY4675 | ysaU::TnMod-lacZYA-RKm′ ysrS::pEP185.2 | This study |
| GY4676 | ysaU::TnMod-lacZYA-RKm′ rcsB::pEP185.2 | This study |
| GY4677 | ysaR::TnMod-lacZYA-RKm′ ysrS::pEP185.2 | This study |
| GY4678 | ysaR::TnMod-lacZYA-RKm′ rcsB::pEP185.2 | This study |
| GY4684 | pYV8081−ysaV::pEP185.2 | This study |
| GY4737 | rcsB::pEP185.2 | This study |
| GY4742 | ysrS::mTn5-Km2-lacZYA | This study |
| GY4887 | ysrS::pEP185.2 | This study |
| GY4888 | ysaE::TnMod-lacZYA-RKm′ rcsB::pEP185.2 | This study |
| GY4891 | orf6 orf7::TnMod-lacZYA-RKm′ rcsB::pEP185.2 | This study |
| GY4892 | ysaE::TnMod-lacZYA-RKm′ ysrS::pEP185.2 | This study |
| GY4893 | rcsB::TnMod-lacZYA-RKm′ ysrS::pEP185.2 | This study |
| GY4894 | ysrS::pFUSE (ysrS-lacZYA) | This study |
| GY4899 | orf6 orf7::TnMod-lacZYA-RKm′ ysrS::pEP185.2 | This study |
| GY5019 | sycB::mTn5-Km2-lacZYA | This study |
| GY5022 | sycB::mTn5-Km2-lacZYA ysrS::pEP185.2 | This study |
| GY5021 | sycB::mTn5-Km2-lacZYA rcsB::pEP185.2 | This study |
| GY5124 | rcsB::aacC1 Gmr | This study |
| GY5196 | rcsB::aacC1 ysrS::pFUSE (ysrS-lacZYA) | This study |
| YVM356 | yscR::mTn5-Km2 | 9 |
| E. coli S17-1 λpir | recA thi pro hsdR−hsdM+ RP4::2-Tc::Mu::Km Tn7 λpir | 27 |
| Plasmids | ||
| p34s-Gm | Gmr plasmid with antibiotic resistance cassette, accC1 | 11 |
| pCR-Blunt II-TOPO | Kmr | Invitrogen |
| pEP185.2 | Cmr, suicide vector | 21 |
| pFUSE | Cmr, suicide vector for transcriptional fusions to lacZYA | 1 |
| pTM100 | mob+oriR6K Tetr Cmr | 24 |
| pTnMod-lacZYA-RKm′ | TnMod-lacZYA-RKm′ in a delivery vector, Kmr | Andrew Darwin, NYU |
| pREV10 | mTn5-Km2-lacZYA′ in a delivery vector, Kmr | Virginia Miller, Washington University |
| pGY100 | Pcat-yplAB derivative of pTM100 | 39 |
| pGY348 | Ca. 0.9-kb fragment of ysaV in pCR-Blunt II-TOPO | This study |
| pGY352 | Ca. 0.9-kb fragment of ysaV in pEP185.2 | This study |
| pGY431 | Ca. 0.5-kb fragment of rcsB in pCR-Blunt II-TOPO | This study |
| pGY435 | Ca. 0.5-kb fragment of rcsB in pEP185.2 | This study |
| pGY454 | ysrRS in pCR-Blunt II-TOPO | This study |
| pGY482 | ysrRS in pTM100 | This study |
| pGY512 | Ca. 0.5-kb fragment of ysrS in pCR-Blunt II-TOPO | This study |
| pGY526 | Ca. 0.5-kb fragment of ysrS in pEP185.2 | This study |
| pGY529 | Ca. 1-kb fragment of ysrS in pCR-Blunt II-TOPO | This study |
| pGY530 | Ca. 1-kb fragment of ysrS in pFUSE | This study |
| pGY551 | rcsB in pCR-Blunt II-TOPO | This study |
| pGY552 | ysrRS in pTM100 with cat removed | This study |
| pGY553 | rcsB in pTM100 | This study |
| pGY556 | pTM100 with cat removed | This study |
| pGY557 | rcsB::accC1 in pCR-Blunt II-TOPO | This study |
| pGY561 | rcsB::accC1 in pEP185.2 | This study |
Preparation of extracellular proteins and SDS-PAGE analysis.
Extracellular proteins were prepared as described previously (38, 39) with minor modifications. Y. enterocolitica was grown overnight in Luria broth and subcultured to an optical density at 600 nm (OD600) of 0.1 in 5 ml of an appropriate medium. To induce Ysp secretion by the Ysa TTS system, cultures were grown for 6 h at 26°C in L medium supplemented with 290 mM NaCl. To induce Yop secretion by the Ysc TTS system, cultures were grown for 6 h at 37°C in Ca2+-depleted L medium. Following the incubation period, the OD600 of each culture was determined and then bacterial cells were removed by centrifugation. Proteins from culture supernatants were concentrated by precipitation with 10% (wt/vol) ice-cold trichloroacetic acid and washed with ice-cold acetone. Each of the protein samples was resuspended in sample buffer containing 2-mercaptoethanol, and sample volumes were adjusted according to the OD600s of the cultures. Samples were heated to 95°C for 5 min and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 12.5% polyacrylamide. Proteins were visualized by staining with silver (2).
Transposon mutagenesis.
Y. enterocolitica pGY100 was mutagenized by conjugation of the plasmid pTnMod-lacZYA-RKm′ (a gift of Andrew Darwin, New York University) or pREV10 (a gift of Virginia Miller, Washington University) by using conditions previously described (32, 39). Twenty-five separate matings were collected and plated on L agar containing nalidixic acid, kanamycin, and tetracycline. The plates were incubated for 48 h at 26°C to recover a collection of transposon insertion mutants. Approximately 20,000 of these mutants were patched onto PLA medium containing 290 mM NaCl and incubated for 48 h at 26°C. Strains that exhibited a phospholipase-negative phenotype were then patched onto MacConkey lactose agar to identify the subset of strains that exhibited a Lac+ phenotype.
Characterization of transposon insertions sites and DNA sequencing.
Chromosomal DNA was isolated from mutants that contained TnMod-lacZYA-RKm′ insertions and digested with EcoRI as described previously (32, 39). The digested DNA was ligated overnight, and replicating plasmids were recovered by electroporation of E. coli S17-1λpir+, followed by selection for kanamycin resistance. Plasmids were isolated and analyzed by restriction digestion to confirm the integrity of the transposon sequences. The sequence of the chromosomal DNA immediately adjacent to the transposon was determined by using primers that anneal near the ends of TnMod-lacZYA-RKm′ (primer KM1, 5′-CCCCGAGCTCTTAATTAA-3′ and primer KM2, 5′-GAACACTTAACGGCTGAC-3′). DNA sequence was obtained by using an Applied Biosystems DNA sequencing system and the BigDye Terminator cycle sequencer kit (Biosystems) according to the manufacturer's instructions.
Strain and plasmid construction. Construction of the strains containing ysrS.
The pFUSE allele was generated by introduction of the suicide plasmid pGY530 (1). Strains carrying a cointegrated copy of pGY530 have a functional copy of ysrS transcriptionally fused to the lacZYA operon. A ca.-1-kb region at the 3′ end of ysrS was amplified by PCR with Pfu polymerase (Stratagene) using primers YSRSLACZ-1, 5′-TCAGTCATGTTCTTTTTCTTAG-3′, and YSRSLACZ-2, 5′-CAGAACCTTTTTATCAGGGCAA-3′. The template DNA for the reaction was purified genomic DNA originating from Y. enterocolitica JB580v. The resulting PCR product was initially cloned into pCR-Blunt II-TOPO according to the manufacturer's instructions (Invitrogen) to create pGY529 and then was subcloned as a SpeI-EcoRV fragment into SmaI-XbaI of pFUSE to create pGY530.
Some mutants carry mutations due to insertion of a suicide plasmid targeted to a specific gene by homologous recombination (30). Construction of strains containing the rcsB::pEP185.2 allele was done by introduction of pGY435 into Y. enterocolitica strain JB580v by conjugation (30). A ca.-0.5-kb internal fragment of rcsB was amplified by PCR with Taq polymerase (QIAGEN) using primers RCSB-1, 5′-GGATCCATTACTGACCTCTCCATG-3′, and RCSB-2, 5′GATCCACCTTCAGCAATAACCG −3′. The resulting PCR product was cloned into pCR-Blunt II-TOPO, creating pGY431. Subsequently, a 0.5-kb EcoRV-KpnI fragment was cloned into complementary sites of the suicide vector pEP185.2, creating pGY435. Construction of the strains containing the ysrS::pEP185.2 allele and ysaV::pEP185.2 allele were done by similar techniques using primers YSRSRNA-1, 5′-GTGCCGGTGTCTATCACCTCAA-3′, and YSRSRNA-2, 5′-TCTATCGCTACCGGCGATTAAA-3′, for ysrS and YSAV-1, 5′-GCTCTAGAGCGTTAGCGTTATCGATCAG-3′, and YSAV-2, 5′-GGATATCCTTGTCGCTTGGTACCAC-3′, for ysaV. The PCR products were cloned into pCR-Blunt II-TOPO to create pGY512 and pGY348, respectively, which were then subcloned as EcoRV-KpnI fragments into pEP185.2 to create pGY526 and pGY352, respectively.
Construction of the strains containing a gentamicin resistance cassette within rcsB was completed by allelic-exchange mutagenesis (30). The gentamicin cassette consisting of aacC1 from p34s-Gm was subcloned as a ScaI fragment into the ScaI site of rcsB residing on pGY551 to create pGY557. The rcsB::aacC1 allele was subcloned as an XhoI-SpeI fragment into the XhoI-XbaI site of pEP185.2 to create pGY561. Plasmid pGY561 was introduced into Y. enterocolitica JB580v by conjugation and resolved using cycloserine enrichment. Mutants carrying the rcsB::aacC1 allele were identified as strains that were gentamicin resistant and chloramphenicol sensitive. The location and integrity of the mutation were confirmed using PCR (data not shown).
The generation of plasmid-borne copies of the ysrRS locus and rcsB for genetic complementation analysis was as follows: DNA corresponding to ysrRS was amplified by PCR with Pfu polymerase (Stratagene) using oligonucleotide primers YSRS-1, 5′-GTGCGGCAACCAGATCAC-3′, and YSRS-2, 5′-CATTGTCCTCACAGCTCTTAA-3′. The fragment of DNA generated by PCR was cloned into pCR-Blunt II-TOPO to create pGY454. Subsequently, a 2-kb SpeI-XbaI fragment was subcloned into the XbaI site of pTM100 to create pGY482. This plasmid was then modified by removing a ca.-2-kb XmnI-NcoI fragment to form pGY552. The chromosomal region containing rcsB was amplified by PCR with Pfu polymerase (Stratagene) using oligonucleotide primers RCSBCOMP-1, 5′-CCGCACATCGATTGAAAGGCGT-3′, and RCSBCOMP-2, 5′-CAGCAAGCGAATGAATCCCGAT-3′, and was cloned into plasmid pCR-Blunt II-TOPO (Invitrogen) as previously described to create pGY551. A 1-kb EcoRI fragment was subcloned into the EcoRI site of pTM100 to create pGY553. Plasmids pGY552 and pGY553 were introduced into Y. enterocolitica strains by conjugation.
Measurements of β-galactosidase activity.
Y. enterocolitica cells were grown overnight at 26°C in L medium and subcultured to an OD600 of 0.1 in 5 ml of the appropriate medium as indicated below. Cells were harvested and assayed for β-galactosidase activity as previously described (26).
Cell culture and cytotoxicity assay.
Mouse monocyte/macrophage-like J774A.1 cells (ATCC) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (Cellgro) at 37°C under 5% CO2. The J774A.1 macrophages were plated at a concentration of 5 × 104 cells/well in 24-well tissue culture plates and incubated at 37°C for 17 h before infection. Y. enterocolitica strains were grown overnight at 26°C in L broth and subcultured to an OD600 of 0.2 into L medium supplemented with 290 mM NaCl. Bacterial cells were then incubated at 26°C for 1 h. Prior to infection, the OD600 was again determined and bacteria were added to the macrophages at a multiplicity of infection of 80. Plates were centrifuged for 5 min at 1,000 × g to synchronize the infection. Cells were incubated at 26°C under 5% CO2 for 2 h. After this infection period, gentamicin was added at a final concentration of 100 μg/ml and cells were incubated at 37°C under 5% CO2 for an additional 3 h. Cytotoxicity was determined by release of lactose dehydrogenase (LDH). The amount of LDH release was determined using CytoTox96 as described by the manufacturer (Promega). Calculation of percent cytotoxicity is as follows: percent cytotoxicity = 100 × (experimental release -spontaneous release)/(total release -spontaneous release), where experimental release is that caused by infection with bacteria, spontaneous release is the background level of LDH release by untreated cells, and total release is the amount of LDH present in cells lysed with Triton X-100.
Animal experiments.
The 50% lethal doses (LD50) of selected bacterial strains were determined as previously described (31). Groups of six female, 5- to 7-week-old BALB/c mice (Charles River Laboratories) were infected perorally with 0.2 ml of successive 10-fold dilutions of a bacterial suspension ranging from 105 to 109 CFU/ml. Mice were monitored twice a day for 14 days and allowed to drink and feed ad libitum. Calculation of the LD50 was done by the method of Reed and Muench (33).
Survival of bacteria in host tissues was examined by perorally infecting groups of mice with ∼1 × 107 bacteria. At the times specified in the text, mice were euthanized and the numbers of bacteria present in selected tissues were determined. Tissues were aseptically removed, weighed, and then homogenized in phosphate-buffered saline. Serial dilutions of these tissue suspensions were then plated on LB medium containing nalidixic acid. The number of bacteria present in a given tissue was then normalized and expressed as the recoverable number of CFU per gram of weight of infected tissue.
The competitive indexes (C.I.) of selected strains of Y. enterocolitica were completed by adaptation of previously described protocols (15, 22). Initially, the in vitro C.I. was determined for a 1:1 mixture of JB580v and each mutant strain grown in LB medium (LB). Each strain was cultured overnight in LB medium at 26°C. The next day, a 1:1 mixture of wild-type and mutant bacteria was subcultured to an OD600 0.03 in fresh LB medium and cultured for 8 h at 26°C. To determine the actual ratio of the two strains, dilutions of the inoculum were spread onto LB medium plus nalidixic acid (total number of CFU) and colonies were patched onto LB with nalidixic acid and either kanamycin or chloramphenicol, which was dependent upon the mutant being evaluated (total number of mutant CFU). Alternatively, dilutions were plated directly onto the different selective media. Both methods of bacterial recovery yielded similar results. Dilutions of the culture following the 8-h growth period were similarly analyzed. The competitive index is defined as the output ratio (mutant to wild-type cells) divided by the input ratio (mutant to wild-type cells).
To determine the in vivo C.I., mice were infected with a mixture of bacteria containing approximately equal numbers of the test strain and the wild-type strain JB580v. Infections were initiated by perorally infecting mice with doses of 2 × 108 or 2 × 109 CFU as specified below. To determine the actual ratio of the two strains, dilutions of the inoculum were spread onto LB medium plus nalidixic acid (total number of CFU) and colonies were patched onto LB with nalidixic acid and either kanamycin or chloramphenicol, which was dependent upon the mutant being evaluated (total number of mutant CFU). We also plated dilutions directly onto the different selective media. Both methods of bacterial recovery yielded similar results. Mice were euthanized 24 h after infection, and infected tissues were aseptically recovered and homogenized in phosphate-buffered saline. Dilutions were plated separately onto LB with nalidixic acid (total number of CFU), LB with nalidixic acid, and either kanamycin or chloramphenicol when appropriate (number of mutant CFU). The C.I. is defined as the output ratio (mutant/wild type) divided by the input ratio (mutant/wild type).
All procedures were approved by the Animal Use and Care Administrative Advisory Committee of the University of California, Davis.
Statistical analysis.
Data were analyzed by using Student's two-tailed t test for competition assays. The Mann-Whitney test (a two-tailed nonparametric analysis of variance) was used for analysis of pure culture infection assays. This test assigns a statistically significant P value if P is 0.02 or less. Statistically significant comparisons are indicated where appropriate.
RESULTS
Isolation of Ysa TTS system mutants.
The goal of this set of experiments was to identify genes that play an important role in the expression and function of the Ysa TTS system. In addition, we wished to construct a set of transcriptional fusions between YSA-related genes and the reporter operon lacZYA. We previously established that protein secretion by the Ysa TTS system can be phenotypically assessed by monitoring the export of YplA, a phospholipase that is activated only when it is secreted and an activity that can be detected with PLA supplemented with NaCl (39). This allowed a genetic screen to be developed to isolate mutants defective for secretion of YplA by the Ysa TTS system (39). Y. enterocolitica JB580v carrying pGY100 was subjected to transposon mutagenesis with TnMod-lacZYA-RKm′ or mTn5Km2-lacZYA to identify additional genes required for YSA-dependent TTS and to generate transcriptional reporter strains that could be used to investigate environmental conditions that affect the expression of Ysa TTS-related genes. Using this genetic approach, greater than 20,000 transposon insertion mutants were screened, resulting in the identification of 48 mutants that were phenotypically negative for YplA activity on PLA-NaCl medium. In a secondary screen, these mutants were tested for the ability to utilize lactose on MacConkey lactose agar, with the NaCl concentration adjusted to 290 mM. These conditions were necessary to induce the Ysa TTS system and allowed for the identification of 12 strains that were not only affected in Ysa TTS system-dependent export of YplA, but also contained transcriptional fusions of the inactivated genes to lacZYA.
To identify the location of the transposon insertion carried by each mutant, the regions of the chromosomal DNA immediately adjacent to the transposon insertions were cloned and sequenced. The DNA sequence was then compared to the recently completed genomic sequence of Y. enterocolitica 8081 (www.sanger.ac.uk/Projects/Y_enterocolitica/). DNA sequencing also established in each case that the transposon insertion was positioned such that the lacZYA operon was in the same transcriptional orientation as the inactivated gene. This analysis revealed that 9 of the 11 mutants contained transposon insertions located within the ∼32-kb YSA PI and mapped to the open reading frame (ORF) 6 gene (orf6), sycB, ysaC, ysaE, ysaR, ysaU, and ysrS (Fig. 1). The remaining two mutants each contained a transposon insertion that mapped outside the YSA PI. One of these strains is a mutant which displayed reduced levels of Ysp secretion and had an insertion that mapped to ddhB, a gene that encodes CDP-glucose 4,6-dehydratase and is involved in O-antigen biosynthesis (42). This mutant was not further characterized since the effect of this mutation caused only a moderate effect on the amount of Ysp secretion (Fig. 2, lane 6). However, the other mutant contained a transposon that was inserted within YE1398, an annotated ORF predicted to encode a protein with 91% amino acid identity to the response regulator RcsB of the RcsC-YojN-RcsB phosphorelay system of Salmonella enterica known to affect virulence (12, 28). Two adjacent ORFs, YE1396 and YE1399, are predicted to encode proteins that have high degrees of identity to YojN and RcsC, respectively. Given the high degree of conservation between these Y. enterocolitica and S. enterica genes, we have renamed these annotated ORFs rcsB, rcsC, and yojN.
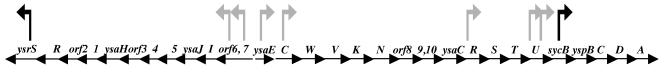
FIG. 1.
YSA pathogenicity island of Y. enterocolitica. Schematic diagram of the YSA PI of Y. enterocolitica (not to scale). Gray flags indicate the locations of TnMod-lacZYA-RKm′ insertions, and black flags indicate the locations of the mTn5-Km2-lacZYA insertion. The orientation of a flag corresponds to the direction of lacZYA transcription. The organization of the ysa locus is based on the published DNA sequence (GenBank accession no. AF005744) and edited based on the recently released genomic sequence of Y. enterocolitica strain 8081 (Sanger Center [www.sanger.ac.uk/Projects/Y_enterocolitica]).
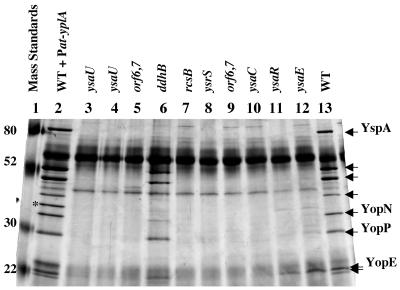
FIG. 2.
Ysp secretion defects of various Ysa TTS mutants. Y. enterocolitica strains were grown under Ysa TTS-inducing conditions. Proteins secreted into culture supernatants were collected, separated by 12.5% SDS-PAGE, and visualized by staining with silver. Lanes: 1, molecular mass standards (Mass Standards); 2, JB580v/pGY100 (wild type [WT] + Pcat-yplA); 3, GY4664 (ysaU); 4, GY4572 (ysaU); 5, GY4573 (orf6 orf7); 6, GY4574 (ddhB); 7, GY4613 (rcsB); 8, GY4742 (ysrS); 9, GY4668 (orf6 orf7); 10, GY4665 (ysaC); 11, GY4639 (ysaR); 12, GY4641 (ysaE); 13, JB580v (WT). Each lane contains protein harvested from culture supernatants equivalent to 1 ml of culture at an OD600 of 3.0. The locations of molecular mass standards are indicated in kilodaltons on the left side of the panel. Arrows on the right side of the panel mark the locations of various Ysps exported by the Ysa TTS system. The asterisk marks the location of YplA secreted by JB580v/pGY100.
Characterization Ysa TTS system mutants.
To establish that the candidate Ysa TTS system mutants were affected in their export of Ysp proteins, culture supernatants were collected from mutants grown under conditions that promote protein secretion. Eleven of the mutants exhibited a defect in the secretion of Ysp proteins, and one mutant secreted reduced levels of Ysp proteins (Fig. 2 and data not shown). Each of the mutants was also examined for effects on the secretion of Yops and Fops (flagellar outer proteins). These two groups of proteins, secreted by other TTS systems under different growth conditions (38, 39), were released by the mutants at levels equivalent to the amounts released by wild-type Y. enterocolitica (data not shown). These results suggested that the function of the Ysa TTS system in the collection of mutants isolated in this study was specifically affected.
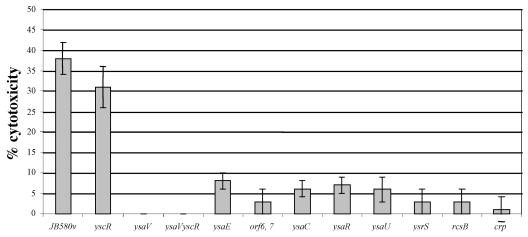
Each mutant was then examined for its ability to induce cytotoxicity when infecting J774A.1 macrophages. It had previously been shown that the Ysa TTS system contributes to the ability of Y. enterocolitica to down-regulate TNF-α release by J774A.1 macrophages (38). This phenotype is due to targeting of the effector YopP. Using the collection of mutants isolated in this study, we wanted to determine if the Ysa TTS system also was capable of having a cytotoxic effect on J774A.1 cells since this is another activity associated with YopP. To examine this possibility, we first tested whether Y. enterocolitica JB580v induces cytotoxicity in J774A.1 macrophages. Cytotoxicity was evaluated by measuring the release of the cytosolic enzyme LDH. The results indicate that Y. enterocolitica JB580v did cause the release of significant levels of LDH (Fig. 3). In addition, J774A.1 cells infected with Y. enterocolitica displayed altered morphologies characterized by rounding, followed by release from the surface of the culture flask (data not shown), and displayed a reduced ability to exclude trypan blue, which is indicative of a loss of cytosolic membrane integrity (data not shown). Under these experimental conditions, the cytotoxic effect was not due to functions related to the plasmid-encoded Ysc TTS system since a yscR mutant and wild-type Y. enterocolitica caused similar levels of LDH release (Fig. 3). In contrast, cytotoxicity did require the Ysa TTS system because there was a sharp reduction in cytotoxicity when J774A.1 cells were infected with strains GY4668, GY4667, GY4639, GY4664, GY4665, GY4613, and GY4531 (Fig. 3). We also tested strain GY4428, which carries an insertion mutation that inactivates ysaV, to eliminate the possibility that the reduction in cytotoxicity might be due to the presence of lacZYA in the other mutant strains. This strain also displayed a reduced capacity to induce a cytotoxic outcome from the infection of J774A.1 cells (Fig. 3).
FIG. 3.
Cytotoxicity of J774A.1 macrophages caused by selected Y. enterocolitica mutants under conditions that promote the function of the Ysa TTS system. Mutations in genes involved in Ysa TTS affect the ability of Y. enterocolitica to induce cytotoxicity in J774A.1 macrophages. Following the initiation of infections, macrophages were incubated at 26°C under 5% CO2 for 2 h. After this infection period, gentamicin was added at a final concentration of 100 μg/ml and cells were incubated at 37°C under 5% CO2 for an additional 3 h. Cytotoxicity was determined by release of LDH. Strains of Y. enterocolitica (described in Table 1) are indicated from left to right: JB580v (wild type), YVM356 (yscR), GY4428 (ysaV), GY1087 (yscR, ysaV), GY4641 (ysaE), GY4668 (orf6 orf7), GY4665 (ysaC), GY4639 (ysaR), GY4664 (ysaU), GY4742 (ysrS), GY4613 (rcsB), and GY4531 (crp). A detailed protocol and description of calculations are presented in Materials and Methods. The results are displayed as means ± standard deviations of results of four individual assays preformed in triplicate.
Ysa TTS system-related genes are induced early in the logarithmic phase of growth.
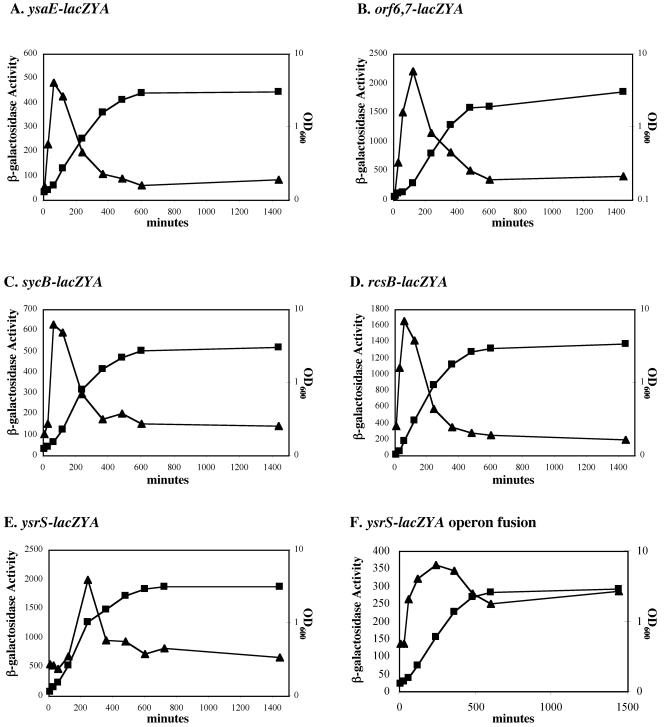
Previous studies have shown that Ysp secretion by the Ysa TTS system and transcription of some genes within the YSA locus are promoted by growth at 26°C in a nutrient-rich medium that contains a concentration of NaCl equal to, or greater than, 290 mM (17, 32, 39). However, a systematic survey designed to better define these and other environmental conditions that influence the expression of these genes has not been reported. Likewise, it is assumed that the lack of Ysp secretion at 37°C was due to transcriptional repression of Ysa TTS system-related genes, but this assumption has not been formally tested. To investigate these topics, we took advantage of the fact that each of the strains described above harbored a transcriptional fusion between a Ysa TTS-related gene and lacZYA. Individual strains were cultured under a variety of conditions at 26°C and 37°C. Levels of transcription of each gene were then inferred by measuring β-galactosidase activity. Initially, bacteria were grown in a rich medium (L medium) containing NaCl adjusted to a concentration of 290 mM. At 37°C, none of the genes examined were induced, indicating that under these laboratory conditions the lack of protein secretion by the Ysa TTS system is indeed due to a reduction of gene expression (data not shown). In contrast, at 26°C, expression of orf6-lacZYA, sycB-lacZYA, ysaE-lacZYA, ysrS-lacZYA, and rcsB-lacZYA increased early during the logarithmic phase of bacterial growth, and then expression dropped rapidly to baseline levels as the cultures of these strains entered into the stationary phase of growth (Fig. 4). This pattern of expression was also exhibited when lacZYA fusions to orf7, ysaC, ysaU, and ysaR were examined (data not shown). These results indicate that expression of many Ysa TTS-related genes occurs in response to temperature and growth phase. However, we note that the change in gene expression that appears to be growth phase dependent may be due to the depletion of a nutrient available in L medium. In addition, induction of these genes is not necessarily coupled to the function of the Ysa TTS apparatus, since each of the reporter stains is defective for Ysp secretion.
FIG. 4.
Effects of growth phase on the expression of various genes involved in the Ysa TTS system. Ysa TTS system-related genes are induced during the logarithmic phase of growth. The genotypes of individual strains are described in Table 1. (A) GY4641 (ysaE-lacZYA); (B) GY4668 (orf6 orf7-lacZYA); (C) GY5019 (sycB-lacZYA); (D) GY4613 (rcsB-lacZYA); (E) GY4742 (ysrS-lacZYA); and GY4894 (ysrS-lacZYA operon fusion). For each panel, growth of the culture was followed by measuring the OD600 (▪) and levels of expression were monitored by determining β-galactosidase activity (▴). Each strain examined was cultivated overnight in L medium plus 90 mM NaCl and then subcultured into L medium plus 290 mM NaCl to an OD600 of ∼0.1. Subsequently, samples were harvested at various time points and assayed for β-galactosidase activity. Each data point represents the mean for two independent experiments performed in duplicate, with a standard deviation of less than 10 percent.
Environmental regulation is affected by the ionic strength and pH of the growth medium.
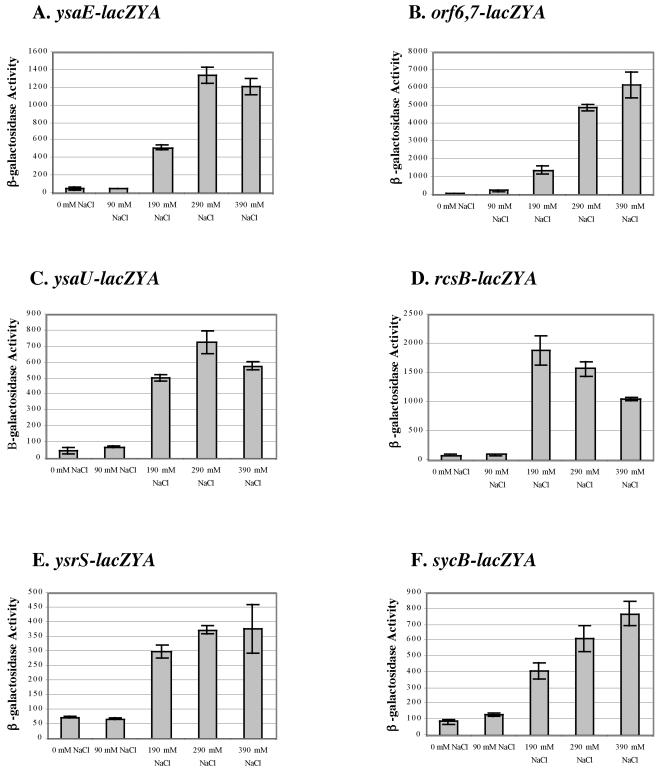
The role of NaCl as a stimulus of transcription for genes located within the YSA PI was then more specifically examined. First, the level of Ysp proteins secreted by wild-type Y. enterocolitica was analyzed for cultures grown in LB medium adjusted to contain various NaCl concentrations. The results showed that Ysp proteins are secreted by Y. enterocolitica when the NaCl concentration of the L medium exceeds ∼180 mM and reaches a maximal level at 290 mM NaCl (data not shown). As noted previously, increasing the NaCl concentration above 290 mM did not further increase Ysp secretion but did negatively affect the growth rate of the culture (39; data not shown). Expression of various genes within the YSA PI was similarly dependent on the concentration of NaCl (Fig. 5). In agreement with Ysp secretion analysis, expression of rcsB, orf6, sycB, ysaE, ysaR, ysaU, and ysrS was highest in L medium with greater than 290 mM NaCl (Fig. 5). When NaCl was replaced with KCl, similar expression patterns were observed, indicating that the regulatory effect is not specific to NaCl (data not shown). To determine if expression was induced in response to the presence of salt or was more generally due to changes in osmolarity of the medium, NaCl was replaced with similar or greater osmolar amounts of the nonmetabolizable sugar galactose. Two strains, GY4641 (ysaE-lacZYA) and GY4668 (orf6 orf7-lacZYA) were grown at 26 or 37°C in L medium supplemented with 200 mM, 600 mM, and 800 mM galactose at 26°C and at 37°C. Expression of ysaE-lacZYA and orf6-lacZYA was low at all galactose concentrations tested, suggesting that the ionic strength of the medium rather than overall osmolarity plays an important role in the induction of the Ysa TTS system (data not shown).
FIG. 5.
Effects of NaCl concentration on the expression of genes involved in Ysa TTS. Levels of β-galactosidase expression were measured for selected strains cultivated at 26°C in L medium containing various concentrations of NaCl as indicated below each panel. (A) GY4641 (ysaE-lacZYA); (B) GY4668 (orf6 orf7-lacZYA); (C) GY4664 (ysaU-lacZYA); (D) GY4613 (rcsB-lacZYA); (E) GY4894 (ysrRS-lacZYA); (F) GY5019 (sycB-lacZYA). β-Galactosidase expression was determined when cultures reached the early to mid log phase of growth. The results represent the means ± standard deviations from at least two independent experiments performed in duplicate.
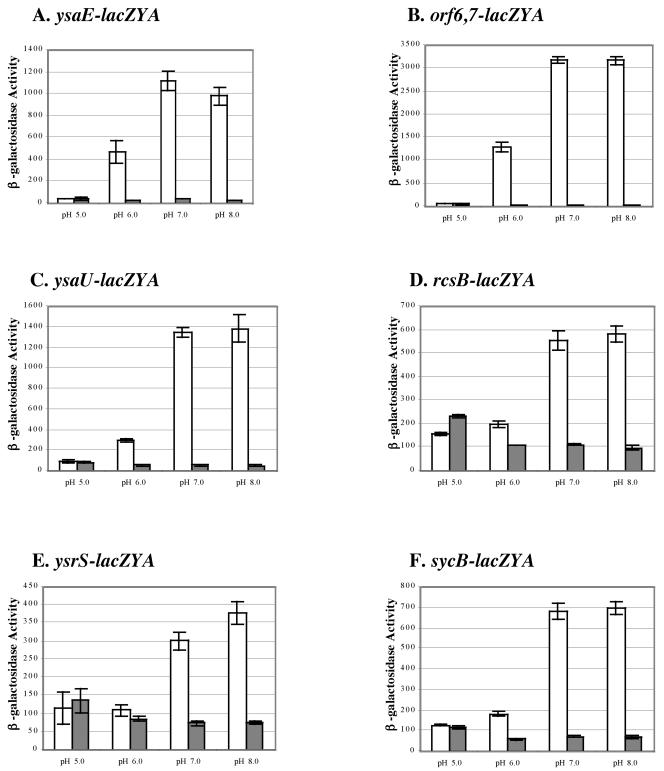
Other conditions known to affect the expression of virulence genes of Y. enterocolitica include nutrient availability, calcium and iron availability, oxygen tension, and pH. We tested a series of strains harboring different lacZYA fusions to determine the effects of varying these conditions on the expression of Ysa TTS system-related genes. For each parameter tested, cultures were incubated at 26°C and 37°C and β-galactosidase levels were determined. This survey was completed using modified L medium because the expression of Ysa TTS-related genes was not induced when strains were cultured in defined minimal media such as M9 and M63 with glucose or glycerol as the carbon and energy sources. At 26°C, induction of expression in L medium with 290 mM NaCl was not affected by calcium limitation or iron limitation (data not shown). In contrast, the pH of the medium did influence the expression of each lacZYA fusion, with the greatest levels observed at a pH above 7.0 and low levels occurring at pH 6.0 or less (Fig. 6). There was no apparent induction of any Ysa TTS-related gene tested at 37°C under the surveyed culture conditions. However, an interesting and important aspect of these results is the observation that the expression of ysrS strongly responds to temperature, NaCl, and pH, much like the other genes within the YSA PI.
FIG. 6.
The expression of genes involved in Ysa TTS is affected by pH. Levels of β-galactosidase expression were measured for selected strains cultivated at 26 and 37°C in L medium plus 290 mM NaCl buffered at various pH values as indicated below each panel. (A) GY4641 (ysaE-lacZYA); (B) GY4668 (orf6 orf7-lacZYA); (C) GY4639 (ysaR-lacZYA); (D) GY4664 (ysaU-lacZYA); (E) GY4894 (ysrS-lacZYA); (F) GY5019 (sycB-lacZYA). Media were buffered at pH 5.0 with 100 mM citrate, at pH 6.0 with 100 mM MES, at pH 7.0 with 100 mM MES, and at pH 8.0 with 100 mM MOPS. White bars represent results for cultures cultivated at 26°C, and gray bars represent results for cultivation at 37°C. β-Galactosidase expression was determined when cultures reached the early to mid log phase of growth. The results represent averages ± standard deviations from two independent experiments performed in duplicate.
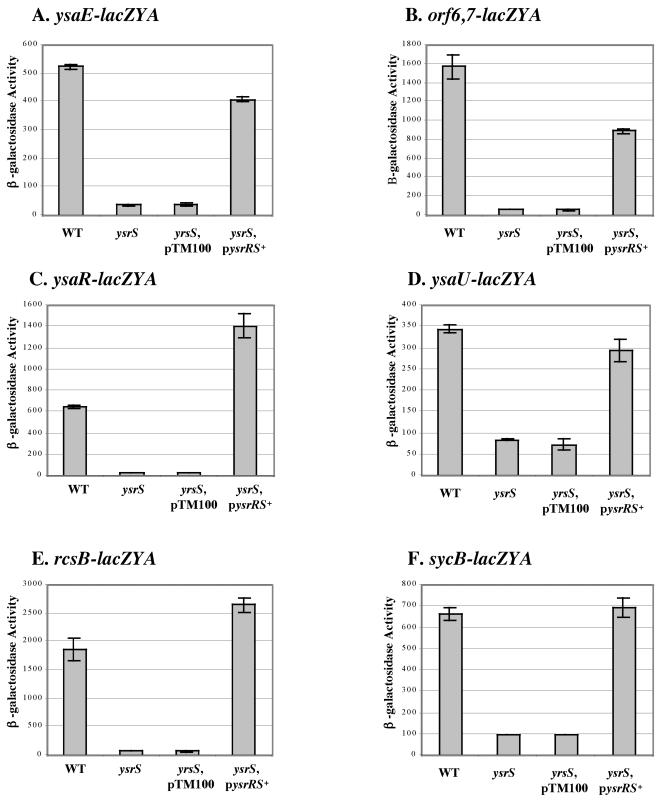
YsrS is required for expression of genes located within and outside of the YSA locus.
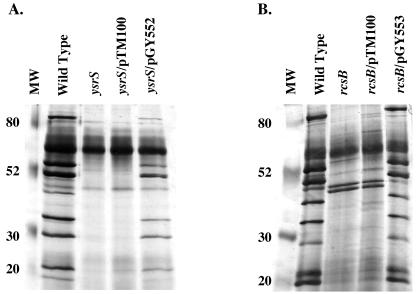
The proteins encoded by ysrR and ysrS are predicted to form a phosphorelay system. YsrS is homologous to membrane-bound sensor kinases, and YsrR is homologous to several response regulators. A transposon insertion mutation in ysrS blocked the production of Ysps (Fig. 1). To confirm this result, we reconstructed a mutation in ysrS and found that this mutant was similarly defective for Ysp secretion (Fig. 7A). This suggested that ysrS plays an important role in controlling the expression of genes located within and, possibly, outside of the YSA PI. Consistent with this hypothesis, during the course of this study another group reported that ysrS affects the transcription of a promoter originating upstream of ysaE and sycB (37). To confirm those results and to further extend our understanding of the regulatory role of ysrS, an insertion mutation of ysrS was introduced into strains GY4641, GY4668, GY4639, GY4664, GY4613, and GY5019. Levels of β-galactosidase produced by each strain were determined under Ysa TTS system-inducing conditions. The results show that inactivation of ysrS causes a reduction in the expression of the corresponding gene for each reporter strain (Fig. 8). These data indicate that ysrS is required for the expression of genes located throughout the YSA PI (orf6, ysaE, ysaR, and sycB) and genes located at distant chromosomal locations, such as rcsB. In agreement with this interpretation, restoration of ysrS function by complementation in trans with plasmid pGY552 (ysrRS+) restored expression of the lacZYA fusion of each strain (Fig. 8).
FIG. 7.
Ysp export is affected in ysrS and rcsB mutants. Analysis of secreted proteins from Y. enterocolitica strains grown under Ysa TTS-inducing conditions. Proteins secreted into culture supernatants were collected, separated by 12.5% SDS-PAGE, and visualized by staining with silver. (A) Lanes: 1, JB580v (wild type); 2, GY4887 (ysrS); 3, GY4887/pTM100 (vector control); 4, GY4887/pGY552 (ysrRS+). (B) Lanes: 1, JB580v (wild type); 2, GY5124 (rcsB); 3, GY5124/pGY556 (vector control); 4, GY5124/pGY553 (rcsB+). Each lane contains protein harvested from culture supernatants equivalent to 1 ml of culture at an OD600 of 3.0. MW, molecular weight (in thousands).
FIG. 8.
Effects of ysrS on the expression of selected genes involved in Ysa TTS. Genes involved in Ysa type III protein secretion require YsrS for expression. Selected reporter strains and isogenic derivatives are shown. (A) ysaE-lacZYA. (B) orf6 orf7-lacZYA. (C) ysaR-lacZYA. (D) ysaU-lacZYA. (E) rcsB-lacZYA. (F) sycB-lacZYA. For each panel, levels of expression are displayed for different genetic backgrounds: wild type (WT), ysrS (ysrS), plasmid control (ysrS, pTM100), and in trans complementation (ysrS pysrRS+). β-Galactosidase activity was determined for each strain cultivated in L medium plus 290 mM NaCl at 26°C. The results are averages ± standard deviations from at least two independent experiments performed in duplicate.
We also examined whether a functional YsrS-YsrR regulatory system might influence the expression of ysrS. This was an important experiment because it has been proposed that the environmental signal sensed by YsrS-YsrR is NaCl (37), a model that may be too simplistic considering that ysrS expression increases in response to salt concentrations even in a ysrS-deficient background (Fig. 4E). Therefore, strain GY4894, which carries an operon fusion between ysrS and lacZYA, was constructed. This strain was confirmed to have a functional YsrS-YsrR regulatory system by demonstrating that Ysps were produced and secreted only when cultures were grown under inducing conditions (data not shown). The induction pattern of ysrS expression in GY4894 was similar to the pattern of expression displayed in GY4742, which carries a transposon insertion-derived fusion to lacZYA (Fig. 4E and F). For both strains, induction of ysrS occurred in response to temperature, pH, and NaCl (data not shown). Also, the magnitudes of ysrS induction during the logarithmic phase of growth were comparable (Fig. 4E and F). As the cultures entered into stationary phase, however, there was a discernible difference in the relative decreases in ysrS expression. Assuming this difference is not due to secondary affects related to the alternative types of transcriptional fusions used, these data may indicate that the YsrS-YsrR system contributes sustained expression of ysrS once the culture enters into the stationary phase of growth.
RcsB affects expression of genes located within the YSA locus.
Results from transposon mutagenesis suggested that rcsB is important for Ysp secretion. In addition, expression of rcsB-lacZY is affected by a mutation in ysrS. Therefore, the necessity of this gene for Ysa TTS was further explored. Insertion mutagenesis was used to inactivate rcsB in a wild-type genetic background of strain JB580v to generate strain GY4737. Ysa TTS system-dependent secretion of proteins was then evaluated and found to be completely blocked (Fig. 7B). Complementation of the rcsB mutation in trans with a plasmid-borne copy of the gene restored Ysp secretion by the Ysa TTS system to levels similar to those of wild-type Y. enterocolitica (Fig. 7B). The rcsB mutation had no effect on Yop and Fop secretion, indicating that function of the Ysc and flagellar TTS systems is independent of RcsC-YojN-RcsB regulation (data not shown).
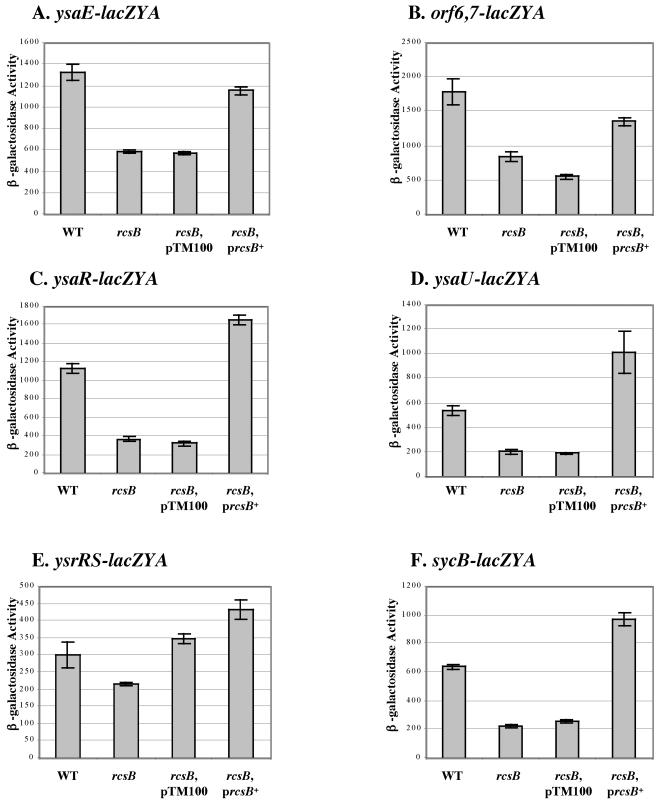
Given that rcsB encodes a response regulator, we hypothesized that a mutation in this gene would affect the expression of genes within the YSA PI. To test this possibility, an rcsB mutation was introduced into a number of different reporter strains (Table 1 and Fig. 9). Levels of β-galactosidase produced by each strain were then determined under Ysa TTS system-inducing conditions. The results revealed that a mutation in rcsB reduced the expression of orf6-lacZYA, sycB-lacZYA, ysaE-lacZYA, ysaR-lacZYA, and ysaU-lacZYA (Fig. 9). When a plasmid-based copy of rcsB was introduced into each of the reporter strains, expression of each gene fusion was restored (Fig. 9). Loss of rcsB function or the presence of multiple copies of rcsB in the cell resulted in a discernible effect on ysrS-lacZYA expression (Fig. 9E). These data indicate that rcsB influences the expression of many genes within the YSA PI, including the regulatory gene ysrS.
FIG. 9.
Effects of rcsB on the expression of selected genes involved in Ysa type III protein secretion. RcsB affects the expression of the genes involved in Ysa TTS. (A) ysaE-lacZYA. (B) orf6 orf7-lacZYA. (C) ysaR-lacZYA. (D) ysaU-lacZYA. (E) ysrRS-lacZYA. (F) sycB-lacZYA. For each panel, levels of expression are displayed for the following different genetic backgrounds: wild type (WT), rcsB (rcsB), plasmid control (rcsB, pTM100), and in trans complementation (rcsB prcsB+). β-Galactosidase activity was determined for each strain cultivated in L medium plus 290 mM NaCl at 26°C. The results are averages ± standard deviations from at least two independent experiments performed in duplicate.
Effect of the Ysa TTS system on the disease mortality rate.
The collection of Ysa TTS mutants provided the opportunity to explore how this system affects host-pathogen interactions. Measurements of disease mortality can provide information regarding the likelihood that a Y. enterocolitica infection will progress systemically. Previously it was reported that inactivation of the Ysa TTS system increased the oral LD50 of Y. enterocolitica 10-fold for BALB/c mice (17). Given that this was a rather modest difference, we decided to utilize our panel of mutants to reevaluate this result. Therefore, the oral LD50 of strains JB580v (wild type), GY4742 (ysrS), GY4641 (ysaE), GY4613 (rcsB), and GY4664 (ysaU) were determined for BALB/c mice. The LD50 of each mutant strain and that of its wild-type parental strain were found to be nearly identical, ranging between 5 × 106 and 1 × 107 CFU. These data indicate that mutational inactivation of genes required for the expression and function of the Ysa TTS system does not significantly alter the disease-induced mortality rate during Y. enterocolitica infection of mice.
Ysa TTS does not contribute to colonization of deep tissues during the systemic phase of infection.
While the Ysa TTS system does not affect the mortality rate of a Y. enterocolitica infection in mice, there is the potential that this system affects colonization of a specific tissue. Within the first 24 h of Y. enterocolitica infection of BALB/c mice, only gastrointestinal system-associated tissues become colonized, including the terminal ileum, the cecum, and Peyer's patches (6, 18). Occasionally bacteria are found in the mesenteric lymph nodes. Infections lasting beyond 3 days consistently progress to a systemic phase in which deep tissues such as the spleen and liver become fully infected with high numbers of bacteria (6, 18). Therefore, mice were perorally infected with various Ysa TTS mutants, including those with mutations mapping to ysrS, ysaE, ysaU, and rcsB. All of the strains displayed the ability to colonize the spleen and the liver at days 3 and 5 to levels equal to that observed in mice infected in parallel experiments with wild-type Y. enterocolitica (data not shown). Consistent with the disease mortality analysis, the Ysa TTS system appears to be dispensable for the systemic phase of infection.
The Ysa TTS system plays a role in the colonization of the terminal ileum.
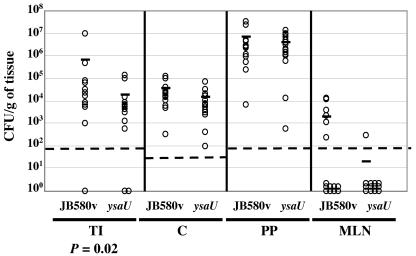
To assess whether Ysa TTS may be important during the gastrointestinal phase of infection, mice were perorally infected with a representative mutant, strain GY4664 (ysaU). Gastrointestinal system-associated tissues from infected mice were examined 24 h later to quantify the level of bacterial colonization. This time point following the initiation of infection was selected because it is early enough to allow for an assessment of the gastrointestinal phase of infection. In addition, this time point exceeds the 3- to 7-h retention time of the gastrointestinal tract, so the results from the analysis would not likely reflect limited passive colonization by the bacterium (18). This set of experiments revealed a small, but significant, reduction (P = 0.02) in the number of Ysa TTS-deficient bacteria colonizing the terminal ileum (Fig. 10). Compared to the wild-type strain, the ysaU mutant showed no significant difference in its colonization of other tissues by this experimental method.
FIG. 10.
The Ysa TTS system plays a role in the colonization of the terminal ilea of mice. The numbers of bacteria recovered from tissues of BALB/c mice perorally infected with Y. enterocolitica are shown. Groups of BALB/c mice (n = 16) were infected with 1 × 108 CFU of JB580v (wild type) or GY4664 (ysaU), and 24 h later, mice were sacrificed and the terminal ileum (TI), cecum (C), Peyer's patches (PP), and mesenteric lymph nodes (MLN) were aseptically removed. Each tissue was then homogenized in phosphate-buffered saline, and appropriate dilutions were plated onto L medium plus NaCl. The dashed line indicates the limit of detection. Data points represent the number of recoverable CFU per gram of tissue from individual animals. The horizontal bar indicates the arithmetic mean for each data set. Data were analyzed by using the Mann-Whitney two-tailed analysis of variance, and P values are indicated where appropriate.
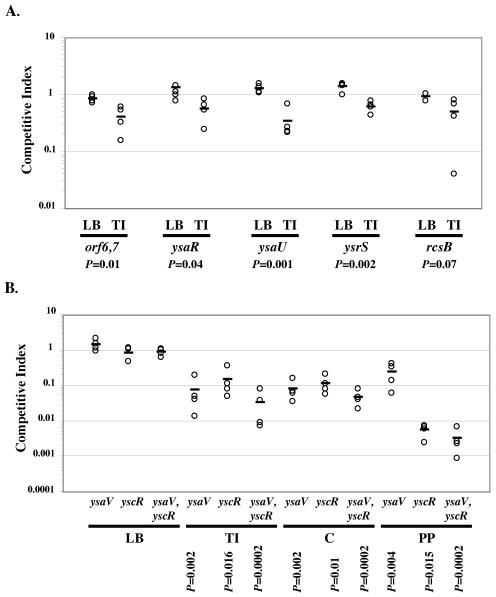
To further test the hypothesis that the Ysa TTS system has a role in colonization of the terminal ileum, the competitive indexes (C.I.) of several of the Ysa TTS mutants (orf6,7, ysaR, ysaU, ysrS, and rcsB mutants) were determined. This type of analysis was used based on previous studies that have established that it allows for the assessment of a virulence defect of a variety of mutants without the need to infect large numbers of mice. Additionally, it can reveal differences which are relatively small. The C.I. of each mutant examined was completed by initially infecting mice orally with equal amounts of wild-type and mutant bacteria (Fig. 11A). Twenty-four hours later, the ratio of mutant to wild-type bacteria colonizing the terminal ileum was determined. A C.I. less that 1.0 indicates that the mutant strain is less virulent than the wild-type strain. Importantly, in vitro competition experiments established that none of the mutants tested in mice have a general growth defect when cultured in LB medium (Fig. 11A, LB samples). The results revealed that all of the Ysa TTS mutants tested displayed a reduced in vivo C.I. for colonization of the terminal ileum, an outcome consistent with the idea that the Ysa TTS system plays an important role during the earliest stages of infection (Fig. 11A, TI samples).
FIG. 11.
Ysa TTS mutants are affected in the competitive colonization of gastrointestinal tissues. Assays of competitive colonization were conducted between wild-type Y. enterocolitica and selected mutants defective for Ysa and Ysc TTS. (A) To determine the in vivo C.I., mice were perorally infected with 2 × 109 CFU, which consisted of a 1:1 mixture of the wild-type strain JB580v and an isogenic mutant strain. The terminal ileum (TI) from each mouse was removed 24 h following inoculation, and the numbers of CFU of the mutant and wild type were determined. The C.I. was calculated as described in Materials and Methods. Each data point represents the C.I. for a single mouse, and the mean C.I. is indicated by the horizontal bar. The C.I. is calculated as follows: (mutant/wild type output ratio)/(mutant/wild type input ratio), where a C.I. of less than 1.0 indicates that a mutant is compromised in its ability to colonize a tissue. As a control, the in vitro C.I. was also determined for a 1:1 mixture of JB580v and an isogenic mutant strain grown in LB medium (LB). The in vitro C.I. and in vivo C.I. for each strain combination were compared by Student's two-tailed t test, and the P value is shown below each set of results. The following strains of Y. enterocolitica affected in Ysa TTS were examined for their ability to colonize Peyer's patch tissue: GY4668 (orf6 orf7), GY4639 (ysaR), GY4664 (ysaU), GY4742 (ysrS), and GY4613 (rcsB). (B) Competitive colonization of the terminal ileum (TI), cecum (C), and Peyer's patches (PP) was measured for strains with mutations that block Ysa TTS, Ysc TTS, or both systems: GY4428 (ysaV), YVM356 (yscR), and JB580v (WT) and GY1087 (yscR ysaV). The in vitro C.I. was also determined for a 1:1 mixture of JB580v, and each mutant strain grown in LB medium (LB). The in vitro C.I. in LB was compared to each in vivo C.I. for each strain/tissue by Student's two-tailed t test, and the P value is shown below each result.
Both the Ysa and Ysc TTS systems contribute to the colonization of gastrointestinal system-associated tissues.
The experiments described above suggest that the Ysa TTS system influences colonization of the terminal ileum, but it remains possible that the significance of this observation might be obscured by activities of the plasmid-encoded Ysc TTS system. This possibility is especially important to consider given the recent finding that YopE, YopN, and YopP are secreted by both the Ysa and Ysc TTS systems (38). Therefore, additional competition experiments were conducted, and the study was expanded to provide an initial evaluation of the relative contribution of each of these two distinct contact-dependent TTS systems to host colonization. To provide some additional perspective, several different gastrointestinal system-associated tissues were examined, including the terminal ileum, cecum, and Peyer's patches. For these assays, we used a set of previously characterized Y. enterocolitica strains with mutations that affect one (ysaV or yscR) or both (ysaV and yscR) contact-dependent TTS systems (38). Interestingly, after 24 h of infection, the ysaV mutant displayed a reduced C.I. for colonization of the terminal ileum as well as the cecum and Peyer's patches (Fig. 11B). The yscR mutant displayed a reduced capacity to colonize the terminal ileum and cecum that was similar in magnitude to the reduction seen for the ysaV mutant. Consistent with the known role of the Ysc TTS system in virulence and colonization during the invasive stages of infection, the yscR mutant was more severely affected for competitive colonization of the Peyer's patches (Fig. 11B). As expected, the C.I. of the double mutants revealed that this strain was compromised for the ability to colonize each of the three different gastrointestinal tissues examined (Fig. 11B). The trends correlate with the possibility that defects in the two systems have an additive effect on competitive colonization. It appears that both the Ysa and Ysc TTS systems enhance colonization of the terminal ileum and the cecum early during infection. While both TTS systems can affect the colonization of Peyer's patches, it is clear that the Ysc TTS system plays a dominant role once bacteria have invaded this tissue.
DISCUSSION
This exploratory study was conducted to provide insight on environmental factors that influence the expression of the Ysa TTS system and to gain some perspective on how the Ysa TTS system affects interactions between Y. enterocolitica biovar 1B and a susceptible host. To accomplish these initial goals, an experimental approach was used that yielded an extended collection of Y. enterocolitica strains that harbored transcriptional fusions between Ysa TTS-related genes and the reporter operon lacZYA. A systematic survey of environmental parameters that influence the expression of these genes revealed important information, some of which is new and some of which confirms or extends observations from other studies. First, expression of a variety of genes distributed throughout the YSA PI was examined, and all of the genes displayed similar regulatory patterns regardless of their predicted functions. The common pattern of expression may reflect that many of the genes within the YSA PI form operons or individual transcriptional units that are coregulated. All of the genes examined were induced during the early logarithmic phase of growth and respond to temperature, pH, and NaCl, including the regulatory genes ysrS and rcsB. The way that these environmental cues are sensed by the cell and mechanistically lead to changes in gene expression remains to be defined. At this point we can only speculate on the exact environmental cues sensed by the YsrS-YsrR and RcsC-YojN-RcsB signal transduction systems. Changes in temperature, pH, and salt concentrations might be specifically monitored. Alternatively, these chemical and physical conditions have a profound effect on cell membrane structure or integrity; the membrane itself may serve as a common site for signal integration. There is some evidence that this is the case for the activation of the RcsC-YojN-RcsB system in E. coli and S. enterica (7, 29, 35). Passage from the terrestrial environment through the gastrointestinal tract exposes the bacterium to a variety of conditions that assault the cell membrane, including changes in pH, temperature, ionic strength, membrane-perturbing antimicrobial peptides, and bile salts.
Genetic analysis demonstrated that ysrS is required for the expression of many YSA PI genes and that rcsB is important for full expression of the same genes. This suggests that both the YsrR-YsrS and RcsC-YojN-RcsB phosphorelay systems control the expression of the Ysa TTS system. Interestingly, inactivation of ysrS also affected the transcription of rcsB, but inactivation of rcsB had only a small effect on ysrS. The dominant effect of the ysrS mutation is consistent with YsrR-YsrS acting upstream of RcsC-YojN-RcsB in controlling the expression of the Ysa TTS system. We propose that the YsrR-YsrS phosphorelay system directly or indirectly affects genes within the YSA PI and of the RcsC-YojN-RcsB phosphorelay system. Changes in the levels of the RcsC-YojN-RcsB system and its activity in turn exert additional modulatory effects on YSA PI genes. A complete regulatory model must also include the cyclic AMP-CRP regulatory system, which was previously shown to be required for the expression of the Ysa TTS system (32). Testing a regulatory model will provide a foundation for understanding the molecular mechanisms used by Y. enterocolitica to integrate environmental cues that affect the expression of the Ysa TTS system.
Expression of YSA-related genes was best induced when strains were cultured in a relatively-high-ionic-strength, nutrient-rich growth medium. Curiously, we observed that Ysa TTS-related genes were best induced at 26°C, but not at 37°C. This is consistent with previous studies that documented that Ysp secretion by the Ysa TTS system in vitro is promoted at low temperatures. This finding may seem to strike a blow against the possibility that the Ysa TTS system has a role in the colonization of a mammalian host by Y. enterocolitica. However, this is not the only occasion where a putative virulence factor of a pathogenic species of Yersinia was observed to be best expressed in vitro at a temperature that is lower than that of a warm-blooded animal. Other genes, such as inv, yplA, yst, the urease gene cluster, and the O-antigen gene cluster, display similar levels of temperature-dependent regulation (10, 19, 25, 34, 36). Only inv and yst have been further shown to require specialized conditions that allow for induction at higher temperatures (25, 30). Yet, all of these genes affect pathogenic outcomes of a Y. enterocolitica infection in experimentally infected mice. Furthermore, the O-antigen gene cluster notwithstanding, these genes promote colonization or progression of disease in gastrointestinal system-associated tissues. This set of results therefore helped to guide the second part of this exploratory study, which focused on evaluating whether the Ysa TTS system affected bacterial-host interactions and, in particular, played a role during early stages of infection when bacteria are predominantly found in gastrointestinal tissues.
The first set of experiments that were conducted to confirm and extend the previously reported observation that Ysa TTS-defective mutants displayed a modest 10-fold reduction in the ability to cause the mortality of orally infected BALB/c mice (17). The collection of mutants tested here included strains that had insertion mutations in regulatory genes (rcsB, ysaE, ysrS) and a structural gene (ysaU) that map to the YSA PI or a distant location in the chromosome. All of the mutants tested displayed abilities to cause mortality that were similar to that of the isogenic wild-type strain of Y. enterocolitica. It is not clear why we were unable to reproduce the previously reported result. Nonetheless, the two studies are consistent in that it appears that the Ysa TTS system is not required by Y. enterocolitica biovar 1B to cause a progressive systemic infection in BALB/c mice that leads to mortality. As a follow-up analysis, the same mutants were tested for the ability to colonize tissues that commonly have large numbers of bacteria during the systemic phase of infection, such as Peyer's patches, mesenteric lymph nodes, liver, and spleen. None of the mutants displayed differences in their potentials to multiply in these tissues. This is consistent with the disease mortality analysis indicating that the Ysa TTS system does not influence systemic phases of infection in mice.
This led us to address whether the Ysa TTS system has a role during the gastrointestinal phase of infection. We were also encouraged to refocus the analysis in this way since the regulatory experiments revealed that Ysa TTS-related genes respond to many of the same environmental conditions as other genes involved in this stage of infection. Mice were infected with Y. enterocolitica or a ysaU mutant, after which colonization of the terminal ileum, cecum, and Peyer's patches was evaluated 24 h postinfection. This time point was selected because it ensured that the infection had not disseminated to deep tissues but exceeded the normal retention time for ingested material (18). This investigation revealed that the Ysa TTS system affects colonization of the intestinal terminal ileum. While this analysis was informative, it also required a large number of animals, so to further test the validity of this result, we utilized in vivo competition assays. This experimental approach measures the ability of a mutant, when mixed with wild-type bacteria, to competitively colonize the host. This type of analysis required fewer animals per experiment and provided the opportunity to investigate many different Ysa TTS mutants for defects in the competitive colonization of host tissues. Importantly, this alternative experimental approach also provided evidence indicating that the Ysa TTS system contributes to colonization of the terminal ileum.
To provide additional perspective on the magnitude of the colonization defect of Ysa TTS mutants, we measured the competitive colonizations of an isogenic set of mutants consisting of strains whose Ysa TTS pathway, Ysc TTS pathway, or both secretion pathways were blocked. It turned out that blockage of individual TTS systems reduced the ability of Y. enterocolitica to competitively colonize the terminal ileum, cecum, and Peyer's patches. Blockage of both TTS systems exacerbated the effect in each case. The data are consistent with previous observations that the Ysc TTS system has an important role in the colonization of Peyer's patches even at an early time point of infection. Importantly, this study revealed the Ysa TTS system influences Y. enterocolitica colonization of the gastrointestinal tract and provides a foundation for future studies. It will be interesting to understand which type of host cells is targeted by the Ysa and Ysc TTS system effector proteins. Understanding this facet of bacterial-host interactions should shed some light on why Y. enterocolitica biovar 1B maintains two distinct contact-dependent TTS systems.
Acknowledgments
We thank Nicole Sunseri and Briana Young for technical and experimental contributions to this study. We are also grateful to members of the laboratory for critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health, R21 AI156042 (G.M.Y.). K.V. was partially supported by an ASM Robert Watkins Minority Scholarship.
Editor: V. J. DiRita
REFERENCES
- 1.Baumler, A. J., R. M. Tsolis, A. W. van der Velden, I. Stojiljkovic, S. Anic, and F. Heffron. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207-213. [DOI] [PubMed] [Google Scholar]
- 2.Blum, H., H. Beier, and H. J. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 3.Bottone, E. J. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1:323-333. [DOI] [PubMed] [Google Scholar]
- 4.Brubaker, R. R. 1991. Factors promoting acute and chronic diseases caused by yersiniae. Clin. Microbiol. Rev. 4:309-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 6.Carter, P. B. 1975. Oral Yersinia enterocolitica infection of mice. Am. J. Pathol. 81:703-705. [PMC free article] [PubMed] [Google Scholar]
- 7.Conter, A., R. Sturny, C. Gutierrez, and K. Cam. 2002. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J. Bacteriol. 184:2850-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover, T. L., and R. C. Aber. 1989. Yersinia enterocolitica. N. Engl. J. Med. 321:16-24. [DOI] [PubMed] [Google Scholar]
- 9.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 10.de Konig-Ward, T. F., and R. M. Robins-Browne. 1997. A novel mechanism of urease regulation in Yersinia enterocolitica. FEMS Microbiol. Lett. 147:221-226. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, J. J., and G. J. Zylstra. 1998. Plaposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez-Bernal, G., M. G. Pucciarelli, F. Ramos-Morales, M. Garcia-Quintanilla, D. A. Cano, J. Casadesus, and F. Garcia-del Portillo. 2004. Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol. Microbiol. 53:1437-1449. [DOI] [PubMed] [Google Scholar]
- 13.Foultier, B., P. Troisfontaines, S. Muller, F. R. Opperdoes, and G. R. Cornelis. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J. Mol. Evol. 55:37-51. [DOI] [PubMed] [Google Scholar]
- 14.Foultier, B., P. Troisfontaines, D. Vertommen, M.-N. Marenne, M. Rider, C. Persot, and G. R. Cornelis. 2003. Identification of substrates and chaperone from the Yersinia enterocolitica 1B Ysa type III secretion system. Infect. Immun. 71:242-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, R. C., and A. J. Darwin. 2004. PspG, a new member of the Yersinia enterocolitica phage shock protein regulon. J. Bacteriol. 186:4910-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gripenberg-Lerche, C., L. Zhang, P. Ahtonen, P. Toivanen, and M. Skurnik. 2000. Construction of urease-negative mutants of Yersinia enterocolitica serotypes O:3 and O:8: role of urease in virulence and arthritogenicity. Infect. Immun. 68:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 18.Hanski, C., U. Kutschka, H. P. Schmoranzer, M. Naumann, A. Stallmach, H. Hahn, H. Menge, and E. O. Riecken. 1989. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O:8 with intestinal mucosa during experimental enteritis. Infect. Immun. 57:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isberg, R. R., A. Swain, and S. Falkow. 1988. Analysis of expression and thermoregulation of the Yersinia pseudotuberculosis inv gene with hybrid proteins. Infect. Immun. 56:2133-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isberg, R. R., D. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769-778. [DOI] [PubMed] [Google Scholar]
- 21.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 22.Longsdon, L., and J. Mecsas. 2003. Requirement of the Yersinia pseudotuberculosis effectors YopH and YopE in colonization and persistence in intestinal and lymph tissues. Infect. Immun. 71:4595-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marenne, M.-N., L. J. Mota, and G. R. Cornelis. 2004. The pYV plasmid and the Ysc-Yop type III secretion system, p. 319-348. In E. Carniel and B. J. Hinebusch (ed.), Yersinia: molecular and cellular biology. Horizon Press, Norfolk, Va.
- 24.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikulskis, A. V., I. Delor, V. Ha Thi, and G. R. Cornelis. 1994. Regulation of the Yersinia enterocolitica enterotoxin Yst gene. Influence of growth phase, temperature, osmolarity, pH and bacterial host factors. Mol. Microbiol. 14:905-915. [DOI] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 27.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mouslim, C., M. Delgado, and E. A. Groisman. 2004. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54:386-395. [DOI] [PubMed] [Google Scholar]
- 29.Mouslim, C., and E. A. Groisman. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol. Microbiol. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 30.Pepe, J. C., J. L. Badger, and V. L. Miller. 1994. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol. Microbiol. 11:123-135. [DOI] [PubMed] [Google Scholar]
- 31.Pepe, J. C., and V. L. Miller. 1993. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc. Natl. Acad. Sci. USA 90:6473-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen, S., and G. M. Young. 2002. An essential role for cAMP and its receptor protein in Yersinia enterocolitica virulence. Infect. Immun. 70:3665-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed, L. J., and H. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 34.Schmiel, D. S., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiba, Y., Y. Yokoyama, Y. Aono, T. Kiuchi, J. Kusaka, K. Matsumoto, and H. Hara. 2004. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J. Bacteriol. 186:6526-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skurnik, M., and P. Toivanen. 1993. Yersinia enterocolitica lipopolysaccharide: genetics and virulence. Trends Microbiol. 1:148-152. [DOI] [PubMed] [Google Scholar]
- 37.Walker, K. A., and V. L. Miller. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young, B. M., and G. M. Young. 2002. Evidence for targeting of Yop effectors by the chromosomally encoded Ysa type III secretion system of Yersinia enterocolitica. J. Bacteriol. 184:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young, G. M., D. Amid, and V. L. Miller. 1996. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J. Bacteriol. 178:6487-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, L., J. Radziejewska-Lebrecht, D. Krajewska-Pietrasik, P. Toivanen, and M. Skurnik. 1997. Molecular and chemical characterization of lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol. Microbiol. 23:63-76. [DOI] [PubMed] [Google Scholar]