Abstract
Selenium can be metabolized for protein synthesis by two major pathways in vivo. In a specific pathway it can be inserted into polypeptide chains as the amino acid selenocysteine, as directed by the UGA codon. Alternatively, selenium can be substituted for sulfur to generate the free amino acids selenocysteine and selenomethionine, and these are incorporated nonspecifically into proteins in place of cysteine and methionine, respectively. A mutant strain of Escherichia coli was constructed that is deficient in utilization of inorganic selenium for both specific and nonspecific pathways of selenoprotein synthesis. Disruption of the cysK gene prevented synthesis of free cysteine and selenocysteine from inorganic S and Se precursors. Inactivation of the selD gene prevented synthesis of selenophosphate, the reactive selenium donor, required for the specific incorporation pathway. As expected, the double mutant strain, RL165ΔselD, when grown anaerobically in LB + glucose medium containing 75SeO32−, failed to synthesize selenium-dependent formate dehydrogenase H and seleno-tRNAs. However, it incorporated 24% as much selenium as the wild-type strain. Selenium in the deficient strain was bound to five different proteins. A 39-kDa species was identified as glyceraldehyde-3-phosphate dehydrogenase. It is possible that selenium was bound as a perselenide derivative to the reactive cysteine residue of this enzyme. A 28-kDa protein identified as deoxyribose phosphate aldolase also contained bound selenium. These 75Se-labeled proteins may have alternate roles as selenium delivery proteins.
In many biological systems selenium can be metabolized in a specific pathway dedicated to the biosynthesis of proteins which contain the amino acid selenocysteine as directed by the UGA codon. In Escherichia coli this pathway requires the products of four genes, selA, selB, selC, and selD. The selC gene product was identified as a selenocysteyl-tRNASec that cotranslationally delivers selenocysteine at the in-frame UGA codon (1, 2). Initially tRNASec is aminoacylated with serine and this is converted to selenocysteyl-tRNASec by the selA gene product, selenocysteine synthetase (3). The selenium is derived from an activated selenium donor identified as selenophosphate that is generated by the selD gene product, selenophosphate synthetase (4, 5). Selenophosphate also is required as the donor form of selenium required for the conversion of 2-thiouridine to 2-selenouridine in tRNA (5). The selB gene product, a specialized translation factor binds guanine nucleotides, selenocysteyl-tRNA, and the SECIS element, a unique stem loop structure in the mRNA located immediately downstream of UGA in E. coli formate dehydrogenase H mRNA (6). The binding of the selB translation factor complex to the stem loop stalls the ribosome at the UGA and prevents termination of protein synthesis long enough for the insertion of the selenocysteine residue to occur (2). In contrast to the specific pathway selenium can substitute for sulfur in many sulfur metabolic pathways because of the chemical similarity of these elements (7, 8). Thus, the enzymes involved in the cysteine biosynthetic pathway can generate free selenocysteine. The cysteyl-tRNA synthetase can charge the tRNAcys with either cysteine or selenocysteine and the latter is inserted into proteins nonspecifically in place of cysteine (9).
Several proteins have been identified as sulfur transferases or delivery proteins in specific sulfur metabolic pathways. These include the thiI gene product, a sulfur transferase required for thiazole and S4U formation (10), 3-mercaptopyruvate sulfurtransferase (11), and rhodanese (12). Considering that in vivo concentrations of sulfur are much greater than selenium, transport proteins may exist to ensure the effective delivery of selenium required in specific pathways of metabolism. These proteins should have a high affinity toward selenium and relatively low binding affinity toward sulfur.
In an attempt to identify potential selenium transport proteins able to bind selenium with high affinity, a double mutant of E. coli was generated. This mutant contains a selD deletion, as well as a cysK mutation, rendering it deficient both in specific and nonspecific pathways of selenium metabolism. The disruption of the two genes results in a strain unable to synthesize selenophosphate required for specific selenium incorporation into proteins and tRNA and free selenocysteine for nonspecific replacement of cysteine in proteins. Inability to fully metabolize selenium could facilitate the detection of various selenium transport proteins containing bound selenium. For example, prevention of normal selenium incorporation into proteins and tRNAs greatly decreases the number of radioactive bands visualized on gels, thus increasing the sensitivity of detection of precursor forms of selenium bound to potential delivery proteins. The inability to fully metabolize selenium, in fact, may lead to accumulated levels of the selenium-bound forms of these proteins. This work reports the characterization of E. coli strain RL165ΔselD.
Materials and Methods
Materials.
E. coli strain RL165 (cysK511) (13) and WL400 (ΔselD) (4) were obtained from the E. coli Genetic Stock Center. Bacteriophage P1 was purchased from America Type Culture Collection. [75Se]selenite was purchased from The University of Missouri Research Reactor Facility, Columbia. The rich medium used was LB medium supplemented with 0.1 μM SeO32− and 0.5% glucose.
Generation of E. coli Strain RL165ΔselD.
The chloramphenicol resistance marker inserted into the selD gene in WL400 was transduced by bacteriophage P1 into RL165. Colonies were selected on LB chloramphenicol plates. Standard methods used for genetic manipulation were according to Miller (14) and Sambrook et al. (15).
Polyacrylamide Gel Electrophoreses Analysis of Cell Extract.
Fifteen-milliliter cultures of E. coli strains MC4100, WL400, RL165, and RL165ΔselD were grown overnight anaerobically at 37°C in LB supplemented with 0.1 μM SeO32−, 10 μCi 75SeO32−, and 0.5% glucose. After growth cells were harvested, resuspended in 100 mM potassium phosphate buffer (pH 7.0), and lysed by freeze–thawing in liquid N2. Aliquots of supernatants containing 11–12 μg of protein were analyzed by SDS-polyacrylamide (10%) electrophoresis and PhosphorImager detection of radioactivity (16).
Identification of Proteins with Bound Selenium.
One-liter cultures of RL165ΔselD were grown in LB supplemented with 0.1 μM SeO32−, 1 mCi 75SeO32−, and 0.5% glucose. After growth cells were harvested, resuspended in 100 mM potassium phosphate buffer (pH 7.0), and lysed by freeze–thawing in liquid N2. Supernatants were applied to a HPLC DEAE column. A linear gradient 100 mM Tricine/KOH (pH 7.9; Buffer A)/100 mM Tricine-KOH (pH 7.9)/1 M KCl (Buffer B) was run at 2 ml/min: 0–20 min 0% Buffer B, 20–40 min 0–5% Buffer B, 40–50 min 5–10% Buffer B, 50–60 min 10–15% Buffer B, 60–70 min 15–20% Buffer B, 70–90 min 20–30% Buffer B, 90–100 min 30–50% Buffer B, 100–110 min 50–100% Buffer B, and 110–130 min 100% Buffer B. Protein separation during the gradient was monitored by absorbency measurements at 215 and 280 nm. 75Se-labeled proteins were determined by on-line scintillation counting. Peak fractions that contained 75Se were pooled, dialyzed against Buffer A, concentrated, and applied to a HPLC 2000SW gel filtration column run as described above. 75Se-labeled proteins were identified by PAGE and PhosphorImager analysis. Labeled protein bands were cut from gels and eluted by overnight incubation in 100 mM Tris⋅HCl (pH 8.5)/0.1% SDS, and N-terminal amino acid sequences were determined by automated Edman degradation (Hewlett Packard G1000A, Palo Alto, CA).
Results and Discussion
Characterization of RL165ΔselD.
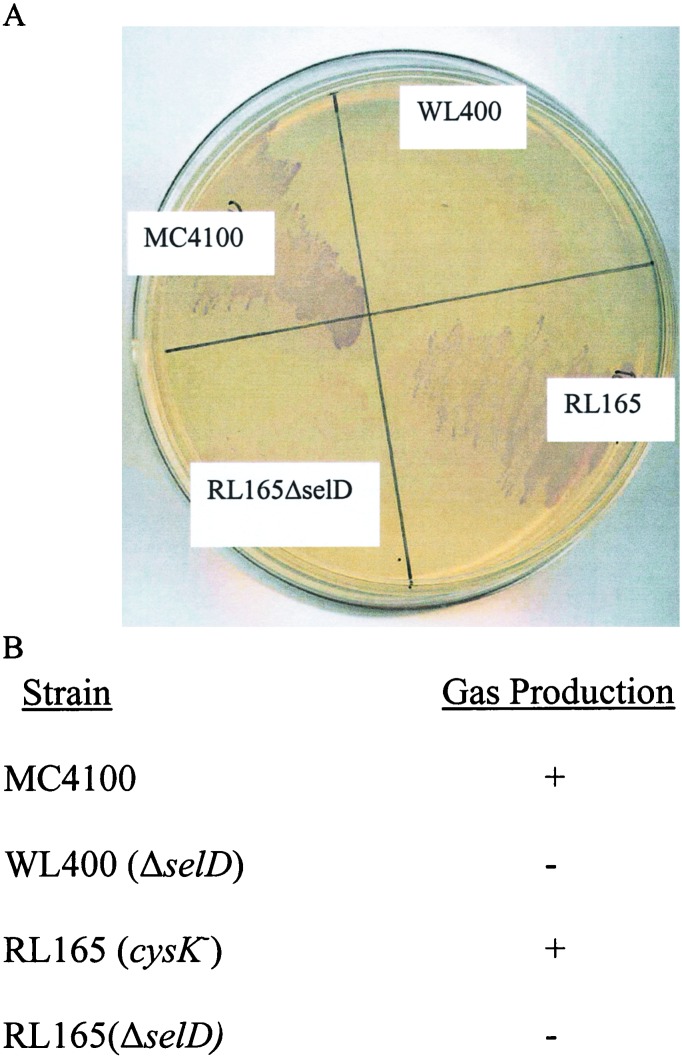
To confirm that the selD gene was inactivated, RL165ΔselD was assayed for ability to synthesize selenium-dependent formate dehydrogenase H in two routinely used assays. RL165ΔselD like WL400 (ΔselD) failed to produce hydrogen gas anaerobically in a glucose medium and was unable to reduce benzyl viologen in agar overlay plates, indicating the absence of formate dehydrogenase activity due to the lack of production of selenophosphate (Fig. 1).
Figure 1.
In vivo assay of the selenocysteine-containing FDHH protein. (A) E. coli strains MC4100 (wild type), WL400 (ΔselD), RL165 (cysK), and RL165ΔselD were grown overnight anaerobically on a LB agar plate. The plate was overlaid with agar containing benzyl viologen. Colonies with active FDHH reduced benzyl viologen resulting in a blue color. (B) Gas production was monitored in all four strains grown in LB overnight anaerobically in the presence of 0.5% glucose.
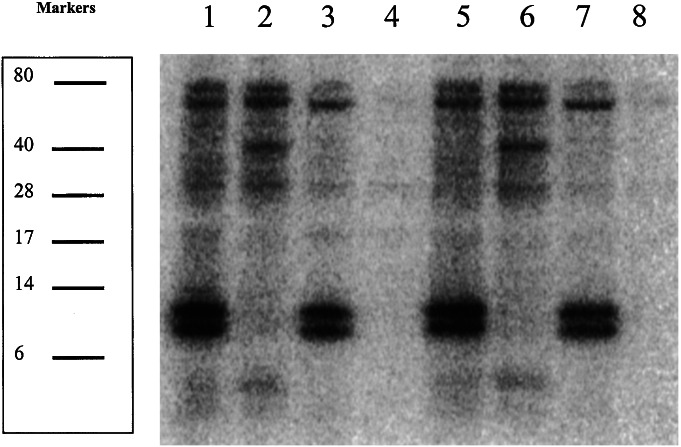
To establish the inability of RL165ΔselD to synthesize selenocysteine from selenite in the nonspecific pathway, cell extracts were examined for extent of selenium incorporation into proteins and tRNAs (Fig. 2). Cultures were grown overnight in media containing [75Se]selenite, cells were lysed, proteins were separated on SDS/10% PAGE gels, and radioactivity was detected by PhosphorImager analysis. Failure of the RL165ΔselD double mutant strain to incorporate radioactivity is shown in lanes 4 and 8 of Fig. 2. In lanes 1 and 5, which contain extract from wild-type MC4100, both nonspecific and specific incorporation can be visualized. The extract from WL400, a selD mutant (lanes 2 and 6), shows nonspecific incorporation into proteins and no specific incorporation into tRNA. The RL165, cysK mutant extract (lanes 3 and 7) shows specific incorporation only. The selenium incorporation in all samples was not affected by the presence of reducing agents in the treatment buffer.
Figure 2.
PhosphorImager analysis of a SDS/10% PAGE gel containing 75Se-labeled cell extract. E. coli strains MC4100 (lanes 1 and 5), WL400 (lanes 2 and 6), RL165 (lanes 3 and 7), and RL165ΔselD (lanes 4 and 8). Samples in lanes 5–8 were prepared in the presence of β-mercaptoethanol.
A comparison of the amount of selenium incorporated into each strain under the growth conditions stated above is reported in Table 1. Cells were lysed and the amount of [75Se]selenium incorporated is reported as the amount of radioactivity determined per microgram of extract protein. WL400, which is not able to insert selenium specifically (4), and RL165 (13), which is not able to insert selenium nonspecifically, show a 35% decrease in selenium incorporation compared with wild-type MC4100 where both pathways are intact. The lack of ability of RL165ΔselD to synthesize selenoproteins by either pathway resulted in a significantly greater decrease (76%) in the amount of 75Se found in cell extracts as compared with MC4100. The residual amount of radioactivity detected in the extract of the double mutant might represent selenium bound to transport or delivery proteins. Because both pathways of selenium metabolism are blocked, turnover of selenium bound to these proteins should be limited and this may explain the extent of selenium binding observed in the present study.
Table 1.
Quantitation of the amount of selenium incorporated into mutant strains of E. coli compared to wild type
| Strain | cpm/μg | % 75Se incorporation |
|---|---|---|
| MC4100 | 2,689 | 100 |
| WL400 (ΔselD) | 1,729 | 64 |
| RL165 (cysK−) | 1,739 | 65 |
| RL165 (ΔselD) | 657 | 24 |
Identification of Selenium-Bound Proteins.
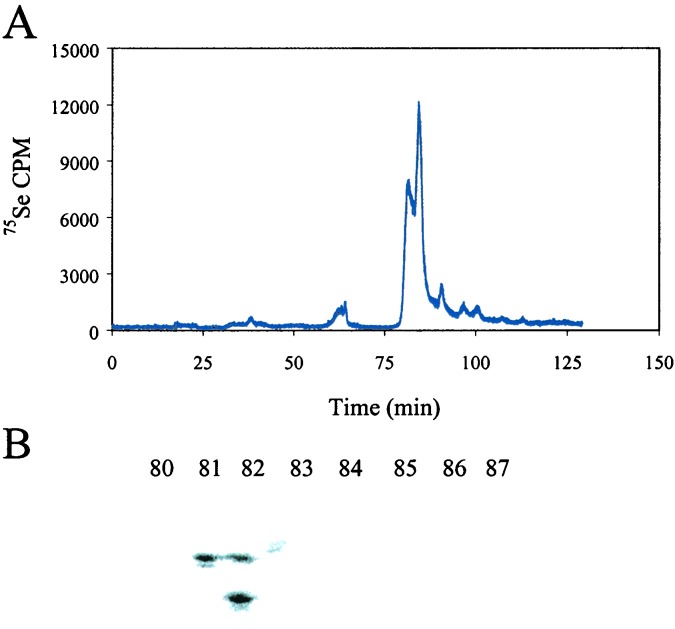
When wild-type E. coli is grown anaerobically in the presence of glucose, formate is produced as a fermentation product, and the formate induces the expression of the selenocysteine-containing FDHH enzyme. To mimic these conditions, 1-liter cultures of RL165ΔselD were grown in LB medium supplemented with 30 mM formate, 0.1 μM SeO32−, and 1 mCi 75SeO32− (1 Ci = 37 GBq). Although expression of selenophosphate synthetase and formate dehydrogenase H are prevented, synthesis of other proteins involved in the specific biosynthesis of selenocysteine and its insertion at the UGA codon may be unaffected. In particular, proteins that bind selenium and normally serve as delivery proteins for selenophosphate synthesis may accumulate as highly labeled derivatized species because of lack of turnover. After growth of the mutant strain in the [75Se]selenite medium, cells were harvested and extracts were analyzed. Residual labeled low molecular weight forms of selenium were separated by passage of extracts through Centricon-10 concentrators (nominal mass species cut off of 10,000 Da). Up to 34% of the total radioactivity passed into the filtrate. This labeled material migrated as low molecular weight inorganic selenium species when fractionated on a calibrated HPLC gel filtration column. The amount of this material was greater if extracts were frozen before analysis or otherwise denatured, suggesting release of protein-bound radioactivity. The radioactive protein fraction retained on the Centricon-10 membranes was fractionated on a DEAE-HPLC column as described in Material and Methods. The protein elution profile revealed a major peak containing 75Se that emerged at 80 min (Fig. 3A). Fractions containing radioactivity were concentrated and applied to a gel filtration 2000SW column. The elution profile revealed two proteins containing 75Se. One protein was eluted with an estimated size of 39 kDa and a second protein with an estimated size of 28 kDa. These protein fractions were subjected to SDS/10% PAGE gel analysis (Fig. 3B). Protein bands containing 75Se were cut out, eluted from the gel, and subjected to automated Edman degradation analysis. The N-terminal amino acid sequence of the 39-kDa protein matched that of GAPDH and the 28-kDa protein was identified as deoxyribose-phosphate aldolase (DPA) (Table 2). To determine the effect of the different growth conditions on the appearance of the 75Se-labeled proteins, 30 mM formate was omitted from the medium and formate generated only as the anaerobic fermentation product of 0.5% glucose was present. Under these conditions fractionation of the 75Se-labeled cell extract revealed the presence of five proteins that contained bound selenium (Table 2). The 39-kDa protein again was identified as GAPDH and the 28-kDa protein was identified as DPA. A smaller 7.3-kDa protein was identified as YDFZ and this exhibited homology to a nitrogenase protein from Clostridium pasteurianum. Two proteins of 90 and 60 kDa could not be identified by N-terminal sequencing or by mass spectral analysis.
Figure 3.
Cell extracts from RL165ΔselD grown in LB containing 0.1 μM SeO32−, 1 mCi 75SeO32−, and 30 mM formate. (A) 75Se elution profile of proteins eluted from DEAE column on HPLC as described in Materials and Methods. (B) PhosphorImager analysis of a SDS/10% PAGE gel containing fractions that eluted from the DEAE column at 81 and 82 min.
Table 2.
Identities of proteins isolated with bound 75Se
| Mass (kDa) | Identified protein |
|---|---|
| 90 | ND |
| 60 | ND |
| 39 | GAPDH* |
| 28 | Deoxyribose-5-phosphate aldolase (DPA)† |
| 7.3 | YDFZ hypothetical protein‡ |
In one experiment the growth medium contained 30 mM formate, and GAPDH and DPA were isolated as 75Se proteins. In a second experiment formate was omitted, 0.5% glucose was present, and GAPDH, DPA, and YDFZ were isolated as selenium-labeled proteins. ND, not determined.
35,401 Da; 330 amino acids; three cysteine residues, Cys-149, Cys-153, and Cys-288. Cys-149 is highly reactive. GAPDH from E. coli K12, TIKVGINGFGRI. 39-kDa [75Se] protein, TIKVGINGIG.
27,733 Da; 259 amino acids; four cysteine residues, Cys-33, Cys-47, Cys-125, and Cys-129. DPA from E. coli K12, MTDLKASSLRALKLMDLT. 28-kDa [75Se] protein, TDLKASSLRALKLM.
7,300 Da; 67 amino acids; one cysteine residue, Cys-52; sequence similar to nitrogenase iron protein from Clostridium pasteurianum. YDFZ from E. coli K12, MTTYDRNRNAITTG. 7.3-kDa [75Se], TTYDRNRNA.
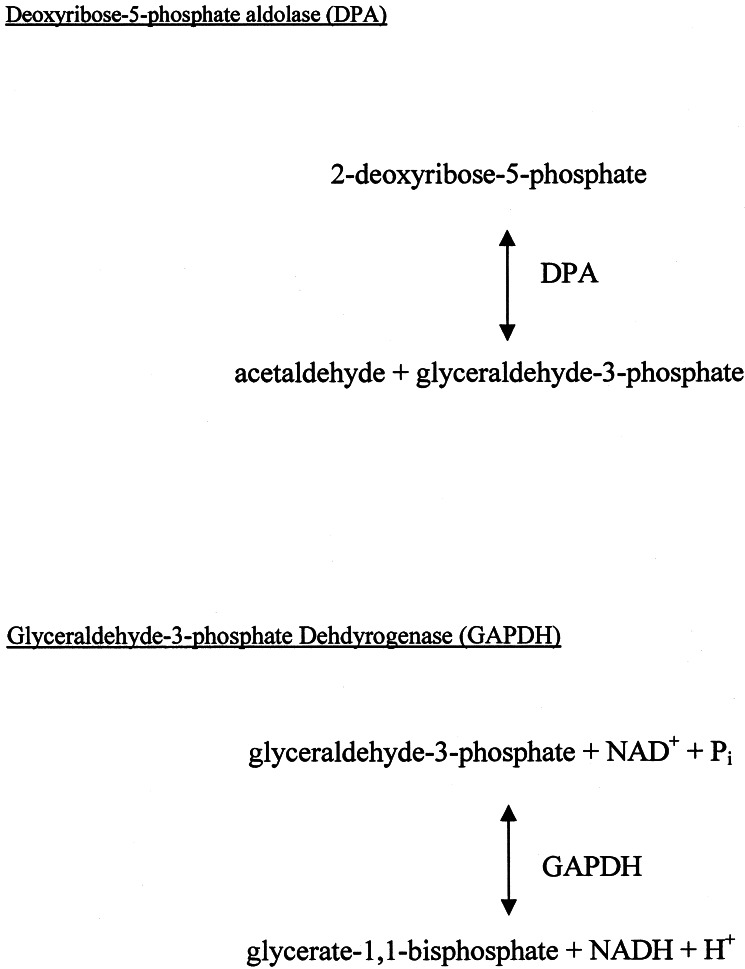
Interestingly, both GAPDH and DPA were isolated as selenium-bound proteins from two separate cultures grown under different conditions for the induction of FDHH. Both of these proteins bind glyceraldehyde-3-phosphate, GAPDH as a substrate and DPA as a product (Scheme S1). Perhaps the binding site for 3-glyceraldehyde phosphate is an ideal binding site for selenium. The presence of a low pKa reactive cysteine residue in GAPDH also could be important for selenium binding. Although the known enzymes identified here may serve as protein delivery agents, at least under certain conditions, other proteins may be involved in the normal delivery of selenium. The latter may not form adducts of sufficient stability to allow their isolation from RL165ΔselD under the conditions of the present study.
Scheme 1.
Reactions catalyzed by DPA and GAPDH.
The exact role of proteins such as GAPDH in selenium metabolism remains unknown. The fact that selenium is retained after denaturation by boiling in SDS and β-mercaptoethanol is indicative of its tight binding to proteins. This type of binding, whether noncovalent or in the form of a perselenide derivative, is consistent with a role as a selenium delivery protein. The well known important roles of GAPDH in both glycolysis and gluconeogenesis have been expanded recently to include diverse activities including phosphotransfer, nuclear RNA transport, microtubule bundling, DNA replication and repair, apoptosis, viral and bacterial pathogenesis, neurodegenerative disease, and prostate cancer (17). Thus, the ability of GAPDH to bind selenium may represent an additional biological role rather than a coincidence. The protein rhodanese, a sulfur transferase that binds sulfur from thiosulfate to a specific cysteine residue as a persulfide, recently was shown to bind selenium in the presence of glutathione (18). The selenium was tightly bound, presumably as a perselenide to the active cysteine, and could be transferred and used as substrate by selenophosphate synthetase if DTT was added to the reaction mixture. The utilization of reactive cysteine residues as sites for sulfur binding and transfer has been documented in several proteins. A similar mechanism in selenium transport may also be used.
Selenium binding and transport proteins have essential roles in the biosynthesis of selenophosphate. Considering that in vivo concentrations of sulfur are much higher compared with selenium and that sulfur pathways freely metabolize selenium as if it were sulfur, a mechanism must exist to effectively accumulate and deliver selenium to selenophosphate synthetase. The lack of the specific delivery of selenium would result in the inability to effectively synthesize selenocysteine-containing proteins. The proteins identified in this work may actually function as specific selenium delivery proteins. Considering that both pathways of selenium metabolism are blocked in RL165ΔselD, the ability of these proteins to bind selenium may indicate new roles as selenium delivery proteins.
Abbreviation
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
References
- 1.Leinfelder W, Zehelein E, Mandrand-Berthelot M-A, Bock A. Nature (London) 1988;331:723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- 2.Commans S, Bock A. FEMS Microbiol Rev. 1999;23:335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 3.Forchhammer K, Bock A. J Biol Chem. 1991;266:6324–6328. [PubMed] [Google Scholar]
- 4.Leinfelder W, Forchhammer K, Veprek B, Zehelein E, Bock A. Proc Natl Acad Sci USA. 1990;87:543–547. doi: 10.1073/pnas.87.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veres Z, Tsai L, Scholz T D, Politino M, Balaban R S, Stadtman T C. Proc Natl Acad Sci USA. 1992;89:2975–2979. doi: 10.1073/pnas.89.7.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forchhammer K, Leinfelder W, Bock A. Nature (London) 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- 7.Kramer G F, Ames B N. J Bacteriol. 1988;170:736–743. doi: 10.1128/jb.170.2.736-743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller S, Heider J, Bock A. Arch Microbiol. 1997;168:421–427. doi: 10.1007/s002030050517. [DOI] [PubMed] [Google Scholar]
- 9.Young P A, Kaiser I J. Arch Biochem Biophys. 1975;171:483–489. doi: 10.1016/0003-9861(75)90057-0. [DOI] [PubMed] [Google Scholar]
- 10.Kambampati R, Lauhon C T. J Biol Chem. 2000;275:10727–10730. doi: 10.1074/jbc.275.15.10727. [DOI] [PubMed] [Google Scholar]
- 11.Nagahara N, Ito T, Minami M. Histol Histopathol. 1999;4:1277–1286. doi: 10.14670/HH-14.1277. [DOI] [PubMed] [Google Scholar]
- 12.Westley J. In: Enzymatic Basis of Detoxication. Jacoby W B, editor. Vol. 2. New York: Academic; 1980. pp. 245–262. [Google Scholar]
- 13.Fimmel A L, Loughlin R E. J Gen Microbiol. 1977;103:37–43. doi: 10.1099/00221287-103-1-37. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 15.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 16.Laemmli U K. Nature (London) 1970;227:16068–16074. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Sirover M A. Biochim Biophys Acta. 1999;1432:159–184. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 18.Ogasawara Y, Lacourciere G, Stadtman T C. Proc Natl Acad Sci USA. 2001;98:9494–9498. doi: 10.1073/pnas.171320998. [DOI] [PMC free article] [PubMed] [Google Scholar]