Abstract
Prp8 is the largest and most highly conserved protein in the spliceosome yet its mechanism of function is poorly understood. Our previous studies implicate Prp8 in control of spliceosome activation for the first catalytic step of splicing, because substitutions in five distinct regions (a–e) of Prp8 suppress a cold-sensitive block to activation caused by a mutation in U4 RNA. Catalytic activation of the spliceosome is thought to require unwinding of the U1 RNA/5′ splice site and U4/U6 RNA helices by the Prp28 and Prp44/Brr2 DExD/H-box helicases, respectively. Here we show that mutations in regions a, d, and e of Prp8 exhibit allele-specific genetic interactions with mutations in Prp28, Prp44/Brr2, and U6 RNA, respectively. These results indicate that Prp8 coordinates multiple processes in spliceosome activation and enable an initial correlation of Prp8 structure and function. Furthermore, additional genetic interactions with U4-cs1 support a two-state model for this RNA conformational switch and implicate another splicing factor, Prp31, in Prp8-mediated spliceosome activation.
Keywords: pre-mRNA splicing‖small nuclear RNA‖retinitis pigmentosa‖RNA helicases‖Prp31
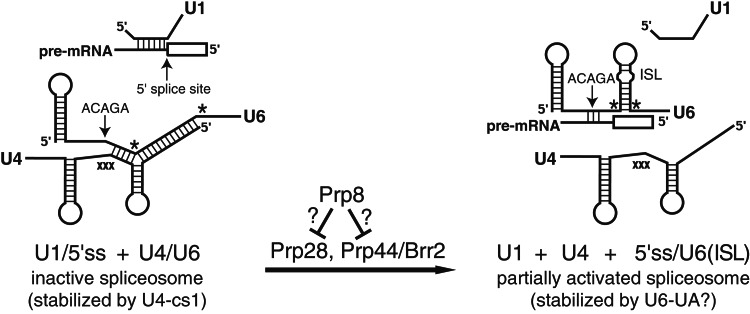
The removal of introns from nuclear pre-mRNA is catalyzed by a macromolecular complex called the spliceosome, which consists of five RNAs (U1, U2, U4, U5, and U6) and more than 70 proteins. Pre-mRNA splicing is thought to be RNA-catalyzed and involves two transesterification reactions (1). The accuracy and efficiency of splicing is achieved by means of a high degree of regulation, imposed by a complex series of protein–protein, protein–RNA, and RNA–RNA interactions. The spliceosome either assembles on the intron by way of stepwise recruitment of the constituent small nuclear ribonucleoproteins, each containing one RNA and several proteins (2), or a fully assembled spliceosome binds the intron as a unit (3). After the complete spliceosome has engaged an intron, it must undergo extensive conformational rearrangements to become catalytically active. Early steps in catalytic activation include unwinding of the U1 RNA/5′ splice site helix and unwinding of the U4/U6 helices (Fig. 1). These unwinding events are thought to be catalyzed by two DExD/H-box RNA helicases, Prp28 (4, 5) and Prp44/Brr2 (6–8), respectively.
Figure 1.
A two-state model of the early stages of spliceosome activation. Shown is a schematic of the RNA rearrangements that are proposed to be inhibited by the U4-cs1 mutation (position indicated by “xxx”) and stimulated by the U6-UA mutation (positions indicated by asterisks). Only the 5′ ends of U1 RNA and the pre-mRNA are shown; the open box represents the 5′ exon. Short lines indicate base pairs. The location of the U6 RNA “ACAGA-box” (see text) is indicated. “ISL” indicates the U6 RNA intramolecular stem-loop implicated in catalysis. Prp8 is proposed to coordinate the rearrangements catalyzed by Prp28 and Prp44/Brr2 by repressing the activities of these factors until the spliceosome is completely assembled and properly aligned on an authentic intron.
We identified a cold-sensitive mutation in U4 RNA, called U4-cs1, which blocks catalytic activation at low temperature (9, 10). U4-cs1 has a triple nucleotide substitution that extends the U4/U6 base-pairing, thereby masking the “ACAGA” sequence in U6 RNA that base pairs with the 5′ splice site when U1 RNA is displaced (Fig. 1). We proposed that in the absence of proper U6 RNA/5′ splice site pairing, U4/U6 RNA unwinding is inhibited. In support of this hypothesis, a cold-sensitive mutation in PRP44/BRR2 (brr2-1) exacerbates (enhances) the U4-cs1 mutation (11).
A selection for suppressors of the cold-sensitive growth defect of U4-cs1 yielded a mutation in the highly conserved 280-kDa splicing factor Prp8 (10). Prp8 is a U5 RNA-associated protein that contacts both the 5′ and 3′ splice sites of the intron (12–15) as well as U6 RNA (16). Prp8 has been implicated in regulation of the second transesterification reaction (17–20); our results suggest that Prp8 also functions in the first catalytic step of splicing. Although Prp8 is two-thirds identical between yeast and humans (21), it contains no recognizable structural motifs. To define domains important for the function of Prp8 in catalytic activation for the first transesterification reaction, we isolated over 40 additional single amino acid substitutions in Prp8 that suppress U4-cs1 (11), here collectively called prp8-cat mutations, because of their positive effect on catalytic activation. These mutations cluster in five regions named a–e (see Fig. 4) and are distinct from mutations in Prp8 that suppress defects in the second transesterification reaction.
Figure 4.
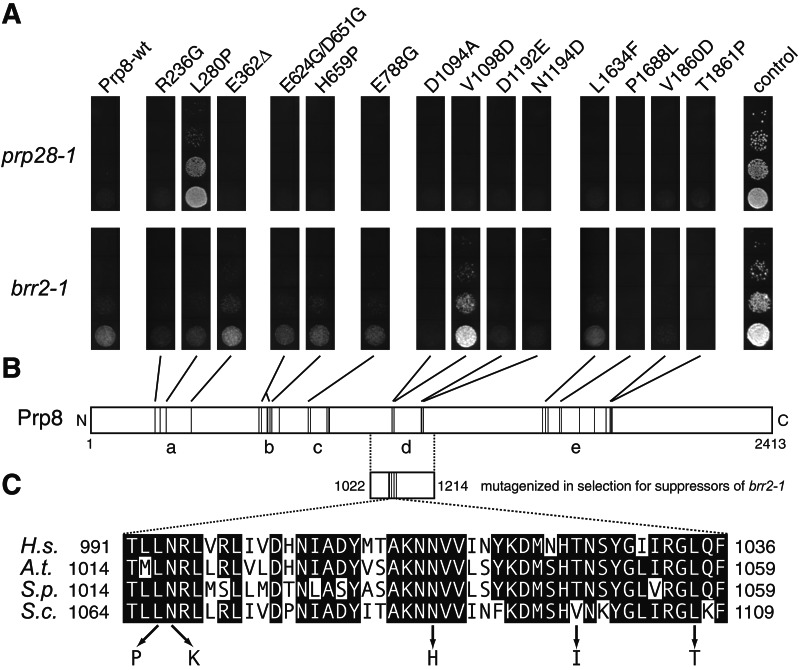
(A) Allele-specific suppression of prp28-1 and brr2-1 by mutations in regions a and d of PRP8, respectively. With cultures of equal cell density, serial dilutions of haploid yeast strains with prp28-1 (Upper) or brr2-1 (Lower) and the PRP8-alleles indicated were spotted onto yeast extract/peptone/dextrose plates and grown at 16°C for 7 (prp28-1) or 5 days (brr2-1). The right-most column (control) shows the growth of the prp28-1 or brr2-1 strain transformed with a plasmid bearing wild-type PRP28 or PRP44/BRR2, respectively. (B) Schematic of the primary structure of Prp8 with the locations of the identified prp8-cat mutations (11) indicated by thin vertical lines. Regions a–e defined by the mutations are labeled. (C) The positions of point mutations in Prp8 that suppress brr2-1 are indicated by thin vertical lines in the fragment that was mutagenized. The lower part shows a sequence alignment of amino acids 1064–1109 of Saccharomyces cerevisiae (S.c.) Prp8 with the corresponding regions of Prp8 orthologues from human (H.s.), Arabidopsis thaliana (A.t.), and Schizosaccharomyces pombe (S.p.). Shadings indicate identity in at least three of the four Prp8 orthologues. The single amino acid substitutions resulting in brr2-1 suppression are listed below the alignment.
We hypothesized that Prp8 regulates spliceosome activation by modulating RNA helicase activity (11). In particular, any mutations in Prp8 that disrupt putative negative regulation of Prp28 or Prp44/Brr2 should act to suppress U4-cs1 (Fig. 1). This hypothesis is supported by results from other labs. An unidentified mutation in Prp8 was shown to suppress a mutation in the helicase domain of Prp28 (22), and Prp8 interacts both physically and genetically with Prp44/Brr2 (23, 24). Furthermore, Prp8, Prp28, and Prp44/Brr2 are all associated with U5 RNA (25, 26).
Here we show that U4-cs1 also interacts genetically with a cold-sensitive mutation in PRP28 (prp28-1) and with the U6-UA mutation, which hyperstabilizes the 3′ intramolecular stem of U6 RNA (27). The pattern of enhancement and suppression further supports the two-state model for the early stages of spliceosome activation illustrated in Fig. 1. Strikingly, U6-UA, prp28-1, and brr2-1 each exhibit genetic interactions with distinct prp8-cat mutations, and in each case the interactions are opposite in sign to their interactions with U4-cs1. This result suggests that prp8-cat mutations suppress the U4-cs1 block to activation by at least three different mechanisms, by using distinct regions in the primary structure of the protein. A targeted selection for suppressors of brr2-1 defined a 42-aa region of Prp8 that influences Prp44/Brr2 function. In addition, we identify Prp31 as another potential target of Prp8 regulation, based on its enhancement of U4-cs1. Altogether, our results imply a remarkably high density of functional domains in Prp8.
Materials and Methods
Yeast Strains.
To delete the chromosomal PRP8 locus in yeast strains with brr2-1 (YSN485; ref. 28), prp28-1 (YES51; ref. 22), or U6-UA (27), the strains were first transformed with YCp50-PRP8, and Ura+ colonies were then transformed with a prp8Δ∷ADE2 fragment (10). Colonies were selected on medium lacking adenine and then tested for 5-fluoroorotic acid (5-FOA) sensitivity, which indicates that YCp50-PRP8 has become essential as a result of deletion of the chromosomal PRP8 locus. Correct disruption of the chromosomal PRP8 locus was verified by PCR. The resulting strains were named ANK821 [MATa brr2-1 prp8Δ∷ADE2 ura3 lys2 his3 ade2 leu2 (YCp50-PRP8)], ANK828 [MATa prp28-1 prp8Δ∷ADE2 trp1 ura3 his3 ade2 lys2 (YCp50-PRP8)], and ANK864 [MATa snr6Δ∷LEU2 prp8Δ::ADE2 trp1 ura3 his3 leu2 lys2 ade2 met2 (pRS317-snr6-UA) (YCp50-PRP8)]. The chromosomal SNR14 gene was disrupted in YES51 with the same strategy. YES51 was first transformed with pRS316-U4-wt (SNR14 URA3 CEN6 ARSH4). A uracil prototroph colony was selected and transformed with the PCR-amplified snr14∷TRP1 fragment from YKS2 (MATa snr14∷TRP1 trp1 ura3 lys2 his3 ade2 [YCp50-SNR14]) (29). Colonies selected on medium lacking tryptophan were tested for sensitivity to 5-FOA, which indicates that pRS316-U4-wt has become essential as a result of deletion of the chromosomal SNR14 locus. One Trp+, 5-FOA-sensitive strain was designated ANK281 [MATa prp28-1 snr14∷TRP1 trp1 ura3 his3 ade2 lys2 (pRS316-U4-wt)]. Strain ANK641 is isogenic to DMF101 [MATa snr6Δ∷LEU2 snr14∷TRP1 trp1 ura3 lys2 his3 ade29)YCp50–39D6) (pRS313-U4-wt)] (27), except that pRS313-U4-wt (SNR14 HIS3 CEN6 ARSH4) is replaced with pRS313-U4-cs1. ANK431 [MATa prp31-1 snr14∷TRP1 trp1 ura3 his3 lys2 (YCp50-SNR14)] was constructed from EWY2857 (30) and YKS2. The two strains were mated, and then the diploid was sporulated. A Trp+, temperature-sensitive, and 5-FOA-sensitive spore was propagated. ANK430 [MATa snr14∷TRP1 trp1 ura3 his3 lys2 (YCp50-SNR14)] was obtained by transforming ANK431 with PCR-amplified wild-type PRP31 and selecting for temperature-resistant colonies. All methods for manipulation of yeasts were performed according to standard procedures (31).
Screen for Mutations in PRP8 That Suppress brr2-1.
The screen for mutations in PRP8 that suppress brr2-1 was based on described procedures (11, 17). The part of PRP8 encoding amino acids 1022–1214 was PCR-mutagenized, and the PCR product was transformed into ANK821 together with SpeI-linearized pRS313-PRP8(SacII) (11), cut within codon 1118 of the PRP8 coding region to allow gap-repair in vivo. Strains with haploviable prp8 alleles were tested for suppression of the cold sensitivity caused by brr2-1 by incubation at 16°C for up to 10 days. Altogether, 12 candidate suppressor strains were selected and tested for linkage of the suppression phenotype to the mutagenized region. The PRP8 regions encoding amino acids 1022–1214 were PCR-amplified with Tfl DNA polymerase (Epicentre Technologies, Madison, WI) and DNA isolated from the candidate suppressor strains. Linkage of the suppression phenotype to the amplified region of PRP8 was analyzed essentially as described for the initial screening. Growth at 16°C was tested for six transformants for each suppressor strain. For all 12 suppressor strains, the suppression phenotype was linked to the PCR fragment as judged by the fact that at least three of the six transformants showed clear suppression of the brr2-1 cold sensitivity at 16°C. The PCR fragments were then sequenced to identify the suppressor mutations.
Results and Discussion
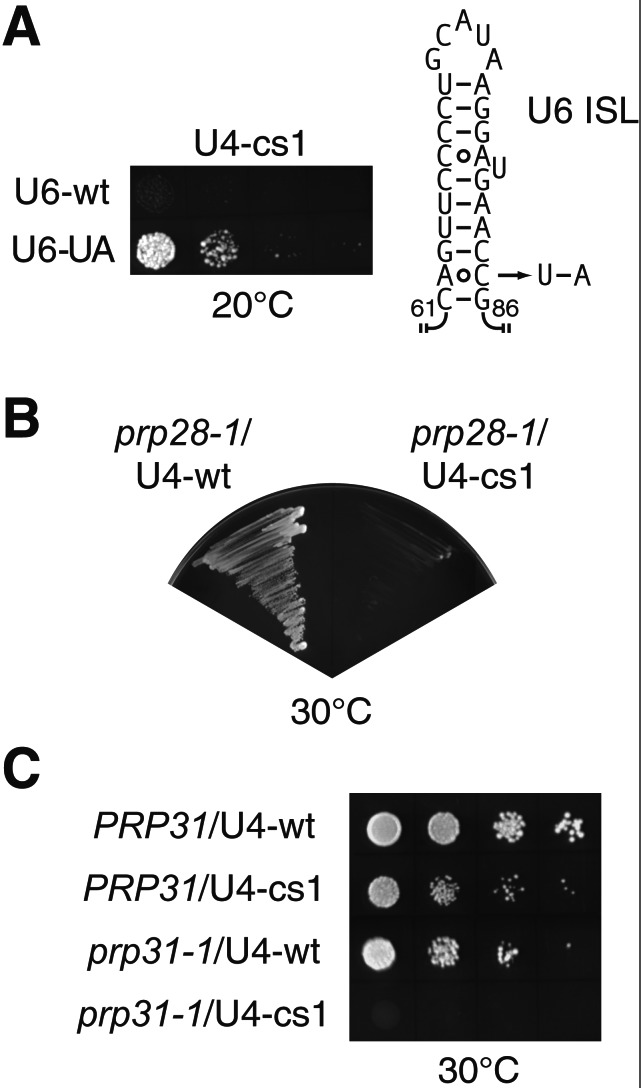
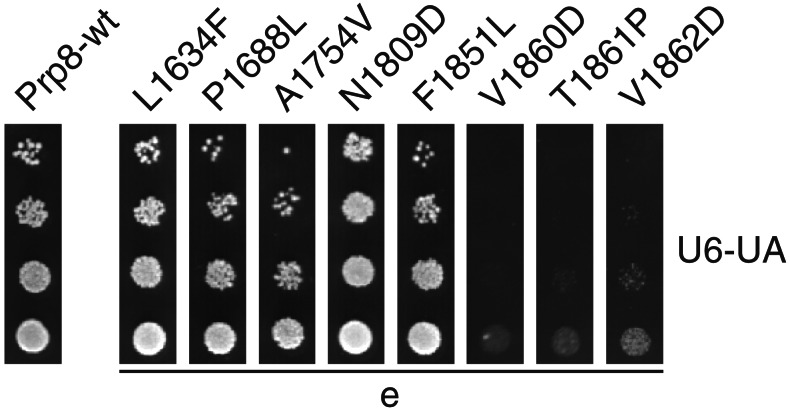
Disruption of U4/U6 RNA base pairing during spliceosome activation allows formation of an intramolecular stem-loop (ISL) structure in U6 RNA that may participate in catalysis (32, 33). In accordance with the two-state model for the early stages of spliceosome activation (Fig. 1), the relative stabilities of U4/U6 vs. free U4 + U6 should influence the steady-state occupancy of the bound and free states of U6 RNA. Therefore, we tested a mutation in U6 RNA that hyperstabilizes the ISL, U6-UA (Fig. 2A), for its effect on U4-cs1. U6-UA confers a cold-sensitive growth defect at 18°C (27) but has little or no effect on U4/U6 complex assembly (34). Therefore, it likely affects subsequent events, such as U4/U6 disassembly. Consistent with the two-state model for early activation, the U6-UA mutation rescues U4-cs1 lethality at 20°C (Fig. 2A; Table 1), presumably by stabilizing the free form of U6 RNA (Fig. 1). The two-state model is supported further by the fact that mutant forms of the splicing factor Prp24 that suppress U6-UA (34) enhance U4-cs1 (11). Thus, one way that Prp8 might regulate spliceosome activation is by modulating the stability of the U6 ISL. We therefore tested for genetic interactions between U6-UA and a representative subset of prp8-cat alleles. Intriguingly, mutation of any one of three adjacent residues at the C terminus of region e is synthetic lethal with U6-UA, whereas all 13 other region a–e mutations tested exhibit no genetic interaction (Fig. 3 and data not shown). This result indicates that the C-terminal part of region e of Prp8 influences formation and/or stability of the U6 ISL.
Figure 2.
Genetic interactions between the U4-cs1 mutation and conditional mutations in other spliceosome components involved in catalytic activation. (A) The U6-UA mutation suppresses the cold-sensitive growth defect of U4-cs1 at 20°C. ANK641, which has U4-cs1 as the sole copy of U4 RNA, a chromosomal disruption of the gene for U6 RNA, and a copy of the gene for wild-type U6 RNA on a counterselectable URA3-marked plasmid, was transformed with plasmids bearing the genes for wild-type U6 or U6-UA RNA as indicated. With cultures of equal cell density, serial 10-fold dilutions were plated to medium containing 5-FOA to select against the plasmid encoding wild-type U6 RNA and incubated at 20°C for 7 days. (Right) A schematic of the ISL of U6 RNA. The U6-UA mutation is indicated with an arrow. (B) U4-cs1 enhances the cold-sensitive growth defect of prp28-1. Plasmids with the genes coding for wild-type U4 RNA or U4-cs1 RNA were introduced into a strain (ANK281) with prp28-1, a chromosomal disruption of the gene for U4 RNA, and a copy of the gene for wild-type U4 RNA on a counterselectable URA3-marked plasmid. Shown are the resulting strains grown on medium containing 5-FOA at 30°C for 3 days. (C) U4-cs1 enhances the temperature-sensitive growth defect of prp31-1. Strains with chromosomal disruption of the gene for U4 RNA, a copy of the gene for wild-type U4 RNA on a counterselectable URA3-marked plasmid, and either PRP31 (ANK430) or prp31-1 (ANK431) at the chromosomal locus were transformed with plasmids bearing the genes for wild-type U4 or U4-cs1 RNA as indicated. With cultures of equal cell density, serial 10-fold dilutions were plated to medium containing 5-FOA to select against the gene for wild-type U4 RNA and incubated at 30°C for 3 days.
Table 1.
Genetic interactions among yeast splicing factors
| Allele | Phenotype | Effect of U4-cs1 | Effect of prp8-cat |
|---|---|---|---|
| U4-cs1 | CS | — | Suppress* |
| U6-UA | CS | Suppress | Enhance† |
| prp28-1 | CS | Enhance | Suppress† |
| brr2-1 | CS | Enhance* | Suppress† |
| prp31-1 | TS | Enhance | Not tested |
CS, little or no growth at 16°C, approximately normal growth at 30°C; TS, little or no growth at 37°C, approximately normal growth at 30°C; Suppress, approximately normal growth at 16°C (20°C for U6-UA/U4-cs1); Enhance, little or no growth at 30°C (synthetic lethal).
Kuhn and Brow (11).
Highly allele-specific with respect to PRP8.
Figure 3.
prp8-cat mutations in three adjacent amino acids in region e enhance a mutation that hyperstabilizes the U6 ISL. Yeast strain ANK864, which has chromosomal disruptions of the genes for Prp8 and U6 RNA, a copy of the gene for U6-UA RNA, and a copy of the gene for wild-type Prp8 on a counterselectable URA3-marked plasmid, was transformed with plasmids bearing the prp8-cat alleles indicated. With cultures of equal cell density, serial 10-fold dilutions were plated to medium containing 5-FOA to select against the wild-type PRP8 plasmid and incubated at 30°C for 3 days. prp8-cat alleles with mutations in regions a–d had no effect on U6-UA.
We found that a cold-sensitive mutation in PRP44/BRR2 (brr2-1) is lethal in combination with the U4-cs1 mutation at the otherwise permissive temperature of 30°C (11). It has been suggested that the activity of Prp44/Brr2 is coordinately regulated with Prp28 during splicing (4, 6, 10). In agreement with this hypothesis, a cold-sensitive mutation in PRP28 (prp28-1) is also synthetic lethal with U4-cs1 (Fig. 2B; Table 1). Enhancement of the U4-cs1 phenotype by prp28-1 and brr2-1 is consistent with the postulated function of Prp28 and Prp44/Brr2 in RNA rearrangements required for catalytic activation. Interestingly, the cold-sensitive growth defect caused by hyperstabilization of the U1 RNA/5′ splice site interaction is enhanced by prp28-1 but not by brr2-1 (4), indicating a somewhat different mechanism of arrest than for U4-cs1.
We considered the possibility that Prp8 coordinates the functioning of Prp28 and Prp44/Brr2 by inhibiting their activity until spliceosome assembly is complete and correct. If this is the case, then at least a subset of the prp8-cat mutations may act by relieving this inhibition and thus may suppress the prp28-1 and brr2-1 mutations. To test this prediction, 14 prp8-cat mutations from regions a–e were combined with prp28-1 or brr2-1. Strikingly, allele-specific suppression for both prp28-1 and brr2-1 was observed (Fig. 4A). The 16°C growth defect of prp28-1 is suppressed by prp8-L280P, a mutation in region a, but by no other prp8-cat mutation tested. Similarly, the brr2-1 cold sensitivity is specifically suppressed by only prp8-V1098D, from region d. These results strongly support the model that discrete domains of Prp8 control both Prp28- and Prp44/Brr2-dependent conformational changes, and that Prp8 may thereby coordinate the activities of these helicases during spliceosome activation.
Although prp8-V1098D is able to suppress the growth defect conferred by brr2-1, a prp8-cat mutation just four residues away (D1094A) cannot. Indeed, this latter mutation seems to slightly enhance the brr2-1 growth defect, an effect observed to varying degrees for a number of prp8-cat alleles (Fig. 4A and data not shown). This fact raises the possibility that suppression of brr2-1 is a fortuitous property of the V1098D mutation. To see whether other mutations in the vicinity of V1098 can suppress brr2-1, we mutagenized the PRP8 region coding for amino acids 1022–1214 and selected for suppressors of the brr2-1 growth defect at 16°C. Of 10 different suppressor alleles obtained, 5 contain a single substitution sufficient for suppression (L1066P, N1067K, N1087H, V1098I, and L1107T; Fig. 4C). Four of these substitutions change phylogenetically invariant amino acids, and the fifth is in the same residue as the interacting prp8-cat mutation (V1098) but is a more conservative change (valine to isoleucine rather than aspartate). Together, the single mutations define a 42-aa block spanning residues 1066–1107, and all five alleles with multiple mutations contain at least one substitution within this span (data not shown). These 42 amino acids correspond to only about one-fifth of the mutagenized region of PRP8, indicating that we have identified a core element that modulates Prp44/Brr2 helicase function. This element occupies only the N-terminal part of region d, which may therefore contain one or more additional functional domains. Indeed, the C-terminal part of region d has been implicated in binding the U1 RNA-associated protein Snp1, the yeast homologue of the human U1-70K protein (35).
The mechanism by which Prp8 may modulate DExD/H-box protein activity is not known. Two possibilities are (i) allosteric repression of helicase activity by direct physical interaction and (ii) masking or stabilization of the RNA substrate. In the case of Prp44/Brr2, the hypothesized allosteric interaction could be mediated by the 42-aa region encompassing the brr2-1 suppressor mutations. However, a directed two-hybrid screen with Prp8 region d as bait, and either a yeast genomic library (11) or segments of PRP44/BRR2 (data not shown) encoding the prey, provided no evidence for such an interaction. Recently, van Nues and Beggs (24) reported yeast–two-hybrid interactions between Prp44/Brr2 and two regions of Prp8, the very N terminus (residues 1–263) and the C terminus (residues 2010–2412). Both regions are far in the primary structure from the 42-aa region encompassing the suppressors of brr2-1 (residues 1066–1107) and therefore may perform a distinct function. With regard to the alternative model, it is interesting to note that region a has weak sequence similarity to eIF4E (11), which binds the 5′ cap of mRNAs. Conceivably, region a could inhibit Prp28-catalyzed unwinding of the U1 RNA/5′ splice site helix by binding to the 5′ cap of U1 RNA and blocking helicase entry or stabilizing the helix.
The prp8-cat mutations that do not interact genetically with prp28-1, brr2-1, or U6-UA may well be involved in additional aspects of spliceosome activation. This interpretation implies a remarkably high density of functional domains in Prp8, consistent with its high degree of sequence conservation. Additional genetic interaction studies are expected to reveal new functions of Prp8. As shown by the results presented here, mutant factors that enhance or suppress U4-cs1 are prime candidates for analysis of allele-specific interactions with prp8-cat mutants. Intriguingly, mutations in human Prp8, Prp31, and Prp3 have recently been found to cause an autosomal dominant form of retinitis pigmentosa (RP) (36–38), a disease that results in progressive degeneration of the retina. The Prp8 RP mutations are in the C-terminal 35 amino acids and do not colocalize in the sequence alignment with any prp8-cat mutations. However, we find that a conditional allele of the yeast PRP31 gene, prp31-1 (39), is synthetic lethal with U4-cs1 (Fig. 2C; Table 1), consistent with a positive function of wild-type Prp31 in spliceosome activation (39, 40). Therefore, the RP mutations may inhibit spliceosome activation and are good candidates for suppression by a subset of prp8-cat mutations.
Acknowledgments
We thank the Guthrie and Woolford labs for yeast strains; members of the Brow lab for discussions; and S. Butcher, N. Käufer, and E. Steinmetz for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM54018 (to D.A.B.) and by a Deutsche Forschungsgemeinschaft fellowship (to A.N.K.).
Abbreviations
- 5-FOA
5-fluoroorotic acid
- ISL
intramolecular stem loop
References
- 1.Collins C A, Guthrie C. Nat Struct Biol. 2000;7:850–854. doi: 10.1038/79598. [DOI] [PubMed] [Google Scholar]
- 2.Burge C B, Tuschl T H, Sharp P A. In: RNA World II. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 525–560. [Google Scholar]
- 3.Stevens S W, Ryan D E, Ge H Y, Moore R E, Young M K, Lee T D, Abelson J. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- 4.Staley J P, Guthrie C. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen J Y, Stands L, Staley J P, Jackups R R, Jr, Latus L J, Chang T H. Mol Cell. 2001;7:227–232. doi: 10.1016/s1097-2765(01)00170-8. [DOI] [PubMed] [Google Scholar]
- 6.Laggerbauer B, Achsel T, Lührmann R. Proc Natl Acad Sci USA. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raghunathan P L, Guthrie C. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 8.Kim D-H, Rossi J J. RNA. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Brow D A. RNA. 1996;2:879–894. [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn A N, Li Z, Brow D A. Mol Cell. 1999;3:65–75. doi: 10.1016/s1097-2765(00)80175-6. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn A N, Brow D A. Genetics. 2000;155:1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyatt J R, Sontheimer E J, Steitz J A. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 13.Teigelkamp S, Newman A J, Beggs J D. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyes J L, Kois P, Konforti B B, Konarska M M. RNA. 1996;2:213–225. [PMC free article] [PubMed] [Google Scholar]
- 16.Vidal V P, Verdone L, Mayes A E, Beggs J D. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umen J G, Guthrie C. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins C A, Guthrie C. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siatecka M, Reyes J L, Konarska M M. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dagher S F, Fu X D. RNA. 2001;7:1284–1297. doi: 10.1017/s1355838201016077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo H R, Moreau G A, Levin N, Moore M J. RNA. 1999;5:893–908. doi: 10.1017/s1355838299990520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strauss E J, Guthrie C. Genes Dev. 1991;5:629–641. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- 23.Achsel T, Ahrens K, Brahms H, Teigelkamp S, Lührmann R. Mol Cell Biol. 1998;18:6756–6766. doi: 10.1128/mcb.18.11.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Nues R W, Beggs J D. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teigelkamp S, Mundt C, Achsel T, Will C L, Lührmann R. RNA. 1997;3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens S W, Barta I, Ge H Y, Moore R E, Young M K, Lee T D, Abelson J. RNA. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- 27.Fortner D M, Troy R G, Brow D A. Genes Dev. 1994;8:221–233. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- 28.Noble S M, Guthrie C. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon K W, Guthrie C. Genes Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 30.Maddock J R, Roy J, Woolford J L., Jr Nucleic Acids Res. 1996;24:1037–1044. doi: 10.1093/nar/24.6.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guthrie C, Fink G R, editors. Guide to Yeast Genetics and Molecular Biology, Methods in Enzymology. New York: Academic; 1991. [PubMed] [Google Scholar]
- 32.Yean S L, Wuenschell G, Termini J, Lin R J. Nature (London) 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huppler A, Nikstad L J, Allmann A M, Brow D A, Butcher S E. Nat Struct Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 34.Vidaver R M, Fortner D M, Loos-Austin L S, Brow D A. Genetics. 1999;153:1205–1218. doi: 10.1093/genetics/153.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Awasthi S, Palmer R, Castro M, Mobarak C D, Ruby S W. J Biol Chem. 2001;276:31004–31015. doi: 10.1074/jbc.M100022200. [DOI] [PubMed] [Google Scholar]
- 36.McKie A B, McHale J C, Keen T J, Tarttelin E E, Goliath R, van Lith-Verhoeven J J, Greenberg J, Ramesar R S, Hoyng C B, Cremers F P, et al. Hum Mol Genet. 2001;10:1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 37.Vithana E N, Abu-Safieh L, Allen M J, Carey A, Papaioannou M, Chakarova C, Al-Maghtheh M, Ebenezer N D, Willis C, Moore A T, et al. Mol Cell. 2001;8:375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 38.Chakarova C F, Hims M M, Bolz H, Abu-Safieh L, Patel R J, Papaioannou M G, Inglehearn C F, Keen T J, Willis C, Moore A T, et al. Hum Mol Genet. 2002;11:87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- 39.Weidenhammer E M, Ruiz-Noriega M, Woolford J L., Jr Mol Cell Biol. 1997;17:3580–3588. doi: 10.1128/mcb.17.7.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makarova O V, Makarov E M, Liu S, Vornlocher H-P, Lührmann R. EMBO J. 2002;21:1148–1157. doi: 10.1093/emboj/21.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]