Abstract
We report the identification of BvfA (for Brucella virulence factor A), a small periplasmic protein unique to the genus Brucella, which is essential for the virulence of Brucella suis. A BvfA knockout mutant was highly attenuated both in in vitro macrophage infection assays and in vivo in the murine model of brucellosis. Fluorescence-activated cell sorting analysis with green fluorescent protein fusions showed that the expression of bvfA is induced within macrophages by phagosome acidification and coregulated with the B. suis virB operon, suggesting that it too may play a role in the establishment of the intracellular replication niche.
Brucella spp. are facultative intracellular gram-negative bacteria. They are pathogenic for many mammalian species, including humans. Brucellosis is a chronic infection causing abortion and sterility in domestic mammals and a chronic undulant fever in humans (6, 42). The key aspect of Brucella virulence is its ability to survive and multiply within professional and nonprofessional phagocytes (6). To do this, Brucella perturbs the maturation of the phagosome, subverts vesicular trafficking within infected eukaryotic cells, and creates a unique intracellular niche in which it multiplies (1, 35, 36). Mutagenesis studies have identified a number of factors, including smooth lipopolysaccharide, stress response proteins, response regulators, and metabolic genes required for virulence in in vitro and animal models (10, 13, 15, 18, 23, 28). Recently, a type IV secretion system (T4SS) encoded by the virB operon has been identified as an essential virulence factor. Mutants lacking the T4SS are unable to survive and multiply in both macrophages and epithelial cells, as well as in the mouse virulence model (15, 20, 33). A possible role of the T4SS in virulence is to inject effector molecules, which induces the establishment of the replication niche, into the host cell (6). They allow the Brucella-containing phagosome to escape from the classical endosomal trafficking pathway and avoid fusion with late endosomes (4, 9, 31).
With the aim of finding the effectors of the virB T4SS, we developed a new strategy using translational fusions to the Yersinia YopP toxin as a reporter. The use of this technique led to the identification of one candidate effector. However, although experiments during its characterization suggested that it is not a VirB effector, it is a novel virulence factor that we named BvfA, for Brucella virulence factor A. A B. suis bvfA null mutant is highly attenuated in in vitro and in vivo virulence models. Expression of bvfA is induced within macrophages by phagosome acidification, suggesting that it too may play a role in the establishment of the intracellular replication niche.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All Brucella strains used in this study were derived from B. suis 1330 (ATCC 23444T) and are listed in Table 1, as are the plasmids and oligonucleotides used in the construction of mutants or fusions with yopP or gfp. Plasmid constructions were performed in Escherichia coli DH5α and B. suis 1330 by standard protocols (2). An isogenic mutant in the bvfA gene was constructed by introduction of a kanamycin resistance cassette and allelic replacement, as described previously (16). Gene inactivation was confirmed by both PCR and Southern blot analysis.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide primers
| Strain, plasmid, or primer | Characteristic(s) or relevant feature(s) | Source or reference |
|---|---|---|
| Brucella strains | ||
| 1330 | B. suis biotype 1, wild type, ATCC 23444T | American Type Culture Collection |
| 1330-GFP | gfp insertion into chromosomal DNA of B. suis 1330, Cmr Nalr | 19 |
| 3G1 | 1330-GFP with pBBR1-3G1 plasmid | This work |
| 1330bvfA::kan | 1330 Kanr; mutant of bvfA gene | This work |
| 1330bvfA::kan(pBBR1-BvfA) | 1330bvfA::kan with pBBR1-BvfA plasmid | This work |
| Plasmids | ||
| pBBR1-YopP | pBBR1MCS4 (22) containing a promoterless yopP | This work |
| pBBR1-3G1 | pBBR1-YopP with BvfA-YopP fusion | This work |
| pBBR1-BvfA | pBBR1MCS containing bvfA gene | This work |
| pBBR1-KGFP | pBBR1MCS containing a promoterless gfpmut3 | 24 |
| pBBR1-KGFPvirB | pBBR1-KGFP with gfpmut3 controlled by pvirB | 5 |
| pBBR1-KGFPcons | pBBR1-KGFP with gfpmut3 controlled by a constitutive promoter | 24 |
| pBBR1-KGFPbvfA | pBBR1-KGFP with gfpmut3 controlled by pbvfA | This work |
| PCR primers (5′-3′)a | ||
| YopEcoU | CGGAATTCGGTGGCTTATCAGAAAAA | This work |
| YopXbaL | GCTCTAGACCATGAGCCAGAAAGAACT | This work |
| BvfA-F | ACCCTTCGTCGATGTGCTGA | This work |
| BvfA-L | CCGCGCTGATTTCATCGCTG | This work |
| PSKU | GCGGTACCCTTCGTCGATGTGCTGA | This work |
| PSBL | GCGGATCCAGAAAAGCCAGGGAATTGA | This work |
The restriction sites are underlined.
Plasmid constructs. (i) pBBR1-YopP.
A 909-bp fragment containing the promoterless yopP gene was amplified from Yersinia enterocolitica pYVO8 (38) using primers YopEcoU and YopXbaL containing, respectively, EcoRI and XbaI sites. The PCR-amplified product was digested with EcoRI and XbaI and cloned into EcoRI/XbaI-digested pBBR1MCS4 (25). This plasmid was used for the library construction.
(ii) pBBR1-BvfA.
A 1,282-bp fragment encompassing the bvfA gene was amplified from B. suis 1330 genomic DNA with BvfA-F and BvfA-L primers with Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) and cloned into HincII-digested pBBR1MCS4 (25).
(iii) pBBR1-KGFPbvfA.
A 546-bp fragment containing the bvfA promoter region (pbvfA) was amplified from B. suis 1330 genomic DNA with the PSKU and PSBL primers, which respectively introduced KpnI and BamHI site ends of the fragment. This fragment was first cloned into pGEM-T (Promega, Madison, WI) and then subcloned into pBBR1-KGFP (24) on a KpnI/BamHI fragment, giving pBBR1-KGFPbvfA.
Tissue culture virulence models.
Three cell lines were used: the human monocyte-like cell line THP1, differentiated for 72 h in the presence of vitamin D3; the murine macrophage-like J774; and HeLa epithelial cells. The cells were cultivated and infected by a standard gentamicin protection assay as described previously (33). Inhibition of phagosome acidification with 100 nM bafilomycin (Sigma, St Louis, MO) was as described previously (37).
In vivo infection assay.
Two groups of 8-week-old female BALB/c mice were infected intraperitoneally with 2 × 105 CFU in 200 μl of phosphate-buffered saline. At 1, 2, 4, and 8 weeks postinfection, five mice were killed, spleens were harvested, and the bacterial load was determined by plating serial dilutions of spleen homogenates on BABSC plates supplemented with Brucella Selectavial antibiotics (MAST Diagnostics, Bootle, United Kingdom). Results are presented as geometric means ± 1 standard error.
FACS analysis.
To determine the effect of pH on bvfA and virB induction, bacteria were grown overnight at 37°C in an orbital shaker in modified minimal E medium (26) at pH 7.0, harvested by centrifugation, resuspended in E medium at pH 4.0 or 7.0, and then incubated further at 37°C to early exponential phase (optical density at 600 nm = 0.1). Bacteria were then harvested, resuspended in phosphate-buffered saline, and subjected to fluorescence-activated cell sorting (FACS) analysis with a FACScalibur using CELLQUEST software (Becton Dickinson, San Jose, CA) as described previously (22). For infection of J774 cells, bacteria were opsonized with a 1:100 dilution of mouse anti-B. suis serum and used at a multiplicity of infection of 100. At 3 h postinfection, monolayers were lysed and analyzed by FACS. Analysis by fluorescence microscopy was performed as described previously (19).
Statistical analysis.
Differences between the means of experimental groups were analyzed by using Student's t test (two tailed; equal variance). Differences were considered significant at P values of <0.05.
RESULTS
Identification of secreted YopP fusions.
In an attempt to detect effectors secreted by the VirB T4SS, we developed a novel reporter system using random translational fusions of Brucella DNA to the Yersinia enterocolitica yopP gene. Our strategy relies on the fact that the YopP reporter will only induce apoptosis when secreted by the bacterium as a fusion protein. In a preliminary experiment, we introduced a plasmid expressing the full-length YopP protein into B. suis 1330. The protein was synthesized by the resulting strain, but it did not induce the death of the infected macrophages, thus ruling out the possibility that it could be secreted directly by Brucella (data not shown). We then constructed a fusion library by insertion of B. suis 1330 partial digested DNA fragments upstream a promoterless yopP gene, and the recombinant plasmids were electroporated in a wild-type strain of B. suis 1330 expressing green fluorescent protein (GFP) constitutively. A bank of 2,400 clones was individually inoculated to THP1 cells, and the intracellular multiplication of the bacteria was verified by fluorescence microscopy at 48 h postinfection (23). Two clones which appeared unable to multiply intracellularly, 17E5 and 3G1, were identified. Their inability to multiply within THP1 was confirmed by viable counting. Several possible reasons could explain the attenuation of the two clones: the predicted induction of apoptosis, blocking of the secretion of a virulence factor by the hybrid protein (27), or a negative effect on bacterial fitness caused by expression of the YopP fusion. Using terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (17), the apoptotic level was still <0.5%, either in uninfected cells or in cells infected by the B. suis 1330 strain. In contrast, the 3G1 clone induced apoptosis, with approximately 10% of cells in an infected monolayer showing terminal nuclei positive by deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling. A similar result was obtained for the 17E5 clone (data not shown). The insert contained in the plasmid of each of the two clones was sequenced, and the sequence was used for BLAST searches of the Brucella melitensis 16 M and B. suis 1330 genomes (11, 34). The 17E5 insert contains the nirV gene, encoding a protein of unknown function, found in a cluster of genes encoding proteins involved in the denitrification process (3). This clone was not characterized further. Analysis of the 384-bp insert in clone 3G1 showed that yopP was fused to a gene (BMEI1366) encoding a hypothetical protein. With the B. suis 1330 annotation of the identical nucleotide sequence, this region is predicted to encode a protein on the complementary strand (BR0567), which would be fused in the wrong sense to yopP. As the gene fusion in the 3G1 clone encodes a chimerical protein with conserved YopP function, we prefer the B. melitensis annotation but we propose that the gene starts at the ATG at bp 52 of BMEI1366 rather than the predicted TTG, giving a 322-bp gene encoding a small basic protein of 111 amino acids with a molecular weight of 14011 and a predicted pI of 9.0. The protein is unique to Brucella, since PSI-BLAST searches failed to demonstrate any homology with any protein sequences in GenBank. Neither conserved domains nor known functional motifs were detected. The protein is most probably periplasmic, as it is predicted to have a Sec-dependent signal sequence with a cleavage site between residues 15 to 16 by the SignalP-2 program (http://www.cbs.dtu.dk/services/SignalP-2.0/). BMEI1366 is located on the larger of the two chromosomes and is present in all the Brucella species we have tested by Southern blotting (data not shown), including the new marine isolates (7).
Interestingly, in our screening, the apoptosis induced by the 3G1 but not the 17E5 fusion was dependent on the presence of a functional VirB system, which led us to believe that this protein was a possible VirB effector. Analysis of the 3G1 fusion suggested the presence of only a Sec-dependent signal sequence; control experiments, where YopP was delivered to the periplasm with an unrelated Sec-dependent signal sequence, also showed similar VirB-dependent induction of apoptosis, suggesting that this could be a nonspecific phenomenon (data not shown). However, this study has permitted the identification and characterization of a novel virulence factor, which we named BvfA, for Brucella virulence factor A.
B. suis bvfA is required for intracellular multiplication.
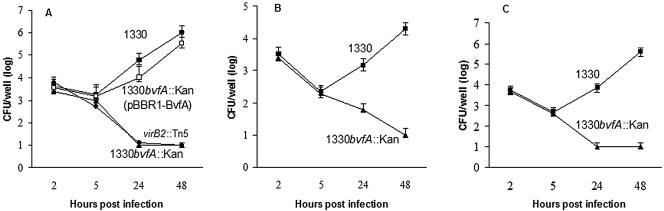
A null mutant (strain 1330bvfA::kan) (Table 1) was constructed in B. suis by allelic replacement (Fig. 1). The virulence of this strain was compared to that of the wild type and of the mutant transformed with plasmid pBBR1-BvfA, which carries the wild-type gene. The number of intracellular bacteria recovered 2 h and 5 h postinfection displayed no significant difference between the wild type and bvfA mutant, indicating that bvfA does not have any apparent role during invasion or resisting early macrophage killing. However, the bvfA mutant demonstrated an attenuated phenotype in both THP1, J774, and HeLa cells, with a significant reduction of intracellular multiplication at 24 h and 48 h postinfection (Fig. 2). The behavior was similar to that of a virB mutant (33). The presence of pBBR1-BvfA in the bvfA mutant fully restored the virulence of the mutant in the three cell lines (Fig. 2A and data not shown).
FIG. 1.
Inactivation of bvfA in Brucella suis by homologous recombination. Schematic representation of double crossing over between the chromosomal bvfA and the bvfA with a kanamycin resistance casette inserted at the unique BspMI site at bp 142108. The black arrow indicates the bvfA gene, and the gray arrows correspond to the neighbor genes. Numbers are the position in the B. suis genome sequence.
FIG. 2.
BvfA is required for the virulence of B. suis in the cell infection virulence models. Monolayers of THP1 macrophages (A) and HeLa (B) and J774 (C) cells were infected with 106 CFU of wild-type (filled squares), 1330bvfA::kan B. suis (filled triangles), 1330bvfA::kan with pBBR1-BvfA (open squares), or virB2::Tn5 (filled circles). Data points are geometric means of three wells with the standard error shown. This data set is representative of at least five independent experiments with similar results.
B suis bvfA mutants are attenuated in mice.
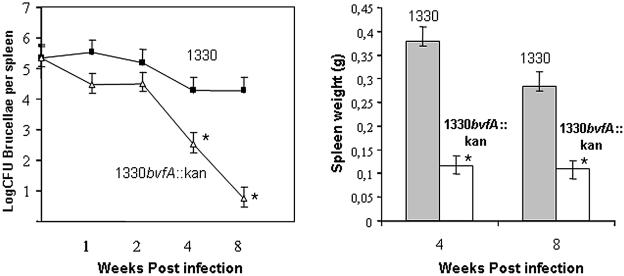
We used the murine model of brucellosis to assess the role of BvfA in vivo, infecting mice intraperitoneally with 2 × 105 CFU of the B. suis wild type or the bvfA mutant. No significant difference was detected between spleen counts of the wild type and the mutant during the first 2 weeks of infection (Fig. 3, left). However, at 4 weeks and 8 weeks postinfection, the numbers of CFU recovered from the spleens of mice infected with bvfA mutant were 1.7- and 3.5-log lower than those of the wild-type strain, respectively. The mutant was rapidly cleared from spleens, while the wild type persisted at high levels, showing that BvfA is required for full virulence. Analysis of spleen weights showed less splenomegaly in mice infected with the bvfA mutant between 2 and 8 weeks (Fig. 3, right). Attempts to complement the mutant were ambiguous due to plasmid instability in mice, a phenomenon previously encountered with the pBBR1-MCS4 plasmids (R. M. Roop II, personal communication).
FIG. 3.
BvfA is required for virulence of B. suis in mice. (Left) Brucella persistence in spleens of mice inoculated with B. suis 1330 wild-type and the 1330bvfA::kan strain. BALB/c mice were inoculated intraperitoneally with 2 × 105 CFU of B. suis 1330 wild types (filled squares) and the 1330bvfA::kan strain (open triangles). Five mice were used for each time point. The number of CFU in spleens was determined at different weeks postinfection (geometric means ± standard deviations of the means). (Right) Spleen weights of mice infected with B. suis 1330 and 1330bvfA::kan at 4 and 8 weeks postinfection.  , significantly different from the wild type (P < 0.05).
, significantly different from the wild type (P < 0.05).
The bvfA promoter is induced intracellularly.
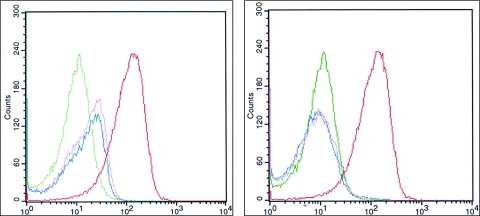
We have recently shown that the B. suis virB operon is induced in acidic minimal medium or by phagosome acidification after uptake by mammalian cells (5, 6). B. suis 1330 was electroporated with plasmid pBBR1-KGFPbvfA and pBBR1-KGFPvirB, which respectively contain the predicted promoter-regulatory region for bvfA and the putative virB promoter region cloned upstream of the gfp gene in a differential fluorescence induction promoter trap vector (23, 40). Transcription was analyzed by fluorescence microsocopy and FACS detecting GFP production. No fluorescence was detected by microscopy with bacteria grown overnight in tryptic soy broth (data not shown) and only very low levels of fluorescence were detected by FACS, showing that neither pvirB nor pbvfA was induced. Induction of pbvfA was observed in acid minimal medium in a manner similar to that of pvirB (data not shown). To determine whether pbvfA was induced within cells, J774 macrophages were infected with bacteria grown overnight in tryptic soy broth (5). At 3 h postinfection, infected cells were lysed, and supernatants containing Brucella were subjected to FACS (5). Here again, there was a clear induction of expression of GFP from both pbvfA and pvirB, with levels of fluorescence approaching that of the constitutive promoter control (Fig. 4, left panel).
FIG. 4.
bvfA is induced intracellularly in response to phagosome acidification. Gene expression was measured by FACS using transcriptional fusions to GFP. The bacteria were recovered from J774 macrophages 3 h postinfection of normal (left) or bafilomycin A (100 nM)-treated (right) macrophages as described previously (5). Expression from the bvfA promoter is shown in pink, pvirB is shown in blue, expression from the empty vector is shown in green, and expression from a constitutive promoter is shown in red.
Phagosome acidification has been shown to be essential for the intracellular multiplication of B. suis (37), and acidification is a stimulus required for virB operon expression within the phagosome (5). We investigated its influence on pbvfA induction. Inhibiting phagosome acidification with the proton pump inhibitor bafilomycin A1 blocked the induction of both pbvfA and pvirB, whereas the constitutive control remained fluorescent (Fig. 4, right). Analysis by fluorescence microscopy confirmed the FACS data (data not shown). Transcription of pbvfA was coregulated, with that of virB induced in acid minimal medium or by phagosome acidification after uptake by macrophages.
DISCUSSION
BvfA is essential for Brucella virulence in both in vitro and in vivo virulence models. How it plays this role is not at all clear; it is a small protein (a mass of 11 kDa is predicted for the mature protein), with no homologues in GenBank and no conserved domains or structural features (Prosite [http://au.expasy.org/prosite/]). The B. suis genome has 3,388 open reading frames, of which 42% are of unknown function, including 528 (15%) which are unique to the genus Brucella (34). To date, >10% of the 180 attenuated mutants reported for Brucella are in genes of unknown function, of which several are unique to Brucella or to the α-Proteobacteria (10). A systematic study is required to assess the role of these “orphans” in virulence.
The gene encoding BvfA (BMEI1366) is located on the large chromosome; neither of the genes surrounding bvfA, encoding a protease II-like protein and a Mn superoxide dismutase (SodA), have been reported to play a role in Brucella virulence. SodA has recently been shown to be required for full virulence of Mycobacterium tuberculosis (12). As the sodA gene downstream of bvfA is transcribed on the opposite strand and as the kanamycin-resistance cassette in 1330bvfA::kan does not interrupt the sodA coding sequence, we can exclude the possibility that the attenuation is due to a polar effect on sodA.
We found that bvfA is tightly regulated, with no expression in rich medium and induction in response to acid shock and within macrophages. While biochemical and differential fluorescence induction studies have identified a wide range of proteins induced by acid shock in vitro and genes induced intracellularly (10, 14, 29), only virB and bvfA have been shown to date to be required for virulence. The coregulation of bvfA with the virB operon, both in vitro and intracellularly, suggests that BvfA may also play a role in the establishment of the intracellular niche in which Brucella replicates.
BvfA was one of two potentially secreted proteins identified with a novel screen using the induction of apoptosis by translational fusions to the Yersinia YopP toxin as a readout. The first, which contained most of the NirV protein, appears to be secreted via the twin-arginine translocation system (J.-P. Lavigne and D. O'Callaghan, unpublished results). In the second, apoptosis was induced by YopP fused to the first 13 residues of BvfA. Induction of apoptosis by the strain expressing this fusion appeared to be dependent on a functional VirB system, suggesting at first glance that this protein was secreted via the T4SS. Analysis of this protein showed that the fusion had created a Sec-dependent signal sequence in the fusion, allowing YopP to be transported to the periplasm with the BvfA moiety cleaved by the signal peptidase. Control experiments, where YopP was delivered to the periplasm with other, unrelated Sec-dependent signal sequences, also showed VirB-dependent induction of apoptosis, suggesting nonspecific release of periplasmic YopP, rather than secretion directed by the fusion protein (data not shown). Nevertheless, how YopP is translocated from the bacterial periplasm to the cytoplasm of the infected cell remains to be elucidated. As most of the T4SS effector proteins studied have been shown to have a C-terminal signal (21, 30, 32, 39, 41), the secretion of the N-terminal YopP fusions argue against the conclusion that BvfA is a VirB effector. The C-terminal signal is thought to interact with the VirD4 coupling protein, which guides it to the secretion system; however, no VirD4 homologue in the Brucella genome has been identified. A similar situation exists in Bordetella pertussis, where the subunits of the pertussis toxin are transported to the periplasm by a Sec-dependent process where they assemble and are then secreted by the Ptl T4SS (8). As BvfA is predicted to be a periplasmic protein, it is not possible to rule out that it is part of a complex secreted from the periplasm by the Brucella T4SS. Further experiments are required to explore this hypothesis.
Acknowledgments
We thank C. Cazevieille for help with the apoptosis assays and V. Jubier-Maurin for help with the FACS analysis.
This work was supported by the Institut National de la Santé et de la Recherche Médicale and the European Community (QLK2-CT-2001-01200), la Région Languedoc-Roussillon, and the Université Montpellier I (BQR).
Editor: D. L. Burns
REFERENCES
- 1.Arenas, G. N., A. S. Staskevich, A. Aballay, L. S. Mayorga. 2000. Intracellular trafficking of Brucella abortus in J774 macrophages. Infect. Immun. 68:4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, E. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Sidman, J. A. Smith, K. Struhl. 1996. Currents protocols in molecular biology Wiley, New York, N.Y.
- 3.Baek, S. H., G. Rajashekara, G. A. Splitter, and J. P. Shapleigh. 2004. Denitrification genes regulate Brucella virulence in mice. J. Bacteriol. 186:6025-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 5.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. USA 99:1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boschiroli, M. L., V. Foulongne, and D. O'Callaghan. 2001. Brucellosis: a worldwide zoonosis. Curr. Opin. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]
- 7.Bricker, B. J., D. R. Ewalt, A. P. MacMillan, G. Foster, and S. Brew. 2000. Molecular characterization of Brucella strains isolated from marine mammals. J. Clin. Microbiol. 38:1258-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 9.Comerci, D. J., M. J. Martinez-Lorenzo, R. Sieira, J. P. Gorvel, and R. A. Ugalde. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159-168. [DOI] [PubMed] [Google Scholar]
- 10.Delrue, R. M., P. Lestrate, A. Tibor, J. J. Letesson, and X. De Bolle. 2004. Brucella pathogenesis, genes identified from random large-scale screens. FEMS Microbiol. Lett. 231:1-12. [DOI] [PubMed] [Google Scholar]
- 11.DelVecchio, V. G., V. Kapatral, N. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, et al. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards, K. M., M. H. Cynamon, R. K. Voladri, C. C. Hager, M. S. DeStefano, K. T. Tham, D. L. Lakey, M. R. Bochan, and D. S. Kernodle. 2001. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 164:2213-2219. [DOI] [PubMed] [Google Scholar]
- 13.Elzer, P. H., R. W. Phillips, M. E. Kovach, K. M. Peterson, and R. M. Roop II. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eskra, L., A. Canavessi, M. Carey, and G. Splitter. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 69:7736-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulongne, V., G. Bourg, C. Cazevieille, S. Michaux-Charachon, and D. O'Callaghan. 2000. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect. Immun. 68:1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foulongne, V., K. Walravens, G. Bourg, M. L. Boschiroli, J. Godfroid, M. Ramuz, and D. O'Callaghan. 2001. Aromatic compound-dependent Brucella suis is attenuated in both cultured cells and mouse models. Infect. Immun. 69:547-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavrieli, Y., Y. Sherman, and S. A. Ben-Sasson. 1992. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godfroid, F., B. Taminiau, I. Danese, P. Denoel, A. Tibor, V. A. Weynants, Cloeckaert, J. Godfroid, and J. J. Letesson. 1998. Identification of the perosamine synthetase gene of Brucella melitensis 16M and involvement of lipopolysaccharide O side chain in Brucella survival in mice and in macrophages. Infect. Immun. 66:5485-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross, A., A. Terraza, S. Ouahrani-Bettache, J. P. Liautard, and J. Dornand. 2000. In vitro Brucella suis infection prevents the programmed cell death of human monocytic cells. Infect. Immun. 68:342-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong, P. C., R. M. Tsolis, and T. A. Ficht. 2000. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect. Immun. 68:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubber, A., A. C. Vergunst, J. T. Sullivan, P. J. Hooykaas, and C. W. Ronson. 2004. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol. Microbiol. 54:561-574. [DOI] [PubMed] [Google Scholar]
- 22.Jubier-Maurin, V., A. Rodrigue, S. Ouahrani-Bettache, M. Layssac, M. A. Mandrand-Berthelot, S. Kohler, and J. P. Liautard. 2001. Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J. Bacteriol. 183:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kölher, S., V. Foulongne, S. Ouarhani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J. P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. USA 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kölher, S., S. Ouarhani-Bettache, M. Layssac, J. Teyssier, and J. P. Liautard. 1999. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect. Immun. 67:6695-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 26.Kulakov, Y. K., P. Guigue-Talet, M. Ramuz, and D. O'Callaghan. 1997. Response of Brucella suis 1330 and B. canis RM6/66 to growth at acid pH and induction of an adaptive acid tolerance response. Res. Microbiol. 148:145-151. [DOI] [PubMed] [Google Scholar]
- 27.Lee, V. T., and O. Schneewind. 2002. Yop fusions to tightly folded protein domains and their effects on Yersinia enterocolitica type III secretion. J. Bacteriol. 184:3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lestrate, P., R. M. Delrue, I. Danese, C. Didembourg, B. Taminiau, P. Mertens, X. De Bolle, A. Tibor, C. M. Tang, and J. J. Letesson. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543-551. [DOI] [PubMed] [Google Scholar]
- 29.Lin, J., T. A. Ficht. 1995. Protein synthesis in Brucella abortus induced during macrophage infection. Infect. Immun. 63:1409-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, Z. Q., and R. R. Isberg. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc. Natl. Acad. Sci. USA 101:841-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno, E., d aI. Moriyon. 2002. Brucella melitensis: a nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. USA 99:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai, H., E. D. Cambronne, J. C. Kagan, J. C. Amor, R. A. Kahn, and C. R. Roy. 2005. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc. Natl. Acad. Sci. USA 102:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 34.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayan, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pizarro-Cerda, J., E. Moreno, V. Sanguedolce, J. L. Mege, and J. P. Gorvel. 1998. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect. Immun. 66:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pizarro-Cerda, J., S. Meresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porte, F., J. P. Liautard, and S. Kölher. 1999. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect. Immun. 67:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruckdeschel, K., A. Roggenkamp, V. Lafont, P. Mangeat, J. Heesemann, and B. Rouot. 1997. Interaction of Yersinia enterocolitica with macrophages leads to macrophage cell death through apoptosis. Infect. Immun. 65:4813-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schulein, R., P. Guye, T. A. Rhomberg, M. C. Schmid, G. Schroder, A. C. Vergunst, I. Carena, and C. Dehio. 2005. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc. Natl. Acad. Sci. USA 102:856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 41.Vergunst, A. C., M. C. van Lier, A. den Dulk-Ras, T. A. Grosse Stuve, A. Ouwehand, and P. J. Hooykaas. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA 102:832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young, E. J. 1983. Human brucellosis. Rev. Infect. Dis. 5:821-842. [DOI] [PubMed] [Google Scholar]