Abstract
We have used a combination of in vitro selection and rational design to generate ribozymes that form a stable phosphoamide bond between the 5′ terminus of an RNA and a specific polypeptide. This reaction differs from that of previously identified ribozymes, although the product is analogous to the enzyme–nucleotidyl intermediates isolated during the reactions of certain proteinaceous enzymes, such as guanyltransferase, DNA ligase, and RNA ligase. Comparative sequence analysis of the isolated ribozymes revealed that they share a compact secondary structure containing six stems arranged in a four-helix junction and branched pseudoknot. An optimized version of the ribozyme reacts with substrate-fusion proteins, allowing it to be used to attach RNA tags to proteins both in vitro and within bacterial cells, suggesting a simple way to tag a specific protein with amplifiable information.
Keywords: in vitro selection‖nucleic acid tag‖phosphoamide‖Tat‖TAR
In vitro selection has been used in combination with rational design to identify and optimize numerous ribozymes and binding motifs (1–3). The results of these experiments continue to give insight into the capacity of RNA and DNA to catalyze a variety of chemical reactions and the ability of nucleic acids to recognize an array of substrates. Some of these catalysts are being developed into effective tools for use in molecular biology. These include allosteric ribozymes, used as detectors of small molecules and proteins, and DNA enzymes that efficiently cleave target sequences (4–6).
Here we use in vitro selection coupled with rational design to produce a new ribozyme that ligates RNA to protein through the formation of a phosphoamide bond. This is similar to a reaction found in naturally occurring proteins such as guanyltransferase, and DNA and RNA ligases that form phosphoamide bonds as enzyme-nucleotidyl intermediates during cap formation and ligation events (7–9). The selection of this new ribozyme expands the repertoire of RNA-catalyzed chemistry and further illustrates the ability of RNA to chemically modify a variety of substrates. Moreover, this ribozyme might be used to synthesize oligonucleotide-tagged proteins in vitro and in vivo. These attached oligonucleotides might be useful as affinity tags or could function as amplifiable identifying markers. For example, this ribozyme might be used to attach mRNAs to the proteins that they encode. A diverse library of mRNA–protein fusions could be used in conjunction with in vitro selection strategies to identify previously undescribed proteins with unique or enhanced properties, as has been shown previously by using phage display, ribosome display, and mRNA display (10–13).
Materials and Methods
Pool Construction.
The double-stranded DNA pool was constructed from two 110-nt DNA fragments, each containing 76 completely randomized positions (14). Each fragment was amplified separately in 110 ml PCR reactions and ligated through BanI sites to generate a final pool with the sequence: TTCTAATACGACTCACTATAGGACAGCTCCGAGCATTCTCGTGTAGCTCTGACCTTGG-N76-GGCACC-N76-ACGCACATCGCAGCAAC (italics, T7 promoter; underlined nucleotides, TAR site; -N76-, 76 nucleotide random-sequence segment). More than 1015 different sequences were transcribed to generate the initial RNA pool.
Peptide Synthesis.
MSY peptide, MSYSGPRPRGTRGKGRRIRRGG-B–am, and MKY peptide, MKYSGPRPRGTRGKGRRIRRGG-B-am [-B-, Lys (biotinyl-ɛ-aminocaproyl) (Bachem); am, amidate], were synthesized by using standard solid-phase synthesis. Formylated MSY peptide was synthesized from MSY peptide-containing resin by using 2,4,5-Trichlorophenyl formate (Bachem). Peptides were purified by reverse-phase HPLC with a C18 column and an acetonitrile gradient of 0.2% per min in 0.1% trifluoroacetic acid. The masses of the peptides were confirmed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry.
Round 1.
MSY peptide (90 nM) and pool RNA (75 nM) were combined in 112 ml of reaction buffer [50 mM Tris⋅OAc, pH 7.5/20 mM Mg(OAc)2/30 mM NH4OAc/100 mM KOAc). This reaction was incubated overnight and quenched with 20 mM EDTA. The reacted pool was precipitated overnight with 0.3M NaCl and 2.5 volumes of ethanol. RNA was pelleted by centrifugation, resuspended in 10 mM EDTA, and purified on a 5% denaturing polyacrylamide gel. The eluted RNA was precipitated, and reacted RNA was captured on 1.3 ml (1 nmol binding capacity) of streptavidin-coated paramagnetic beads (Promega) at ambient temperature and washed with 0.5× SSC. The final wash was with reverse transcriptase (RT) buffer (50 mM Tris⋅HCl, pH 8.3/75 mM KCl/3 mM MgCl2/10 mM DTT/0.5 mM each dNTP) supplemented with 5 μM primer 17.21, GTTGCTGCGATGTGCGT.
Reverse Transcription on Streptavidin Beads.
Streptavidin beads (Promega) were resuspended in 300 μl of RT buffer with 3,000 units of Superscript II (GIBCO/BRL) and incubated 1 h at 48°C. Beads were washed with 1 ml 0.1× SSC, and the cDNA was released by incubating in 150 mM KOH containing primer 17.21 and 5 μM primer 58.4 TTCTAATACGACTCACTATAGGACAGCTCCGAGCATTCTCGTGTAGCTCTGACCTTGG at ambient temperature for 20 min. The eluate was incubated for an additional 10 min at 90°C. This mix was neutralized with 150 mM Tris⋅HCl, and its final pH adjusted to 8.3. The cDNA was amplified by PCR, then used as a template for T7 transcription to generate RNA transcripts for the next round of in vitro selection. In subsequent rounds, the RNA was treated with DNase I (RQ1 DNase I, 12 units, Promega) before reacting the RNA with peptide. To assay pool activity, peptide and pool RNA were combined, reacted (30°C; 15 h; 50 mM Tris⋅OAc, pH 7.5/15 mM Mg(OAc)2/30 mM NH4OAc/100 mM KOAc/2 mM DTT/75 nM pool RNA/90 nM MSY peptide) and then mixed with an excess of streptavidin (Sigma). Reacted products were resolved on a 6% denaturing polyacrylamide gel.
Doped Pool Synthesis.
The 16.8 and 16.9 degenerate pools were based on the following DNA oligonucleiotides: ggacagctccgagcaTTCTCGTGTAGCTCTGACCATGGGGTAGGCTCT- TCGGAGCCGTATATAGTCACTTTGTCTTCGGACAATGCATCCCCCCAGG-N10-agatagtaagtgcaatct and ggacagctccgagcaTTCTCGTGTAGCTCTGACCATGGAGTACAGCTTCGGCTGGTATATGGCCAAGTACTTCGGTACTCATCCCTCCAAG-N10-agatagtaagtgcaatct (uppercase positions, 20% randomized; -N10-, 10 completely randomized nucleotides). Each pool contained more than 1014 sequences. Primers for PCR and reverse transcription were 18.65, AGATTGCACTTACTATCT and 35.8, TTCTAATACGACTCACTATAGGACAGCTCCGAGCA.
Round 1 Doped Selection.
Each pool was incubated overnight in a 22 ml reaction volume (30°C, 50 mM Tris-OAc, pH 7.5/5 mM Mg(OAc)2/100 mM KOAc/2 mM DTT/75 nM RNA pool/100 nM MKY peptide). The reaction was quenched with 6 mM EDTA (pH 8.0). These reactions were combined and concentrated to 1 ml by ultrafiltration (Amicon). The concentrated product was purified on an 8% denaturing polyacrylamide gel and eluted into 10 ml of 0.5× SSC. The eluate was captured on streptavidin paramagnetic beads and reverse transcribed and PCR amplified, as described for the initial selection.
Gel Selection Method.
In later rounds of selection, reacted products were isolated from 10% denaturing polyacrylamide gels. Ribozyme was incubated with peptide for 14 min followed by the simultaneous addition of Mg(OAc)2 and an excess of a specific RNA competitor (final reaction mixes: 50 mM Tris-OAc, pH 7.5/15 mM Mg(OAc)2 100 mM KOAc/2.5 mM octyl glucoside/2 mM DTT/0.5 mM EDTA, pH 8.0/1 μM tRNA/75 nM RNA pool/100 nM MKY peptide/100 μM 12.3, GGACAGCUCCGAGCACUAGCUCGUGGUAGCUCUGACCAUAGAGUACCAGCUUCGGGCUGGGUAUAUGGCCAAGUAUUUCGUUACUCAUCCCUCCAAG). Reactions were immediately stopped (25 mM EDTA/8 M urea), and streptavidin was added to a final concentration of 2.5 μM before loading onto the gel (10% polyacrylamide/8 M urea; 1.5 mm, 28 Watts, 1 h). The shifted product was visualized with a PhosphorImager (Molecular Dynamics) and excised from the gel. This product was eluted and ethanol-precipitated overnight, then purified from a 10% denaturing polyacrylamide gel. The product was excised, eluted, then reverse transcribed and PCR amplified.
Ribozyme Assays.
DNA encoding ribozymes were generated by PCR using the appropriate primers and DNA templates. RNA for these ribozymes was generated by T7 transcription (15). Unless otherwise noted, the observed rate constants (kobs) for self-ligation (or single-turnover transligation) were determined at saturating peptide concentration (typically 40 nM peptide). MSY or MKY peptide was mixed with radiolabeled ribozyme in reaction buffer (30°C, 50 mM Tris⋅OAc, pH 7.5/15 mM Mg(OAc)2/100 mM KOAc/2 mM DTT/2.5 mM octyl glucoside/1 μM tRNA/40 nM MSY peptide or MKY peptide/0.01 nM or 1 nM ribozyme). Aliquots were removed at the indicated time and the reaction quenched (8 M urea/25 mM EDTA/1 μM 21.4). Products were separated on a 20% denaturing polyacrylamide gel. The Tag-Hisx6-FLAG THF–GFPuv fusion protein (H2N-MKYSGPRPRGTRGKGRRIRRGGQLHHHHHHV- DYKDDDDK fused to the amino terminus of GFPuv, CLONTECH; Fig. 4A; GFP, green fluorescent protein) was expressed in pTHF–GFPuv transformed cells (BL21-Gold, DE3, Stratagene) and purified on a nickel affinity resin (Ni-NTA, Qiagen). In reactions with purified protein, the THF–GFPuv protein was incubated with cordycepin-labeled ribozyme (30°C, 50 mM Tris⋅OAc, pH 7.5/15 mM Mg(OAc)2/100 mM KOAc/2 mM DTT/1 μM tRNA/2.5 mM octyl glucoside/100 nM THF–GFPuv/30 nM ribozyme). Products were resolved on a 7% SDS polyacrylamide gel. Protein assayed for intracellular coupling activity was prepared under denaturing conditions. THF–GFPuv protein expression was induced (0.3 OD, 4 h, 37°C). Cells were pelleted and then lysed in a French press (lysis and capture buffer, 20 mM Tris⋅HCl, pH 7.6/150 mM NaCl/10 mM imidazole/6 M Guanidinium⋅HCl). Clarified lysate was mixed with nickel containing resin (Ni-NTA, 0.3 ml, Qiagen) and protein captured (60 min, 4°C). The resin was loaded onto a column and successively washed (10 column volumes, buffer B: 20 mM Tris⋅HCl, pH 7.6/1 M NaCl/6 M urea) and (10 column volumes, buffer C: 20 mM Tris⋅HCl, pH 7.6/150 mM NaCl/30 mM imidazole). Protein was eluted with buffer D (20 mM Tris⋅HCl, pH 7.6/150 mM NaCl/200 mM imidazole) and buffer was exchanged by ultrafiltration. After the labeling reaction [3 μg THF–GFPuv, yeast poly(A) polymerase (United States Biochemicals), 730 units, 30°C, 30 min] unincorporated cordycepin was removed and buffer exchanged with a combination of size-exclusion chromatography (sephadex G-50, Boehringer Mannheim) and ultrafiltration (50 mM Tris⋅HCl, pH 7.5, Amicon). One-half of the cordycepin-labeled material from cells overexpressing pTHF–GFPuv and 21.4 was treated with 2 μg of Proteinase K (Stratagene) for 1.5 h. Radiolabeled products were resolved on a 7% SDS polyacrylamide gel. The RNA substrate of the 16.trans ribozyme was transcribed from two annealed oligonucleotides (15), gel-purified, and radiolabeled with cordycepin. Ribozyme and substrate were mixed together in water, incubated at 80°C for 2 min, and allowed to cool to ambient temperature. Reaction buffer and peptide were added to start the reaction [40 nM RNA substrate/200 nM 16.trans/200 nM MSY peptide/100 mM KOAc/2 mM DTT/50 mM Tris·OAc, pH 7.5/15 mM Mg(OAc)2]. These conditions may be subsaturating for a construct that contains an altered binding site. Aliquots were removed, quenched with 0.1M EDTA, and products were resolved on a 15% denaturing polyacrylamide gel.
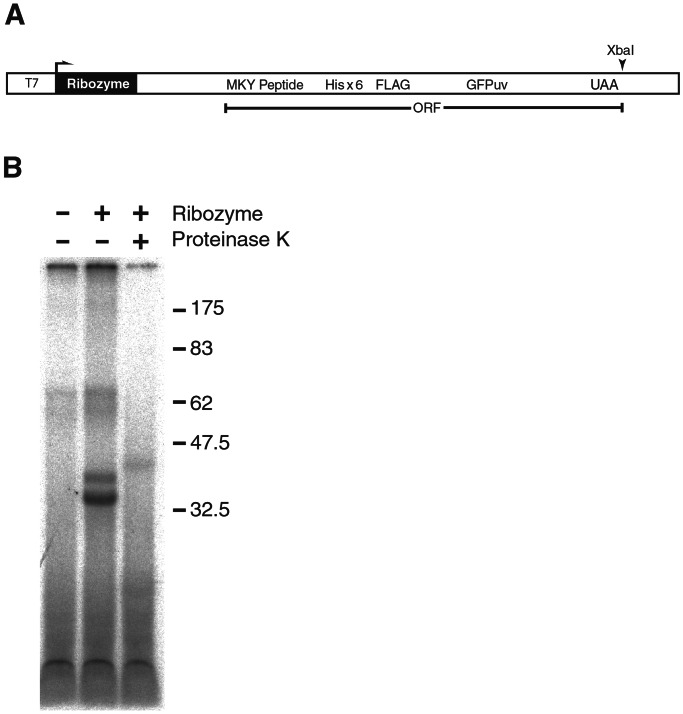
Figure 4.
(A) DNA template to generate RNAs containing both a ribozyme and an ORF encoding the polypeptide substrate of the ribozyme. Plasmid pTHF–GFPuv encodes the ribozyme and MKY peptide fused to six histidine residues, the FLAG epitope, and GFPuv (CLONTECH). The T7 polymerase promoter (T7) facilitated in vitro transcription and over expression in the appropriate bacterial strain. (B) Intracellular RNA-protein ligation. Histidine-tagged protein from E. coli cells overexpressing pTHF–GFPuv encoding both the ribozyme and THF–GFPuv substrate (+Ribozyme) or an analogous plasmid that had the ribozyme deleted (−Ribozyme) were affinity purified, and RNA fragments covalently attached to the protein were labeled at their 3′-end with cordycepin. Half of the cordycepin-labeled material isolated from cells overexpressing 21.4 and THF–GFP protein was treated with Proteinase K.
Pyrophosphate Release Assay.
Ribozyme 21.4 that had been transcribed in the presence of γ-32P GTP was incubated with MKY peptide [30°C, 50 mM Tris⋅OAc, pH 7.5/15 mM Mg(OAc)2/150 nM ribozyme/300 nM MKY peptide], reaction aliquots were removed and spotted onto PEI cellulose at the indicated times, and pyrophosphate was resolved with 0.35 M HCl.
Mass Spectrometry.
Reacted ribozyme was incubated with substrate (2 h, 30°C, 50 mM Tris⋅OAc, pH 7.5/15 mM Mg(OAc)2/4 nmol ribozyme/3 nmol MKY peptide, 20 ml), concentrated by ultrafiltration to 190 μl (10,000 molecular weight, Amicon), and treated with a combination of 0.6 units P1 (Calbiochem) and 10 units T1 ribonuclease (GIBCO/BRL) (4 h, 45°C). The masses of the peptide and the digested reacted product were determined by MALDI-TOF mass spectrometry with a PerSeptive Voyager-DE STR.
Results and Discussion
Pool Design.
The first step toward identifying ribozymes with RNA–protein ligation activity was to design a diverse library of RNA molecules. To favor the binding of a specific polypeptide to each member of the starting pool and to place nucleophiles near a potentially reactive 5′-triphosphate, a high-affinity peptide-binding site was incorporated near the 5′-end of the library. Only a few characterized RNA-peptide interactions have the affinity, specificity, and stoichiometry suitable for this purpose (16). One of these is BIV-1 Tat/TAR (Fig. 1A) (17–19). On binding to the TAR site, the Tat peptide adopts a β-hairpin conformation nestled deep within the major groove of the TAR RNA (20, 21). Using structural information as a guide, a permuted BIV-1 TAR site was placed near the 5′-end of every molecule within an RNA pool. The permuted TAR site was followed by a block containing 152 randomized positions (Fig. 1B) (14, 22).
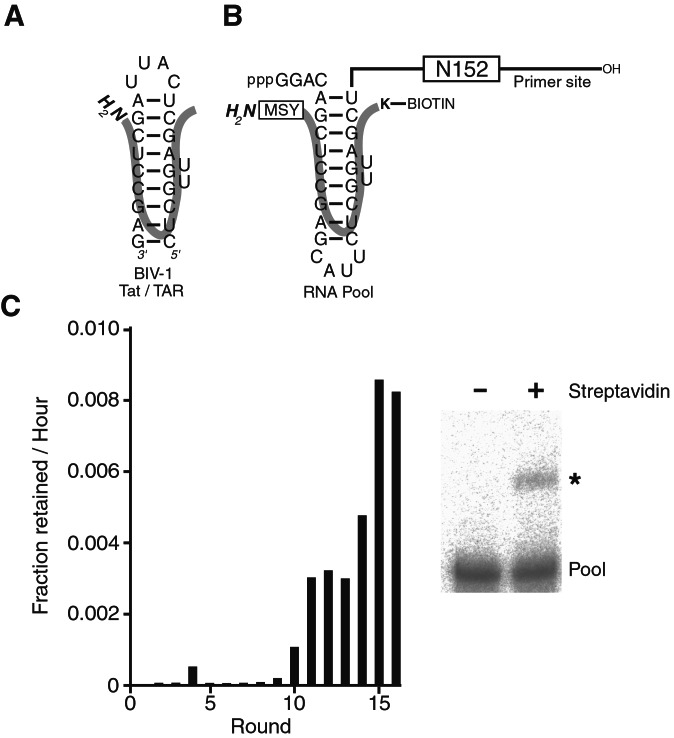
Figure 1.
(A) BIV-1 Tat/TAR. (B) Pool design. The block containing 152 random positions (N152) was flanked by defined segments used as primer-binding sites for reverse transcription and amplification of selected molecules. A circularly permuted BIV-1 TAR site was placed near the 5′-end of the RNA pool. The orientation of the BIV-1 Tat peptide bound to the TAR site is shown (thick line). A biotinylated lysine within the polypeptide served as an affinity tag for the selection of reacted molecules (K-Biotin). Residues were appended to the amino terminus of the wild-type peptide to increase the diversity of potential nucleophiles within the peptide (boxed MSY). (C) Progress of the selection. The amount of RNA retained on the streptavidin beads was quantitated during the course of selection. After 9 rounds, this amount increased to levels where the reacted product could be visualized on a denaturing polyacrylamide gel (asterisk).
The detailed model of the BIV-1 Tat/TAR complex allowed the placement of potential nucleophiles within the peptide proximal to the 5′-triphosphate of the pool RNA, with the idea that one or more of the pool molecules might be capable of promoting the formation of a covalent linkage between a nucleophile of the bound peptide and the α-phosphate of the RNA 5′-triphosphate. Others have attempted to position substrates within pools by using similar strategies (22, 23). Placing the substrate proximal to a leaving group of the pool molecules is advantageous if this eliminates the need to select substrate-binding activity from the random-sequence pool so that the pool diversity can be more fully applied to task of fine-tuning the substrate orientation and promoting the chemical transformation. The risk of this strategy is that the substrate will be inadvertently placed in an orientation incompatible with catalysis. In an effort to allow some flexibility in the orientation of the reacting moieties and to increase the diversity of potential nucleophiles within the peptide, the segment GGAC was inserted between the 5′-triphosphate and TAR stem, and the tripeptide MSY, which contains three potential nucleophiles (the α-NH2 moiety and two hydroxyl groups) was appended to the amino terminus of the Tat peptide (Fig. 1B).
Ribozyme Selection.
The RNA pool was incubated with a BIV-1 Tat peptide that had been biotinylated near its carboxyl terminus. Reacted ribozymes within the pool formed a ribozyme–peptide–biotin product that was captured on streptavidin beads. In subsequent rounds, the reacted RNA pool was reverse transcribed before capture on streptavidin beads. Selection-amplification was repeated until the reacted fraction increased to a level where it was possible to observe ribozyme activity with a denaturing polyacrylamide gel assay (Fig. 1C). The selection was continued for seven additional rounds, and the population was cloned and sequenced.
Isolates from this population fell into two sequence families. Closer examination revealed a set of nucleotides shared between the two families, as seen when sequence families share the same fold (5, 24, 25). Indeed, both ribozyme families have the potential to assume the same secondary structure (Fig. 2). This shared fold simplified the process of establishing the boundaries of the ribozyme structure; segments common to the two families were assumed to be important, whereas divergent elements were deemed extraneous and eliminated. Deleting the 3′-terminal 107 nucleotides and replacing loop sequences with stable tetra-loops had no effect on the reaction rate of ribozyme 16.9 (Fig. 2, 16.min). This finding supported the assertion that the two families shared the proposed secondary structure and yielded a compact molecule for further optimization.
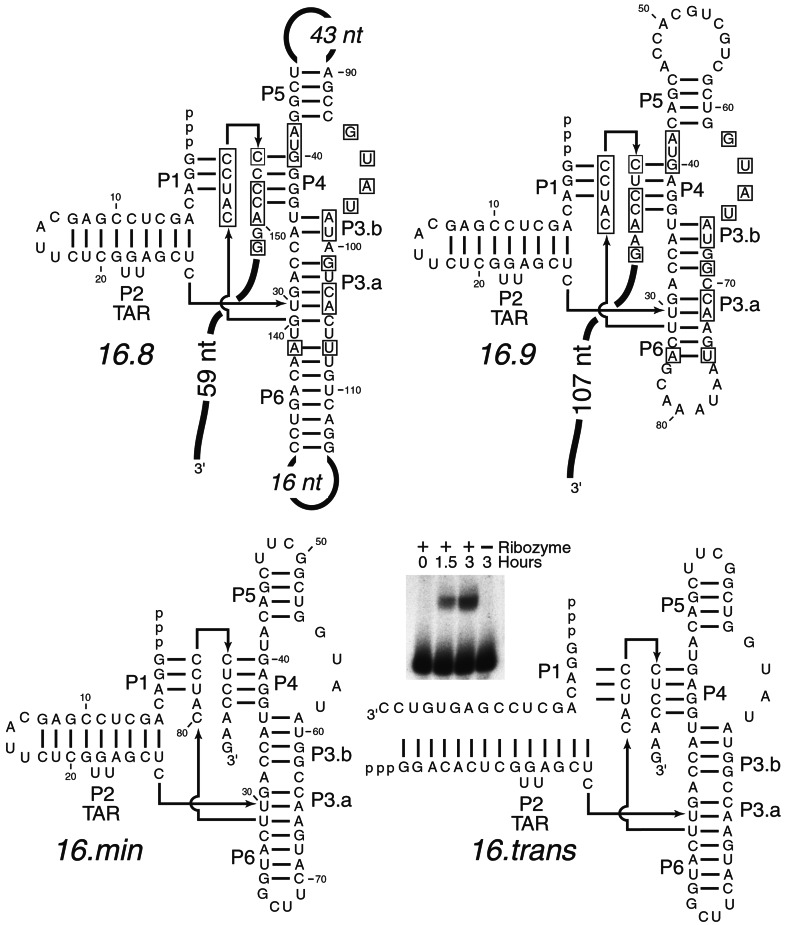
Figure 2.
Secondary structure of the ribozyme. Comparative analysis was used to establish the secondary structure of the ribozyme. Representative clones are shown from each family isolated from round 16 (16.8, 16.9). Nucleotides shared between families are boxed. The 16.9 ribozyme was truncated to a minimal form (16.min) without detectable loss of activity. 16.min was synthesized as a two piece trans-acting ribozyme split within stem P2 (16.trans). Short oligonucleotide-peptide fusions can be detected as retarded products in a denaturing polyacrylamide gel when 16.trans reacts with its polypeptide substrate (Inset).
Some in vitro-selected ribozymes have been engineered to generate trans-acting catalysts (26–28). To test whether 16.min could react in trans, the ribozyme was split in the loop sequence of stem P2 to generate a two-piece molecule (Fig. 2, 16.trans). In this format, any reacted peptides will be ligated to a short oligonucleotide. Oligonucleotide–peptide fusions are synthesized in a time-dependent and ribozyme-dependent fashion (Fig. 2, Inset), although the observed rate constant (kobs) is somewhat reduced (16.min, 0.13 h−1; 16.trans, 0.038 h−1). In the future it might be possible to expand this reaction to include oligonucleotides containing modifications, such as fluorescent tags, or longer oligonucleotides of diverse sequence, which could be used as unique amplifiable identifying tags.
Characterization and Optimization.
One potential reaction catalyzed by this ribozyme is the formation of a phosphoamide bond between the amino terminus of the peptide and the 5′-α-phosphate of the RNA. This is consistent with the observation that a peptide substrate formylated at its amino terminus did not react with the minimized ribozyme within limits of detection (<2.8 × 10−6 min−1). Furthermore, the product of the reaction, while very stable at physiological conditions, is sensitive to acid hydrolysis (80% acetic acid, t1/2 = 10 h) a property indicative of a phosphoamide bond (29). In an effort to enrich for rare ribozymes that react with other nucleophiles within the polypeptide, the selection was continued with pool RNA from round 10 for six additional rounds of selection-amplification using a formylated polypeptide, but no active molecules were isolated. One explanation for this observation is that ribozymes that form a phosphodiester bond by reacting with other nucleophiles, such as the hydroxyl groups of Ser or Tyr at positions 2 and 3 in the polypeptide might not have been present in the initial population. Alternatively, these molecules might have been present but were not substantially enriched, perhaps because of low activity.
Forming a phosphoamide bond between the amino terminus of the substrate peptide and the 5′-α-phosphate would restrict the use of this ribozyme to protein substrates that contained unmodified amino termini. In Escherichia coli and other bacteria, protein synthesis is initiated with formyl methionine, a modification that would prevent the reaction of the polypeptide with the selected ribozyme. In addition, some processing enzymes, namely the methionine amino peptidases, can remove the amino-terminal methionine of many proteins (30). These alterations of the peptide substrate might prevent it from reacting with the ribozyme when expressed in cells or in translation extracts. Thus, to maintain the integrity of the amino terminus, the second residue of the polypeptide substrate was changed from Ser (MSY peptide) to Lys (MKY peptide). This modification allows deformylation in E. coli and inhibits the removal of the terminal methionine in rabbit reticulocyte extracts (31, 32). This substitution was tolerated by the ribozyme, although the kobs of the reaction was reduced 7.3-fold from 0.0022 min−1 (MSY peptide) to 0.0003 min−1 (MKY peptide).
To find ribozyme variants that reacted more readily with the MKY substrate, each of the two ribozyme families was synthesized as a pool of partially degenerate molecules and selected as before, but using the altered peptide substrate. In later rounds, ribozymes that attached themselves to biotinylated substrate were selected based on a streptavidin gel shift rather than affinity to streptavidin-coated beads. After an additional 21 cycles of selection-amplification, the population was cloned, and sequencing identified variants of three different closely related sequences. The fastest isolate assayed, 21.4, was magnesium-dependent and reacted with kobs = 0.013 min−1 (Fig. 3 A and B). This was a 6-fold improvement over the 16.min ribozyme reacting with the original MSY peptide, and a 43-fold enhancement when compared with the activity of 16.min ribozyme with the MKY peptide. The apparent Km for the MKY substrate was 4 nM, with concentrations of peptide greater than 40 nM inhibiting the ribozyme (data not shown).
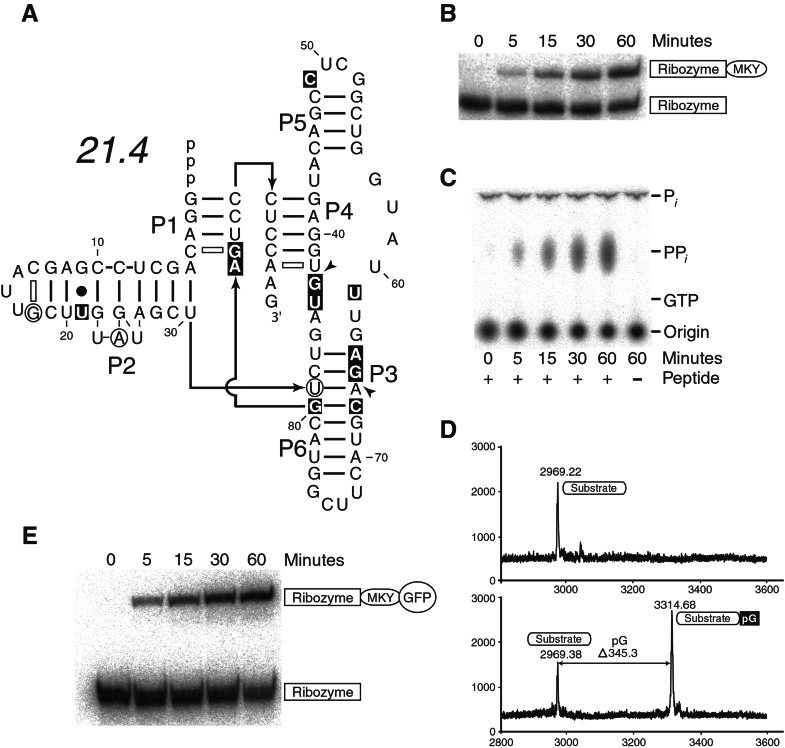
Figure 3.
An improved ribozyme. (A) The secondary structure of the fastest ribozyme variant isolated after randomization and reselection. Circled nucleotides represent insertions; arrowheads indicate positions where deletions have occurred; substitutions are highlighted with shaded boxes. Additional potential pairing is indicated with open dashes. (B) RNA-peptide ligation. MKY peptide was reacted with 21.4, and reacted products were resolved on a denaturing polyacrylamide gel. (C) Ribozyme-mediated pyrophosphate (PPi) release. (D) Mass of reacted product. Peptide and ribozyme were reacted and then treated with a combination of ribonuclease T1 and P1. The masses of the unreacted peptide (Upper) and reacted nuclease-treated peptide (Lower) were determined by MALDI-TOF mass spectrometry. (E) RNA-protein ligation. Purified protein containing the MKY peptide fused to GFPuv protein was reacted with 21.4, and reacted products were resolved on a SDS polyacrylamide gel.
When compared with the 16.min isolate, 21.4 has 16 mutations, including 3 insertions, 2 deletions and 11 substitutions (Fig. 3A). These mutations cluster around the central region of the molecule and within the TAR site. Interestingly, one of these mutations, the insertion of an adenosine at position 24 of the ribozyme, is similar to a modification previously identified in the wild-type TAR site that increases the affinity of the site ≈3-fold (33). The higher affinity was attributed to the creation of a less constrained TAR conformation or increased major groove accessibility (33). Other mutations clustered in the central region of the molecule proximal to stem 3. They might reflect the accommodation of the Lys side chain, although the improved isolate retains a slight preference for the MSY peptide (MSY peptide, kobs = 0.04 min−1, MKY peptide, kobs = 0.013 min−1).
The reaction catalyzed by 21.4 shares the properties of the 16.min ribozyme. Pyrophosphate is released, and the reaction product is sensitive to acid hydrolysis, indicating that the chemistry of the reaction remains unchanged (Fig. 3C). In addition, a peptide substrate formylated at its amino terminus did not react with the ribozyme (formylated MSY peptide, <0.00012 min−1; the trace activity of the substrate, attributed to contaminating nonformylated peptide, represents the upper limit for the reaction rate with formylated peptide).
If a covalent bond forms between the ribozyme and the polypeptide substrate, it should be possible to isolate the modified polypeptide and determine the mass of the product. Accordingly, ribozyme was reacted with polypeptide and the RNA digested with a mixture of P1 and T1 ribonucleases. The modified peptide had a mass consistent with the formation of a covalent phosphoamide bond between the peptide and the α-phosphate of guanosine found at the 5′-terminus of the ribozyme (Fig. 3D). In total, the data support a reaction mechanism where a peptide amine attacks the 5′-α-phosphate of the ribozyme, forming a phosphoamide bond with the concomitant release of pyrophosphate. The inactivity of the fMet containing polypeptide implicates the amino terminus of the substrate as the nucleophile but does not definitively rule out side-chain amines.
To examine whether the peptide substrate was accessible in the context of a larger protein, it was fused to the amino terminus of green fluorescent protein, in a format similar to one that might be encountered when attaching a nucleic acid tag to a protein (34). The GFP-substrate fusion protein (THF–GFPuv) was purified on a nickel affinity resin and reacted with the ribozyme. The fusion protein reacts with the ribozyme with a rate indistinguishable from that of the peptide alone, demonstrating the ability to attach RNA to long heterologous proteins (Fig. 3E). In addition, this experiment demonstrates that the biotin moiety, used earlier for selection and detection of product, is not required for peptide recognition or catalysis.
Alternatively, derivatives of this ribozyme might eventually be used to covalently attach ribonucleic acids to specific proteins in a cell. This could target an RNA sequence to the destination of the protein, alter or suppress the function of the protein, or possibly be used to create an RNA scaffold that could bring two proteins together in a controlled fashion. To test whether this ribozyme could function within a cell, 21.4 was overexpressed as part of an mRNA that encoded its polypeptide substrate, THF–GFPuv (Fig. 4A). Extensive RNA degradation by cellular nucleases was expected under these conditions, and any remaining fragments of RNA covalently attached to the protein that contain a free 3′-hydroxyl were expected to be substrates for radiolabeling by yeast poly(A) polymerase and α-32P-cordycepin triphosphate. THF–GFPuv was purified on a nickel affinity resin under denaturing conditions. Purified protein was radiolabeled and protein resolved on an SDS polyacrylamide gel (Fig. 4B). The mobility of the major product is identical to the that of unmodified protein, implying that a majority of the RNA covalently attached to the protein is being hydrolyzed to short fragments of RNA that have little effect on the mobility of the protein in the SDS gel. The slower migration of the minor product might result from a larger RNA fragment remaining attached to the protein or from an alternative conformer of the GFP protein. Previously, derivatives of naturally occurring ribozymes have been shown to act in vivo, as has an artificial DNA enzyme (35, 36). The observed in vivo activity of the 21.4 ribozyme demonstrates that ribozymes selected de novo by using combinatorial in vitro methods are catalytically active when expressed within cells and suggests that such ribozymes might be useful for the controlled regulation of cellular processes.
Although the formation of a phosphoamide bond between a protein and RNA is unique among ribozymes, this type of bond is found as an intermediate in guanyltransferase and both DNA and RNA ligases. The isolation of this activity from a population of random sequence adds to the diversity of chemical transformations that might have originated as RNA but were then transferred to protein. In addition, the demonstration that a ribozyme can be used to connect the genetic information of nucleic acids to proteins presents an opportunity to apply RNA catalysis toward the affinity tagging, detection, and encoding of polypeptides.
Acknowledgments
We would like to thank Peter Unrau for the single-stranded DNA used for the construction of the initial pool, Mike Burgess for peptide synthesis, and Chuck Merryman and members of the Bartel lab for their many helpful comments and suggestions. S.B. was supported by postdoctoral fellowships from the National Science Foundation/Alfred P. Sloan Foundation and a National Research Service Award from the National Institutes of Health.
Abbreviations
- GFP
green fluorescent protein
- THF
the leader peptide expressed from the GFPuv fusion construct (Tag-Hisx6-FLAG)
- MALDI-TOF
matrix-assisted laser desorption ionization–time-of-flight
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wilson D S, Szostak J W. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D P, Unrau P J. Trends Cell Biol. 1999;9:M9–M13. [PubMed] [Google Scholar]
- 3.Marshall K A, Ellington A D. Methods Enzymol. 2000;318:193–214. doi: 10.1016/s0076-6879(00)18053-x. [DOI] [PubMed] [Google Scholar]
- 4.Carmi N, Balkhi S R, Breaker R R. Proc Natl Acad Sci USA. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson M P, Ellington A D. Nat Biotechnol. 1999;17:62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 6.Robertson M P, Ellington A D. Nucleic Acids Res. 2000;28:1751–1759. doi: 10.1093/nar/28.8.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong P, Shuman S. J Biol Chem. 1993;268:7256–7260. [PubMed] [Google Scholar]
- 8.Fresco L D, Buratowski S. Proc Natl Acad Sci USA. 1994;91:6624–6628. doi: 10.1073/pnas.91.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thogersen H C, Morris H R, Rand K N, Gait M J. Eur J Biochem. 1985;147:325–329. doi: 10.1111/j.1432-1033.1985.tb08753.x. [DOI] [PubMed] [Google Scholar]
- 10.Fujita S, Sawata S Y, Yamamoto-Fujita R, Endo Y, Kise H, Iwakura M, Taira K. J Med Chem. 2002;45:1598–1606. doi: 10.1021/jm010398p. [DOI] [PubMed] [Google Scholar]
- 11.Roberts R W, Szostak J W. Proc Natl Acad Sci USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattheakis L C, Bhatt R R, Dower W J. Proc Natl Acad Sci USA. 1994;91:9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith G P. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 14.Unrau P J, Bartel D P. Nature (London) 1998;395:260–263. doi: 10.1038/26193. [DOI] [PubMed] [Google Scholar]
- 15.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 16.Frankel A D. Curr Opin Struct Biol. 2000;10:332–340. doi: 10.1016/s0959-440x(00)00092-0. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Frankel A D. Proc Natl Acad Sci USA. 1995;92:5077–5081. doi: 10.1073/pnas.92.11.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Frankel A D. Biochemistry. 1994;33:2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- 19.Reyes C M, Nifosi R, Frankel A D, Kollman P A. Biophys J. 2001;80:2833–2842. doi: 10.1016/S0006-3495(01)76250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puglisi J D, Chen L, Blanchard S, Frankel A D. Science. 1995;270:1200–1203. doi: 10.1126/science.270.5239.1200. [DOI] [PubMed] [Google Scholar]
- 21.Ye X, Kumar R A, Patel D J. Chem Biol. 1995;2:827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 22.Bartel D P, Szostak J W. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 23.Lorsch J R, Szostak J W. Nature (London) 1994;371:31–36. doi: 10.1038/371031a0. [DOI] [PubMed] [Google Scholar]
- 24.Ekland E H, Szostak J W, Bartel D P. Science. 1995;269:364–370. doi: 10.1126/science.7618102. [DOI] [PubMed] [Google Scholar]
- 25.Sassanfar M, Szostak J W. Nature (London) 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 26.Breaker R R, Joyce G F. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 27.Carmi N, Breaker R R. Bioorg Med Chem. 2001;9:2589–2600. doi: 10.1016/s0968-0896(01)00035-9. [DOI] [PubMed] [Google Scholar]
- 28.Johnston W K, Unrau P J, Lawrence M S, Glasner M E, Bartel D P. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 29.Venkatesan S, Moss B. Proc Natl Acad Sci USA. 1982;79:340–344. doi: 10.1073/pnas.79.2.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meinnel T, Mechulam Y, Blanquet S. Biochimie. 1993;75:1061–1075. doi: 10.1016/0300-9084(93)90005-d. [DOI] [PubMed] [Google Scholar]
- 31.Boissel J P, Kasper T J, Bunn H F. J Biol Chem. 1988;263:8443–8449. [PubMed] [Google Scholar]
- 32.Meinnel T, Patiny L, Ragusa S, Blanquet S. Biochemistry. 1999;38:4287–4295. doi: 10.1021/bi982622r. [DOI] [PubMed] [Google Scholar]
- 33.Smith C A, Crotty S, Harada Y, Frankel A D. Biochemistry. 1998;37:10808–10814. doi: 10.1021/bi980382+. [DOI] [PubMed] [Google Scholar]
- 34.Crameri A, Whitehorn E A, Tate E, Stemmer W P. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 35.Samarsky D, Ferbeyre G, Bertrand E. Curr Issues Mol Biol. 2000;2:87–93. [PubMed] [Google Scholar]
- 36.Sun L Q, Cairns M J, Gerlach W L, Witherington C, Wang L, King A. J Biol Chem. 1999;274:17236–17241. doi: 10.1074/jbc.274.24.17236. [DOI] [PubMed] [Google Scholar]