Abstract
Vibrio cholerae encodes a small RNA with homology to Escherichia coli RyhB. Like E. coli ryhB, V. cholerae ryhB is negatively regulated by iron and Fur and is required for repression of genes encoding the superoxide dismutase SodB and multiple tricarboxylic acid cycle enzymes. However, V. cholerae RyhB is considerably longer (>200 nucleotides) than the E. coli RNA (90 nucleotides), and it regulates the expression of a variety of genes that are not known to be regulated by RyhB in E. coli, including genes involved in motility, chemotaxis, and biofilm formation. A mutant with a deletion in ryhB had reduced chemotactic motility in low-iron medium and was unable to form wild-type biofilms. The defect in biofilm formation was suppressed by growing the mutant in the presence of excess iron or succinate. The wild-type strain showed reduced biofilm formation in iron-deficient medium, further supporting a role for iron in normal biofilm formation. The ryhB mutant was not defective for colonization in a mouse model and appeared to be at a slight advantage when competing with the wild-type parental strain. Other genes whose expression was influenced by RyhB included those encoding the outer membrane porins OmpT and OmpU, several iron transport systems, and proteins containing heme or iron-sulfur clusters. These data indicate that V. cholerae RyhB has diverse functions, ranging from iron homeostasis to the regulation of biofilm formation.
Iron plays a critical role in the cellular metabolism of almost all living organisms. Iron is required for processes as diverse as the tricarboxylic acid (TCA) cycle, electron transport, DNA metabolism, and response to oxidative stress. Because iron has the potential for catalyzing production of reactive oxygen species, excess iron can also pose a significant problem. The influx and intracellular fate of iron must therefore be tightly regulated. This is achieved in part through the action of the iron-dependent negative regulator Fur, which functions to coordinate the iron status of the cell with the expression of genes involved in iron transport, storage, and metabolism. Under iron-replete conditions, Fur complexes with the ferrous ion and blocks transcription of its regulon by binding to conserved regions termed Fur boxes within the promoter region of these genes. There is another layer of complexity in the scheme of iron- and Fur-dependent regulation. In Escherichia coli, certain genes involved in iron storage, iron metabolism, and antioxidant defense appear to be positively regulated by Fur (6, 36, 42), and this was recently shown to be mediated through the action of a small RNA (sRNA), RyhB (26). RyhB negatively regulates the expression of sodB (encoding superoxide dismutase), ftn and bfr (encoding ferritin and bacterioferritin), and several iron-sulfur cluster-containing TCA cycle enzyme genes, including the sdh operon (encoding succinate dehydrogenase) and acnA (encoding aconitase). Because RyhB is itself negatively regulated by Fur, the net effect is positive regulation of these genes under high-iron conditions, which promote the formation of the active Fur-Fe2+ complex.
Positive regulation by Fur via an sRNA is not unique to E. coli. A similar system, which encodes two functional homologs of RyhB, PrrF1 and PrrF2, is present in Pseudomonas aeruginosa (59). Although there is no detectable sequence conservation between E. coli RyhB and the PrrF sRNAs, the P. aeruginosa sRNAs control many of the same types of targets as RyhB, including sodB, sdh, and a probable bacterioferritin gene, PA4880.
The mechanism of regulation by RyhB (reviewed in reference 12) is posttranscriptional and involves base pairing between RyhB and its mRNA target and subsequent RNase E-mediated degradation of the RyhB-mRNA duplex (25, 33). Like many reactions involving RNA-RNA interactions, this requires the Hfq protein. Hfq is thought to function as an RNA chaperone that facilitates formation of the sRNA-mRNA duplex by controlling access of the sRNA to the complementary regions of the target mRNA (11). Another role of Hfq may be to protect the sRNA, as well as its mRNA targets, from degradation by blocking sites in the RNA that would otherwise be accessible to RNase E (25, 33). Once duplexing between RyhB and its target mRNA has occurred, RNaseE cleavage sites presumably become exposed, since the entire complex is then rapidly degraded (25, 33).
Vibrio cholerae, the causative agent of the diarrheal disease cholera, encodes a homolog of the E. coli Hfq protein (5, 22). One strain of V. cholerae was shown to use an Hfq-dependent sRNA pathway to regulate quorum sensing and virulence gene expression (22). Interestingly, Hfq, although needed for virulence, did not appear to modulate expression of known virulence factors in a different V. cholerae strain (5). It is likely that a complex and strain-specific hierarchy of virulence gene regulation involving Hfq and multiple sRNAs exists in this pathogen.
V. cholerae has been shown to exert iron- and Fur-dependent negative regulation on many genes involved in iron acquisition (3, 17, 45) (A. R. Mey, E. E. Wyckoff, V. R. Kanukurthy, L. R. Fisher, and S. M. Payne, unpublished data). There have also been reports of positive regulation of V. cholerae protein expression by iron-bound Fur (23). In addition, a fur mutant of V. cholerae was unable to use pyruvate, succinate, and fumarate as carbon sources, signaling possible defects in certain TCA cycle pathways (23). This suggests that V. cholerae may use a system analogous to the E. coli RyhB mechanism for regulating genes encoding iron-containing proteins and those involved in iron metabolism. Indeed, Masse and Gottesman (26) found a region exhibiting significant sequence similarity to E. coli ryhB in a BLAST search of the V. cholerae genome sequence (16) using the complete E. coli ryhB sequence. In this study, we demonstrate iron- and Fur-dependent expression of the predicted V. cholerae RyhB sRNA and show that, in addition to recognizing many of the same targets as E. coli RyhB, V. cholerae RyhB has some unique functions, including the iron-dependent regulation of biofilm formation.
MATERIALS AND METHODS
Bacterial strains and plasmids and media.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were maintained at −80°C in tryptic soy broth plus 20% glycerol. The strains were routinely grown at 37°C in Luria (L) broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl) or on L agar; in Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% NaCl) or on LB agar; in EZ Rich Defined Medium (EZ RDM) (http://www.genome.wisc.edu/functional/protocols.htm), a modification of the Supplemented MOPS Defined Medium described by Neidhardt et al. (34); or in EZ RDM with no added iron (iron limited but not iron depleted; it supports growth of V. cholerae while causing induction of the Fur regulon [Mey et al., unpublished]). Sucrose (0.2% final concentration) was added to EZ RDM as the carbon source. APW no. 3 medium was made as described by Ali et al. (1). Iron-replete medium was prepared by the addition of ferrous sulfate (FeSO4), and iron-depleted medium was prepared by the addition of ethylenediamine-di-(o-hydroxyphenylacetic acid) (EDDA), deferrated by the method of Rogers (44). The concentrations of FeSO4 and EDDA added varied depending on the medium and assay conditions used and are listed separately for each experiment in the relevant Materials and Methods section or figure legend. Antibiotics were used at the following concentrations for E. coli: 250 μg of carbenicillin per ml, 50 μg of kanamycin per ml, and 30 μg of chloramphenicol per ml. For V. cholerae, the concentrations were 125 μg of carbenicillin per ml, 25 μg of kanamycin per ml, and 7.5 μg of chloramphenicol per ml. Polymyxin B was used at 10 μg per ml, and streptomycin was used at 75 μg per ml. Electroporation of V. cholerae strains was carried out as described previously (37).
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| O395 | V. cholerae classical biotype | 28 |
| ARM572 | O395 ryhBΔ::kan | This study |
| ARM573 | O395 furΔ::tmp | This study |
| N16961 | V. cholerae E1 Tor biotype | J. Kaper |
| SAC101 | N16961 mshAΔ::kan | This study |
| ARM711 | N16961 ryhBΔ::kan | This study |
| E. coli | ||
| DH5α(λpir) | Cloning strain; host strain for pGP704 derivatives | J. Kaper |
| SM10(λpir) | λpir recA Kanr; host and mobilizing strain for pGP704 derivatives | 32 |
| Plasmids | ||
| pQE-2 | IPTG-inducible expression vector | Qiagen |
| pHM5 | Suicide vector pGP704 carrying sacB; Cbr Sucs | 46 |
| pWKS30 | Low-copy-number cloning vector; Cbr | 53 |
| pQF50 | Promoterless lacZ reporter plasmid | 8 |
| pAML20 | hutA-lacZ transcriptional fusion in pQF50 | 29 |
| pAML23 | ryhB-lacZ transcriptional fusion in pQF50 | This study |
| pAMR70 | Full-length ryhB in pQE-2 | This study |
| pAMR72 | Full-length ryhB in pWKS30 | This study |
| pAMR73 | 5′-truncated ryhB (ryhBΔ5′) in pWKS30 | This study |
| pAMS10 | pHM5 carrying ryhBΔ::kan | This study |
| pAMS12 | pHM5 carrying furΔ::tmp | This study |
| pSAC10C | pHM5 carrying mshAΔ::kan | This study |
PCR.
The oligonucleotide primers for PCR (Table 2) were purchased from IDT Inc. (Coralville, IA). PCR was performed using Taq polymerase (QIAGEN) or Pfu polymerase (Stratagene) according to the manufacturer's instructions. Bacterial cultures grown overnight were used as the template. All clones derived from PCR fragments were verified by sequencing.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Primer sequence (5′-3′)a |
|---|---|
| bfd1 | CGTTTGTTTGTGTCATGGTGTCTC |
| bfd2 | TGAGCAATCAGCGATTCTTCG |
| fur1 | TGGGCGTCGACCAGCCAAGAGCC (SalI) |
| fur2 | GTGAGCGTTAACTTTAAGACCAGC (HpaI) |
| fur3 | GGTCTTAAAGTTAACGCTCACAAGCC (HpaI) |
| fur4 | GGAGGCGCTGATATCCATTTGGC (EcoRI) |
| mshA1 | GGGAGATCTTTCAGCGAAAGCGAATAGTGG (BglII) |
| mshA2 | ATAGTCGACCCATTGCACCAGCAACTGCACC (SalI) |
| mshA3 | AAAGTCGACCCTGCAACGGTTGCTATGC (SalI) |
| mshA4 | CCCTCTAGAGTGGTTACCACCGCAAAGG (XbaI) |
| polyT-ClaI | CCATCGATTTTTTTTTTTTTTTTTT (ClaI) |
| sodB1 | ATTTCCACCACGGTAAGCACC |
| sodB2 | CCGAAGTTGTTGATGGCTGAATC |
| vryhB1 | TGTGTCGACTGATGAAACAACGG (SalI) |
| vryhB2 | ATGTCGGTTAACCTTCACTTGTTCCC (HpaI) |
| vryhB3 | AAGTGAAGGTTAACCGACATCTTCGG (HpaI) |
| vryhB4 | TACCGTAGGTCGACATCAACGC (SalI) |
| vryhB5 | GGAATCCATGGTCGAGAGCC (NcoI) |
| vryhB6 | TTCACTGGATCCCTAAGACG (BamHI) |
| vryhB8 | CACGTCTTAGGGAACAAGTG |
| vryhB10 | GCTAAAAATTACACTGGAAGC |
| vryhB12 | AGAATTCACGTCTTAGGGAACAAGTG (EcoRI) |
| vryhB13 | GCCAAGCTTACGAGGTCAAAGCC |
| vryhB14 | TTCAAGCTTGTCCACTCATTGC |
| vryhB19 | ATCGAATTCCTCGATATGCGGTAGCG (EcoRI) |
| vryhB20 | GCTCGATATCAAAACGTTCTACAGC (EcoRV) |
| vryhB21 | CGCATATCGGTTAATAATAATAGTTCTC |
| vryhB22 | TATTATTAACCGATATGCGGTAGCG |
| vryhB23 | TGATGGATCCGCCAGTGGATGAG (BamHI) |
Any relevant recognition sites for restriction enzymes (listed in parentheses) are underlined.
Promoter fusion studies.
The predicted promoter region for V. cholerae ryhB was amplified by PCR using primers vryhB5 and vryhB6 (Table 2). The PCR fragment was digested with NcoI and BamHI and cloned into pQF50 digested with NcoI and BamHI to yield pAML23. Following transfer of pQF50, pAML20, and pAML23 into O395 and ARM573 by electroporation, the transformed strains were grown to mid-log phase in iron-replete LB broth containing 40 μM FeSO4 or in iron-depleted LB broth containing 12.5 μg EDDA per ml, and β-galactosidase activity was measured as described by Miller (30).
Construction of chromosomal mutations in V. cholerae.
Deletion of the V. cholerae ryhB gene was carried out by overlap extension PCR with primer pairs vryhB1-vryhB2 and vryhB3-vryhB4 to amplify overlapping fragments, followed by amplification of the overlap extension product with primer pair vryhB1-vryhB4 to create the final ryhBΔ allele containing a unique HpaI site. The PCR fragment was digested with SalI and cloned into the SalI site of pWKS30. The resulting plasmid was digested with HpaI, and the kanamycin resistance cassette from pUC4K was inserted into this site as a Klenow-blunted SalI fragment. The ryhBΔ::kan fragment was then excised using SalI and cloned into the λpir-dependent suicide vector pHM5 digested with SalI to create pAMS10. pAMS10 was conjugated from DH5α(λpir) into the relevant V. cholerae strain using the helper strain MM294/pRK2013, and allelic exchange was performed as described previously (29). To create a deletion within the V. cholerae fur gene, primer sets fur1-fur2 and fur3-fur4 were used to synthesize overlapping fragments, and primers fur1 and fur4 were used to amplify the overlap extension product. The furΔ product containing a unique HpaI site was digested with EcoRV and SalI and cloned into pWKS30 digested with EcoRV and SalI. The resulting plasmid was digested with HpaI to allow insertion of an SmaI fragment derived from pMTLtmp (E. E. Wyckoff) containing a trimethoprim resistance cassette. The furΔ::tmp allele was cloned as an EcoRV/XhoI fragment into pHM5 digested with EcoRV and SalI to yield pAMS12, and pAMS12 was transferred to V. cholerae for allelic exchange as described above. To create the deletion in the mshA gene, a fragment containing just the 5′ end of mshA was PCR amplified using primers mshA1 and mshA2, digested with BglII and SalI, and cloned into pHM5 digested with BglII and SalI to create pSAC10A. A second PCR fragment containing just the 3′ end of the mshA gene was synthesized using primers mshA3 and mshA4, digested with SalI and XbaI, and cloned into pSAC10A digested with SalI and XbaI to yield pSAC10B. The kan cassette from pUC4K was inserted as a SalI fragment into the SalI site of pSAC10B to generate pSAC10C. pSAC10C was transferred from SM10(λpir) into N16961 by conjugation, and allelic exchange was carried out as described above. All mutations were verified by PCR.
Construction of V. cholerae RyhB expression plasmids.
The full-length V. cholerae ryhB gene was amplified by PCR using primers vryhB12 and vryhB14. The PCR product was digested with EcoRI and HindIII and cloned into pQE-2 digested with MfeI and HindIII for IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible expression of ryhB in pAMR70. Induction of this construct was achieved using 200 μM IPTG. To express ryhB from its native promoter, a PCR fragment, generated using primers vryhB1 and vryhB4, was digested with PvuII and MscI and cloned into pBlueskript-SK (Stratagene) digested with SmaI. The ryhB insert was then excised using BamHI/EcoRV and cloned into pWKS30 digested with BamHI/EcoRV to yield pAMR72. To express the 5′-deleted ryhB allele (missing 60 nucleotides at the 5′ end compared to full-length ryhB in pAMR72) (Fig. 1A) from its native promoter, primer sets vryhB20-vryhB21 and vryhB22-vryhB23 were used to create overlapping fragments containing the deletion, and primers vryhB20 and vryhB23 were used to amplify the overlap extension product. This fragment was cloned as a BamHI/EcoRV fragment into pWKS30 digested with BamHI/EcoRV to yield pAMR73.
FIG. 1.
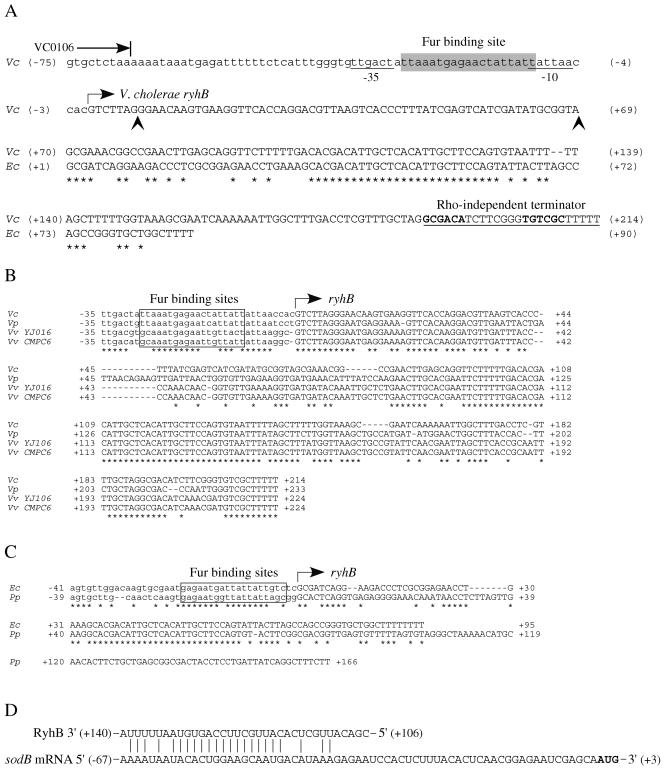
Sequence features of the ryhB locus in V. cholerae. (A) Organization of V. cholerae ryhB and alignment with E. coli ryhB. The end of the upstream open reading frame VC0106 is marked with a blunted arrow. The promoter region, including a −35 and a −10 sequence (underlined), is shown in lowercase letters, while the V. cholerae (Vc) and E. coli (Ec) ryhB genes are shown in uppercase letters. The proposed Fur binding sequence is shaded in gray. The right-angled arrow indicates the start of V. cholerae ryhB, as predicted by sequence analysis and confirmed by 5′ RACE. The vertical arrowheads delineate the sequence deleted from full-length ryhB to create the 5′-deleted ryhB allele (ryhBΔ5′) in pAMR73. The left vertical arrowhead also marks the 3′ end of the sequence included in the lacZ promoter fusion constructs (the 5′ end of that sequence corresponds to the −101 position of the sequence shown). The numbers flanking the sequences indicate the nucleotide position relative to the start of ryhB (+1). Identical residues are marked with asterisks. (B) V. cholerae ryhB sequence features are conserved among members ofthe Vibrionaceae. The ryhB sequences from V. cholerae N16961 (Vc), Vibrio parahaemolyticus (Vp), Vibrio vulnificus YJ016 (Vv YJ016), and Vibrio vulnificus CMPC6 (Vv CMPC6) were aligned. The proposed Fur binding sites are boxed, and the start of ryhB is indicated by the right-angled arrow. The numbers flanking the sequences indicate the nucleotide positions relative to the start of ryhB (+1). Asterisks mark identical residues. (C) The putative Photobacterium profundum ryhB has characteristics of E. coli ryhB. An alignment of the putative P. profundum (Pp) ryhB gene with E. coli ryhB is shown. The predicted Fur binding sites are boxed, and the start of ryhB is indicated with a right-angled arrow. The numbers flanking the sequences show the nucleotide positions relative to +1. (D) V. cholerae RyhB exhibits complementarity with the V. cholerae sodB mRNA upstream region. Complementary nucleotides are marked with vertical lines. The numbers flanking the RyhB sequence correspond to the numbering scheme used for V. cholerae ryhB in panel A. The numbers flanking the sodB mRNA sequence refer to the nucleotide positions relative to the +1 position in the sodB transcript.
Sequence analysis.
DNA sequencing was performed by the University of Texas Institute for Cellular and Molecular Biology DNA Core Facility using an ABI Prism 3700 DNA Sequencer. Analysis of DNA sequences was carried out using MacVector 7.1.
RNA isolation and Northern analysis.
Overnight cultures were diluted 1:200 into fresh LB broth containing 5 μg of EDDA per ml and grown to an optical density at 650 nm of 0.8. Cells were harvested, and RNA was extracted using RNeasy midi-columns (QIAGEN) according to the manufacturer's instructions. Northern hybridization was carried out using the NorthernMax system (Ambion). Briefly, 10 μg of each RNA sample was separated in a 1.8% denaturing agarose gel, transferred to a positively charged nylon membrane (BrightStar-Plus; Ambion) by downward capillary transfer, and subjected to UV light cross-linking (GS Gene Linker UV chamber; Bio-Rad). The ryhB probe was generated by PCR using primers vryhB8 and vryhB13, and the sodB probe was made by PCR using primers sodB1 and sodB2. The DNA probes were labeled using the BrightStar Psoralen-Biotin Nonisotopic Labeling Kit (Ambion). Following hybridization, the biotinylated probes were visualized using the BrightStar Biodetect Nonisotopic Detection Kit (Ambion). Equal loading of the RNA samples was determined by ethidium bromide staining and visualization under UV light.
5′ RACE.
5′ rapid amplification of cDNA ends (RACE) was performed as described by Frohman et al. (9) with modifications. Total RNA was isolated from V. cholerae strains grown to late log phase in EZ RDM with no added iron or in LB broth containing 5 μg of EDDA per ml as described above. Five micrograms of each RNA sample was used as a template for a reverse transcriptase (RT) reaction with primer vryhB13 (25 pmol) using Superscript II RT (Invitrogen) according to the manufacturer's instructions. The resulting cDNA was purified over a Mini Elute column (QIAGEN), and A tailing of the cDNA was carried out using terminal deoxynucleotidyltransferase (Promega) as described previously (9). The postreaction volume was brought to 100 μl with 10 mM Tris-HCl, pH 8.0-1 mM EDTA, and 1 μl of this was used in the subsequent amplification steps. The A-tailed cDNA products were amplified as follows. After denaturation at 94°C for 5 min and annealing of primer polyT-ClaI at 50°C for 2 min, the products were extended at 72°C for 40 min. This was followed by a standard PCR amplification using primers polyT-ClaI and vryhB10 in 30 cycles of 94°C for 30 seconds, 50°C for 30 seconds, 72°C for 1 min, and a final extension at 72°C for 7 min. The amplified products were resolved on a 1.5% agarose gel, purified over QIAquick columns (QIAGEN), and sequenced using primer vryhB10.
Microarrays.
The V. cholerae microarray slides were generated at the University of Texas at Austin Microarray Facility using a set of oligonucleotides (Oligator Custom DNA Synthesis; Illumina) representing the complete genome of V. cholerae N16961 (16) plus V. cholerae ryhB. The arrays were printed by a robotic arrayer onto poly-l-lysine-coated glass slides. A set of V. cholerae microarray slides was also obtained from the Pathogen Functional Genomics Resource Center at the Institute for Genomic Research. To isolate total RNA, bacterial strains were grown to mid- or late log phase in EZ RDM with no added iron (N16961 strains) or in LB plus 5 μg/ml EDDA (O395 strains), and equal numbers of bacteria were harvested from the reference and the test strain by centrifugation. Total RNA was extracted from the cells using RNeasy midi-columns (QIAGEN) according to the manufacturer's instructions. With the use of 15 μg of total RNA per sample, cDNA was synthesized in the presence of amino-allyl deoxyuridine triphosphates in an RT reaction with Superscript II. Cy3 fluorescent dye (Amersham Biosciences) was coupled to the reference sample; Cy5 was coupled to the test sample. Following purification of the two labeled cDNA samples using the Mini Elute PCR purification kit (QIAGEN), a cDNA probe mixture was generated and applied to the array surface for hybridization at 65°C for 4 h. After hybridization, the arrays were washed, dried, and then scanned using a GenePix 4000 B scanner (Axon Instruments). The fluorescence intensities were determined using the GenePix Pro 4.2 software package. The Longhorn Array Database (http://chipmunk.icmb.utexas.edu/ilat/) was used to perform data filtering and analysis (19). Only gene features (“spots”) that passed certain quality control filters, including minimum intensity and pixel consistency, were included for further analysis. Differential expression was considered significant if the | log2R | was ≥1, where R is the normalized red/green ratio.
Biofilm assays.
The biofilm assays were performed as described by O'Toole and Kolter (39) with modifications. Single colonies from freshly streaked LB agar plates were inoculated into LB broth with the appropriate antibiotics and incubated at 37°C with shaking overnight. The overnight cultures were diluted 1:100 into EZ RDM with no added iron and further incubated overnight at 37°C with shaking. The EZ RDM low-iron medium was chosen in order to more closely match the conditions used for the microarrays. These cultures were freshly diluted approximately 1:50 into EZ RDM (with or without additional supplements, as described in the relevant figure legends), diluting each strain proportionally to match the inoculum size of the wild-type strain. For each strain, 200 μl of cell suspension was added per well in a 96-well polystyrene plate (Corning catalog no. 3788) and incubated at 30°C for 20 h without shaking. Following this incubation, the cell suspension within each well was mixed by pipetting, and the optical density was measured to assess the planktonic growth of each strain. To visualize the biofilm, the planktonic cells were removed by aspiration, and 200 μl of freshly made 1% crystal violet solution was added to each well. After incubation for 30 min at room temperature, the crystal violet solution was removed by aspiration, and each well was rinsed twice with distilled H2O. To solubilize the dye for spectrophotometric readings, 200 μl absolute ethanol was added to each well. The plates were incubated for 20 min at room temperature, and the absorbance (A595) was measured in a Microplate Autoreader EL311s (Bio-Tek Instruments).
Chemotaxis assays.
The chemotactic-motility assays were performed as described by Gardel and Mekalanos (10). Briefly, an inoculating wire was used to stab single colonies into chemotaxis agar (L broth plus 0.3% agar). The plates were incubated at 37°C for 8 to 10 h, and the diameter of the test strain swarm zone was compared with the diameter of the parental-strain swarm zone.
Isolation of V. cholerae rugose colonies.
To promote switching of N16961 to the rugose morphotype, cultures were grown statically for 24 h at 37°C in APW no. 3 and then plated on L agar and incubated for 24 h at 30°C, as described by Ali et al. (1). The rugose colonies were identified visually.
In vivo competition assays.
In vivo competition assays were performed using 5-day-old BALB/c mice as described by Taylor et al. (49). Prestarved mice were inoculated intragastrically with 50 μl saline containing 0.5% sucrose, 0.02% Evan's Blue dye, and 2 × 105 CFU of each competing strain grown to mid-log phase. The mice were sacrificed after 24 h, and the small intestines were isolated and homogenized in sterile phosphate-buffered saline. Serial dilutions were plated on media selective for all viable V. cholerae cells and then replica plated on differential media to determine the viable counts for each strain. The output ratios were normalized to the input ratios to determine the competitive index.
RESULTS
Organization and sequence features of the ryhB locus in V. cholerae.
The putative V. cholerae ryhB gene is located on the large chromosome (V. cholerae chromosome I) between open reading frames VC0106 and VC0107, both of which encode hypothetical proteins. Over the 90-nucleotide stretch of V. cholerae ryhB that aligns with E. coli ryhB, the V. cholerae ryhB gene shares 56% identity with E. coli ryhB, and the predicted V. cholerae RyhB sRNA contains the highly conserved 30-nucleotide region that forms the central stem-loop structure (26) in E. coli RyhB (Fig. 1A). Based on an alignment of putative ryhB genes from several gram-negative species, a promoter region, including a Fur binding region, and a transcriptional start site had been assigned to the V. cholerae ryhB gene (26). However, inspection of the nucleotide sequence shows that the proposed Fur binding site is a poor match to Fur boxes found in V. cholerae, as well as in E. coli (40), suggesting that V. cholerae ryhB may not be regulated by Fur or may have a Fur-regulated promoter elsewhere. Analysis of the sequence upstream of ryhB indicates that the latter may be the case. A sequence bearing significant identity to several known Fur boxes in V. cholerae (74% identity to the candidate Fur boxes of vibA [60] and viuP [61]) and in E. coli (79% identity to the fepB Fur box [7]; 74% identity to the iucA Fur box) and flanked by potential −35 and −10 sequences is present nearly 100 bases upstream of the previously predicted +1 site (Fig. 1A). This suggests that V. cholerae ryhB may be transcribed from an iron-regulated promoter upstream of the previously predicted ryhB start site, potentially adding almost 70 nucleotides to the 5′ end of the V. cholerae ryhB transcript compared with the E. coli ryhB transcript. V. cholerae RyhB may also be longer than E. coli RyhB at the 3′ end; the first potential Rho-independent terminator sequence for V. cholerae ryhB commences approximately 33 nucleotides downstream of the previously predicted 3′ end of the ryhB gene (26), suggesting that the V. cholerae RyhB sRNA carries a 3′ extension compared to E. coli RyhB (Fig. 1A). Based on these new predictions for the location of a Fur-regulated promoter upstream of ryhB and a Rho-independent terminator downstream of ryhB, we calculate the length of the V. cholerae RyhB sRNA to be greater than 200 nucleotides, compared with 90 nucleotides for E. coli RyhB.
Among the Vibrionaceae sequenced to date, the longer RyhB seems to be the predominant form. An alignment of V. cholerae ryhB with putative ryhB genes from Vibrio parahaemolyticus and two strains of Vibrio vulnificus, YJ016 and CMPC6, identified in BLAST searches of the completed genomes (4, 20, 24) using V. cholerae ryhB, shows that the promoter region, including the predicted Fur binding sequence, as well as the predicted Rho-independent terminator sequence, is highly conserved among these Vibrio species (Fig. 1B). At 233 nucleotides, the predicted V. parahaemolyticus RyhB is the longest, followed by V. vulnificus RyhB at 224 nucleotides and V. cholerae RyhB at 214 nucleotides. These predicted RyhB sRNAs all contain the highly conserved central region found in E. coli RyhB but also exhibit significant homology throughout the predicted 5′ region, suggesting that there may be a common function for this segment of the RyhB molecule. One member of the sequenced Vibrionaceae, Photobacterium profundum, has a putative ryhB gene (identified in a BLAST search of the completed P. profundum genome [52] using V. cholerae ryhB) that is more similar to E. coli ryhB than to V. cholerae ryhB, with an almost identical Fur box and a very similar organization of the 5′ end (Fig. 1C). It is not clear how large this RyhB sRNA is, however, since no obvious terminator was found downstream of the highly conserved region (Fig. 1C and data not shown). These findings suggest that, while certain functions of RyhB are likely highly conserved among the gram-negative organisms, other functions may be specific to families and even to genera within the same family.
Regulation of V. cholerae ryhB expression by iron and Fur.
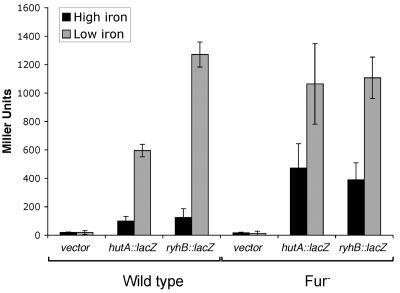
To determine whether the candidate Fur-regulated promoter upstream of the V. cholerae ryhB gene is functional, a 108-base-pair fragment, including the predicted Fur box and −35 and −10 sequences, but none of the downstream sequence (Fig. 1A), was cloned upstream of the promoterless lacZ gene in pQF50 to create pAML23. As a control, a similarly constructed transcriptional fusion between lacZ and the iron- and Fur-regulated promoter for hutA (17), pAML20 (29), was included in the study. V. cholerae strains carrying the promoter fusion constructs were grown to mid-log phase in either high- or low-iron conditions, and β-galactosidase activities were measured. In the wild-type strain O395, the hutA and ryhB promoters were both strongly induced in response to iron starvation, with the hutA promoter fusion exhibiting a 6-fold increase in expression and the ryhB promoter fusion showing a >10-fold increase in expression in iron-limited media (Fig. 2). In the absence of fur, the level of repression in response to high iron was markedly decreased, showing that both of these fusions respond to the presence of iron and Fur. These data indicate that the sequence upstream of ryhB in V. cholerae functions as an iron- and Fur-regulated promoter. Interestingly, loss of fur did not completely abolish regulation of these fusions by iron, indicating that there may be other iron-responsive regulatory elements operating in V. cholerae (Fig. 2). This result is consistent with other V. cholerae studies showing residual iron regulation of Fur-controlled genes in the absence of Fur (17, 23).
FIG. 2.
Regulation of V. cholerae ryhB by iron and Fur. The activity of a hutA-lacZ (pAML20) and a ryhB-lacZ (pAML23) transcriptional fusion in high- and low-iron conditions was measured in the wild-type (O395) and the fur mutant (ARM573) strain backgrounds. Bacterial cultures were grown to mid-log phase in iron-replete media (plus FeSO4; high iron) or in iron-depleted media (plus EDDA; low iron), and β-galactosidase activity was measured as described in Materials and Methods. The data represent the averages of three independent experiments. Standard deviations are indicated by the error bars.
Determination of the size and the transcriptional start site of V. cholerae RyhB.
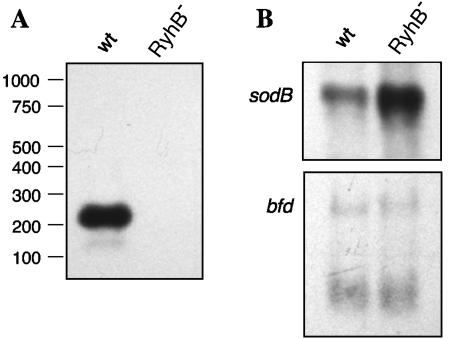
To determine whether V. cholerae RyhB is expressed from the upstream promoter as the full 214-nucleotide sRNA predicted by sequence analysis and promoter studies, O395 (wild type) and ARM572 (O395 ryhBΔ::kan) were grown to late log phase in low-iron media to induce expression of ryhB, and RNA was isolated and analyzed by Northern blotting. The data demonstrate that V. cholerae RyhB is highly expressed under the conditions described and that it is indeed >200 nucleotides in size (Fig. 3A). No hybridizing band was visible at this position in the RNA sample from the ryhB mutant ARM572. An obvious implication of this result is that, because of its greater size, V. cholerae RyhB may have targets that differ from those of E. coli RyhB.
FIG. 3.
Analysis of selected transcripts in O395 (wild-type V. cholerae [wt]) and ARM572 (O395 ryhBΔ::kan [RhyB−]). Total RNA was isolated from O395 and ARM572 grown to late log phase in iron-limited media and analyzed by Northern blotting as described in Materials and Methods. (A) Estimation of V. cholerae RyhB size. Size determinations are based on a comparison with biotinylated Century RNA standards (Ambion) run alongside the RNA samples (not shown). (B) Effect of ryhB deletion on the expression of sodB and bfd. The blots were probed as indicated.
To pinpoint exactly the start of the ryhB transcript, 5′ RACE was carried out. Total RNA was harvested from the classical biotype O395 and from the El Tor biotype N16961 grown under iron-limiting conditions, and 5′ RACE was performed as described in Materials and Methods. The results from four independent RNA samples, two from each strain, demonstrated that the cDNA generated from the ryhB transcript (3′-CAGAATCCC . . .-5′) is in agreement with the start site (5′-GTCTTAGGG . . .-3′) shown for V. cholerae ryhB in Fig. 1A.
Analysis of potential V. cholerae RyhB targets identified by sequence analysis.
Because RyhB in E. coli has been shown to function by base pairing with target mRNAs, a BLAST search of the complete V. cholerae genome was performed using the full-length V. cholerae RyhB to find sequences exhibiting stretches of identity with this sRNA (data not shown). Among the putative targets identified in this way are sodB (sequence complementarity to RyhB in the 5′ untranslated region of the predicted sodB mRNA [Fig. 1D]) and the bfd-bfr putative operon (homology to RyhB within the intergenic region between bfd and bfr [data not shown]), both of which loci have been shown to be regulated by RyhB in E. coli (26). The region of V. cholerae RyhB predicted to hybridize to each of these targets overlaps the region highly conserved between E. coli and V. cholerae RyhB (Fig. 1A and D and data not shown). To test the prediction of regulation by RyhB, Northern analysis of RNA isolated from O395 (wild type) and ARM572 (ryhBΔ::kan) grown to late log phase in iron-limited media was performed using probes specific for sodB and bfd. The sodB transcript was clearly more abundant in the ryhB mutant, consistent with alleviation of RyhB repression in this strain; however, no such effect was seen for the bfd-bfr transcript (Fig. 3B). These results were confirmed by microarray analysis of strains grown under conditions similar to those used for the Northern analyses (Table 3 and data not shown). Although no effect of RyhB on bfd-bfr transcription was seen, these genes are iron regulated in V. cholerae, and expression was increased >2-fold in low-iron medium (Mey et al., unpublished).
TABLE 3.
Genes whose expression is decreased by RyhBa
| Encoded protein | Gene designation(s) (decrease in expression [n-fold]) |
|---|---|
| Products of known RyhB-regulated genes | |
| Superoxide dismutase (SodB)b,c,d | VC2045 (15.6) |
| Succinate dehydrogenaseb,c,d | VC2089 (2.2), VC2090 (2.2), VC2091 (2.0) |
| Fumarasec | VC1304 (2.8) |
| Metabolism, respiration, and other functions | |
| Fumarate reductasec,d | VC2656 (2.5), VC2658 (2.8), VC2659 (6.8) |
| Citrate synthasec | VC2092 (3.3) |
| Ubiquinol-cytochrome c reductaseb,c | VC0573 (2.2), VC0574 (3.6), VC0575 (3.0) |
| RnfEGDB-related proteinsb,d | VC1012 (2.2), VC1013 (2.0), VC1014 (2.0), VC1016 (2.0) |
| Cytochrome c oxidase (CcoPQRN)b,c | VC1439 (2.7), VC1440 (2.5), VC1441 (2.0), VC1442 (2.0) |
| Cytochrome d oxidasec | VC1843 (9.7), VC1844 (2.4) |
| Cytochrome c-type biogenesis proteinb | VC2052 (2.7) |
| Cytochrome b561b | VCA0249 (2.6) |
| 2-Oxoglutarate dehydrogenase E2 E1b | VC2086 (2.0), VC2087 (2.0) |
| NADH-ubiquinone oxidoreductaseb,c,d | VC2205 (2.1), VC2290 (2.0), VC2291 (2.0), VC2292 (2.0) |
| Oxaloacetate decarboxylaseb | VC0550 (2.3) |
| NADP transhydrogenasec,d | VCA0564 (2.0) |
| OmpUb | VC0633 (3.3) |
| OmpWc | VCA0867 (2.8) |
| Maltose ABC transporterc,d | VCA0944 (4.4), VCA0945 (2.5) |
| Maltose operon periplasmic proteinc,d | VCA1027 (3.4), VCA1028 (4.1) |
| Formate transporterb | VCA0540 (2.0) |
| A/G-specific adenine glycosylaseb | VC0452 (4.0) |
| Iron-regulated protein | |
| Hemolysinb | VCA0219 (4.1) |
| Hypothetical proteins | |
| VC1559b (3.9) | |
| VCA0377c (3.8) | |
| VCA0502c (2.1) | |
| VCA0505c (2.5) | |
| VCA0651b (2.3) |
RNAs were isolated from the parental strain and the ryhB mutant or from the ryhB mutant and the ryhB overexpression strain grown to late log phase and analyzed using microarrays.
Mean expression of gene was at least twofold higher in the ryhB mutant of N16961 than in the wild-type parental strain in three arrays.
Mean expression of gene was at least twofold lower in the ryhB mutant of O395 overexpressing ryhB than in the ryhB mutant carrying the vector in three arrays.
Mean expression of gene was at least twofold higher in the ryhB mutant of O395 than in the wild-type parental strain in three arrays.
Identification of V. cholerae RyhB targets by microarray analysis.
Very few targets of V. cholerae RyhB were identifiable by homology searching, including most of the genes previously shown to be RyhB regulated in E. coli. For example, none of the genes encoding the succinate dehydrogenase complex exhibited obvious stretches of homology with RyhB in a BLAST search, yet the sdh operon is among the most highly conserved targets of RyhB regulation identified to date (26, 59). To study the V. cholerae RyhB regulon, microarrays were used to compile the expression profile of a V. cholerae ryhB mutant, as well as a RyhB-overexpressing strain (Tables 3, 4, and 5). Several genes known to be subject to posttranscriptional negative regulation by RyhB in other organisms, including sodB (26, 59), the sdh operon (26, 59), and fumA, encoding fumarate hydratase (26), appear also to be targets for RyhB regulation in V. cholerae (Table 3). The aconitate hydratase gene, acnA, which is repressed by RyhB in E. coli, showed consistent but weak (a <2-fold change) regulation by V. cholerae RyhB under the conditions used for the arrays and is therefore not listed in Table 3. Other enzymes involved in or linked to the TCA cycle were also repressed by RyhB in V. cholerae. These include citrate synthase and oxaloacetate decarboxylase (Table 3).
TABLE 4.
Genes whose expression is increased by RyhBa
| Encoded protein | Gene designation(s) (increase in expression [n-fold]) |
|---|---|
| Iron-regulated proteins | |
| FhuACb | VC0200 (2.0), VC0201 (2.5) |
| FeoABc | VC0207 (2.2), VC2078 (2.8) |
| Enterobactin receptor IrgAb | VC0475 (4.0) |
| Metabolism, transport, and other functions: | |
| Ketol-acid reductoisomeraseb | VC0162 (4.6) |
| Peptide ABC transporterb,d | VC0170 (3.0), VC0171 (2.3), VC0172 (2.0) |
| Arginine repressorb,d | VC0431 (4.0) |
| Fructose repressorb | VCA0519 (3.0) |
| Lipoprotein NlpDb,d | VC0533 (2.8) |
| RNA polymerase sigma-38 (RpoS)b | VC0534 (2.0) |
| TlcRb | VC1470 (2.5) |
| ABC transporterc | VC1524 (5.5), VC1525 (4.1) |
| OmpTb | VC1854 (32.4) |
| OmpAb | VC2213 (3.4) |
| Formate acetyltransferaseb | VC1866 (2.1) |
| Formate acetyltransferase-related | VC2361 (2.6) |
| proteinb | |
| Carbamoyl-phosphate synthetaseb | VC2390 (3.7) |
| Ornithine carbamoyltransferaseb | VC2508 (20.4) |
| Aspartate carbamoyltransferaseb | VC2510 (2.0) |
| Arginosuccinate lyaseb | VC2641 (8.7) |
| Arginosuccinate synthaseb | VC2642 (29.5) |
| Nicotinate phosphoribosyltransferaseb | VCA0098 (3.1) |
| Oxidoreductaseb | VCA0099 (3.0) |
| Hypothetical proteins | |
| VC0628b (2.7) | |
| VC1207b (2.0) | |
| VC1380b (2.1) | |
| VC1538b (2.0) | |
| VC1624b (3.2) | |
| VC1641b (2.6), VC1642b (2.4) | |
| VC1828b (2.3) | |
| VC2717b (4.3) | |
| VCA0076b (2.5) | |
| VCA0087b (2.4) | |
| VCA0125b (2.0) | |
| VCA0152b (2.1) | |
| VCA0568b (2.4), VCA0569b (2.3) |
RNAs were isolated from the parental strain and the ryhB mutant or from the ryhB mutant and the ryhB overexpression strain grown to late log phase and analyzed using microarrays.
Mean expression of gene was at least twofold lower in the ryhB mutant of N16961 than in the wild-type parental strain in three arrays.
Mean expression of gene was at least twofold higher in the ryhB mutant of O395 overexpressing ryhB than in the ryhB mutant carrying the vector in three arrays.
Mean expression of gene was at least twofold lower in the ryhB mutant of O395 than in the wild-type parental strain in three arrays.
TABLE 5.
Possible biofilm- and virulence-related genes whose expression is decreased in a ryhB mutanta
| Encoded protein | Gene designation (decrease in expression [n-fold]) |
|---|---|
| LPS O-antigen transport protein | VC0246 (2.0) |
| RfbL (O-antigen ligase) | VC0249 (2.0) |
| MSHA pilin MshB | VC0408 (2.0) |
| PomAB (chemotaxis) | VC0892 (2.8), VC0893 (2.3) |
| Methyl-accepting chemotaxis proteins | VC1313 (3.8), VC2161 (3.1), VCA0906 (3.2) |
| CheV (chemotaxis) | VC1602 (2.1), VC2202 (2.3) |
| Flagellar sigma factor | VC2066 (2.3) |
| Flagellar protein FlhF | VC2068 (2.0) |
| Flagellar protein FliG,S | VC2134 (2.1), VC2138 (2.3) |
| Flagellar proteins FlaIGBDE | VC2139 (2.3), VC2141 (2.1), VC2142 (2.9), VC2143 (3.6), VC2144 (4.4) |
| Flagellar protein FlaC | VC2187 (4.2) |
| Flagellar protein FlgM | VC2204 (2.1) |
| ToxS | VC0983 (2.2) |
| TagD | VC0824 (2.3) |
| TagE | VCA1043 (2.0) |
RNAs were isolated from the N16961 ryhB mutant and its parental strain grown to late log phase and analyzed using microarrays. Genes whose expression was at least two-fold lower in the mutant strain in each of three arrays are shown.
A major group of V. cholerae genes whose expression is reduced by RyhB contains genes encoding respiration and energy metabolism proteins, particularly components of electron transport. These include cytochrome oxidases, NADH-ubiquinone oxidoreductase, E1 and E2 components of complex I, and the RnfEGDB-related proteins. The rnf genes were identified in Rhodobacter capsulatus as nitrogen fixation genes encoding a putative membrane complex involved in electron transport to nitrogenase (47). Notably, this group includes proteins predicted to contain iron, in either an iron-sulfur cluster or heme. Regulation by RyhB would couple the expression of these proteins to the level of iron in the cell and to the levels of TCA cycle enzymes.
In addition to the metabolic genes, the levels of some genes encoding membrane proteins were influenced by ryhB expression. Expression of the genes encoding the outer membrane proteins OmpT and OmpU was affected by RyhB (Tables 3 and 4). In the ryhB mutant, expression of ompT was reduced 32-fold, making it the most highly regulated target in our arrays, and ompU was increased over 3-fold. Mutations in relA (15) and toxR (32) have also been shown to have reciprocal effects on the regulation of these two genes; a relA mutant increased ompU and decreased ompT expression, while toxR mutants have increased ompT and decreased ompU expression. This suggests that RyhB could be required for RelA synthesis or could be reducing the expression of toxR. However, no significant regulation of either relA or toxR, or of any known downstream targets of these regulators, was seen in the ryhB mutant or in the strain overexpressing ryhB under the conditions tested (data not shown).
Several of the membrane or secreted proteins identified in the microarray analysis are iron-regulated proteins. Components of the ferrichrome (Fhu), ferrous iron (Feo), and enterobactin (IrgA) transport systems were induced when RyhB was expressed (Table 4). This may reflect a slight iron starvation caused by increased expression of iron-containing proteins that are normally repressed by RyhB. In contrast, expression of the hemolysin gene, which is also repressed by iron (48), was reduced by RyhB (Table 3). ABC transporters were identified that were up-regulated (Table 4) or down-regulated (Table 3) by RyhB, but they are not known to contain iron or to have a metabolic role related to iron metabolism.
rpoS expression was lower in a ryhB mutant (Table 4), but reduced RpoS does not appear to be responsible for the changes in gene expression observed in the microarray experiments. Only one of the genes identified by Yildiz et al. (62) as being expressed at a reduced level in microarray analysis of an rpoS mutant was also expressed at a lower level in the ryhB mutant (VCA0568) (Table 4), and the fumarate reductase genes (VC2657 and VC2658) that have reduced expression in the rpoS mutant of V. cholerae (62) are up-regulated in the ryhB mutant (Table 3).
An additional category of genes whose expression was reduced in the ryhB mutant includes those encoding functions related to motility, chemotaxis, and cell surface antigens (Table 5). In particular, expression of a number of genes involved in flagellar synthesis is lower in the ryhB mutant than in the parental strain. Because these genes are associated with biofilm formation, we determined the effect of a ryhB mutation on biofilm production.
RyhB is required for biofilm formation in V. cholerae.
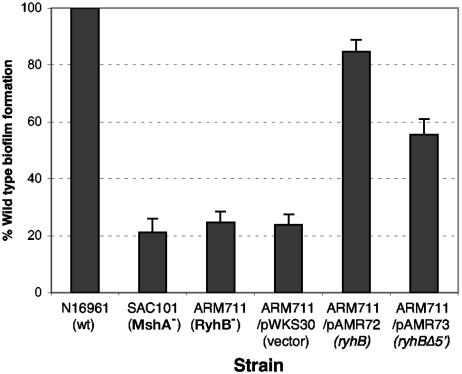
We tested biofilm formation in the classical strain O395 and in the El Tor strain N16961 and found that N16961 forms a much more robust biofilm than does O395 under our assay conditions. We therefore tested the effect of RyhB in biofilm formation in the El Tor strain. The same ryhB deletion mutation in ARM572 was made in the N16961 background, and the El Tor ryhB mutant (ARM711) was compared with the parental strain in a biofilm assay. An mshA mutant of N16961, SAC101, which does not form the mannose-sensitive hemagglutinin (MSHA) pilus necessary for the primary attachment to the substrate and therefore cannot initiate wild-type biofilm growth (55, 56), was used as the negative control in these assays. Figure 4 shows that the ryhB mutant ARM711 is a very poor biofilm producer compared with its parental strain, N16961, exhibiting biofilm growth comparable to that seen in the negative control strain SAC101. Planktonic growth rates, as measured by determining the optical density of the nonadherent cell suspension within the well, were comparable for all three strains (data not shown), indicating that the reduced biofilm was not due to poor growth of the mutant. Biofilm formation was restored when ryhB was supplied on a plasmid, pAMR72. These data indicate that RyhB is necessary for biofilm formation in N16961 under the conditions used. To determine whether the additional 5′ sequences of V. cholerae RyhB, compared with E. coli RyhB, play a role in biofilm formation, a plasmid (pAMR73) carrying an ryhB allele missing 60 nucleotides at the 5′ end was constructed (Fig. 1A). The ryhB mutation was partially complemented by pAMR73, albeit not to wild-type levels. This suggests that the unique 5′ region of V. cholerae RyhB does not contain sequence-specific information required for regulating a target involved in biofilm growth. Rather, failure to entirely complement the biofilm defect may be related to the stability of the 5′-deleted sRNA; it was determined by real-time PCR that the level of the 5′-deleted transcript is approximately 25% of that of the full-length RyhB expressed from the same vector and approximately 60% of that of RyhB expressed from the chromosomal ryhB gene (data not shown).
FIG. 4.
RyhB is required for biofilm formation in the El Tor strain N16961. The amount of biofilm formed by each strain was determined as described in Materials and Methods and is expressed as a percentage of the biofilm formed by the wild-type (wt) parental strain N16961 grown under the same conditions. Means and standard deviations of three independent experiments are shown.
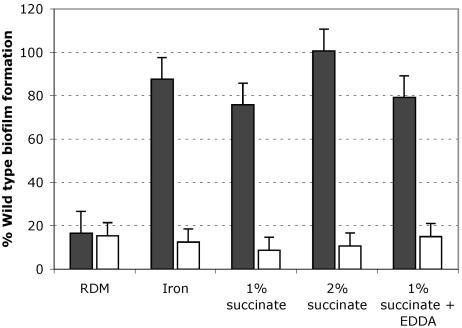
The V. cholerae biofilm defect can be rescued by the addition of excess iron or succinate.
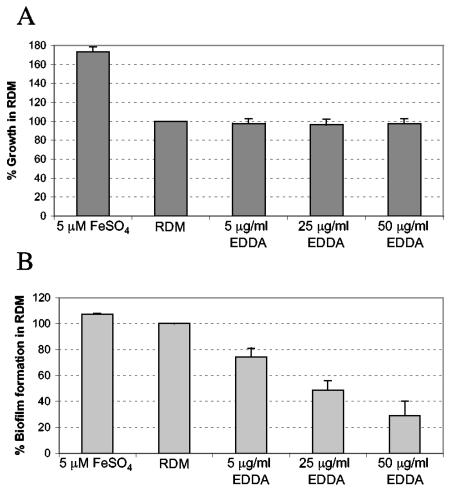
RyhB has been shown to play an important role in cellular iron metabolism in other organisms. Therefore, we decided to study the relationship between iron status and biofilm growth in V. cholerae (Fig. 5). Addition of excess iron to the biofilm assay medium restored biofilm formation in the ryhB mutant strain, suggesting that the presence of iron overrides the biofilm defect associated with loss of RyhB. Iron did not rescue the mshA mutant for biofilm growth (Fig. 5), nor did it greatly enhance biofilm formation in the wild-type strain N16961 (Fig. 6). These data suggest that the ryhB mutant may be somewhat iron stressed in the biofilm assay medium compared with the parental strain. The iron stress experienced by the ryhB mutant is likely to be fairly subtle, since the planktonic growth of the ryhB mutant in this medium was comparable to that of the parental strain (data not shown). Adding iron chelators to the biofilm medium further compromised biofilm growth in the ryhB mutant (data not shown) and adversely affected biofilm formation in the wild-type strain as well, even at concentrations that did not inhibit the planktonic growth of the wild-type strain (Fig. 6). These data are consistent with a role for iron and RyhB in the regulation of biofilm formation.
FIG. 5.
Iron or succinate can restore biofilm formation in the ryhB mutant. Biofilm growth of the ryhB mutant ARM711 (filled bars) and the mshA mutant negative control strain SAC101 (unfilled bars) was measured in EZ RDM with or without added iron (5 μM), succinate (1% or 2%), or succinate (1%) plus EDDA (25 μg per ml) as described in Materials and Methods and is shown as a percentage of the biofilm growth of the wild-type N16961 strain under the same conditions. Means and standard deviations of three independent experiments are shown.
FIG. 6.
Iron depletion inhibits biofilm formation in N16961. Planktonic growth and biofilm formation were measured as described in Materials and Methods and are expressed as percentages of the growth or biofilm produced in EZ RDM without added iron or chelator. Means and standard deviations of three independent experiments are shown. (A) Planktonic growth of N16961 in the biofilm assay with or without added iron or the iron chelator EDDA. (B) Biofilm formation by the cultures shown in panel A.
One of the most conserved functions of RyhB in E. coli, P. aeruginosa, and V. cholerae is the regulation of the succinate dehydrogenase operon (Table 3). Interestingly, adding exogenous succinate to the biofilm medium restored biofilm formation in the ryhB mutant in a concentration-dependent manner (Fig. 5). This does not appear to be due to iron contamination of the succinate preparation, since succinate rescued the biofilm defect of the ryhB mutant even in the presence of high levels of the iron chelator EDDA (Fig. 5). The succinate effect is specific for the ryhB pathway and did not restore biofilm formation in the mshA mutant (Fig. 5).
Roles of iron and RyhB in switching from the smooth to the rugose colony type.
V. cholerae grown under nutrient starvation conditions can exhibit two distinct colony morphologies: smooth and rugose. The rugose morphology is associated with elevated production of exopolysaccharide (EPS) (63), which is also a hallmark of cells growing in biofilms (56). Thus, mutants unable to switch to the rugose morphotype may also have defects in biofilm formation. To test the roles of iron and V. cholerae RyhB in rugose switching, strains were grown under conditions known to promote high-level switching of N16961 (1). The results (Table 6) show that the N16961 ryhB mutant, although able to switch, did so at a consistently lower frequency than the wild-type strain. In addition, iron starvation inhibited the smooth-to-rugose transition, since increasing concentrations of the iron chelator EDDA in the medium reduced and ultimately eliminated rugose switching in either the wild-type or the ryhB mutant strain (Table 6). Supplementation of the medium with excess iron dramatically increased the percentage of rugose colonies in the ryhB mutant strain, consistent with results from biofilm assays showing that iron can rescue the biofilm defect of the ryhB mutant. These results suggest that iron plays a role in regulating EPS production and that RyhB may be involved in some aspect of this regulation. However, this is clearly not an absolute requirement, since rugose switching did occur in the ryhB mutant. The classical biotype strain O395 did not switch to the rugose morphotype in any medium tested, consistent with poor biofilm formation by this strain under our laboratory conditions.
TABLE 6.
Roles of iron and RyhB in rugose switching
| Strain | % Rugose colonies after growth in APW no. 3 witha:
|
|||
|---|---|---|---|---|
| FeSO4 (40 μM) | No additions | EDDA (5 μg/ml) | EDDA (10 μg/ml) | |
| N16961 | 82 | 81 | 14 | 0 |
| ARM711 (ryhB mutant) | 86 | 55 | 7 | 0 |
The strains shown were grown statically at 37°C for 24 hours in the indicated media and then plated on L agar to determine the percentage of rugose colonies. The experiment was repeated three times, and one representative data set is shown.
Iron and succinate do not stimulate rugose switching within biofilms of the N16961 ryhB mutant.
Based on the observation that adding iron to the APW no. 3 medium increased the frequency of rugose switching in the N16961 ryhB mutant strain (Table 6), it is possible that excess iron in the biofilm medium might also promote switching. The presence of even a very small number of rugose cells (fewer than 1% at the outset of the biofilm experiment; fewer than 10% at the time of biofilm harvesting) (14) within a population has been shown to result in wild-type levels of biofilm growth, both because the rugose cells outgrow their smooth counterparts and because the smooth cells become embedded in the copious EPS produced by the rugose cells. To test whether a proportion of the ryhB mutant cells growing in biofilms in the assay medium supplemented with iron had switched to the rugose colony type, adherent as well as planktonic cells were recovered from the assay wells and plated. Despite healthy biofilm growth in the presence of iron, none of the ryhB mutant cells had switched to the rugose morphotype (switching frequency, <10−4). The role of exogenous succinate in the biofilm assay medium was similarly tested and found not to promote rugose switching in the N16961 ryhB mutant strain. These data suggest that, while iron and succinate may affect EPS production, they do so using a mechanism other than the phase-variable genetic change underlying rugose switching.
V. cholerae ryhB plays a role in chemotactic motility.
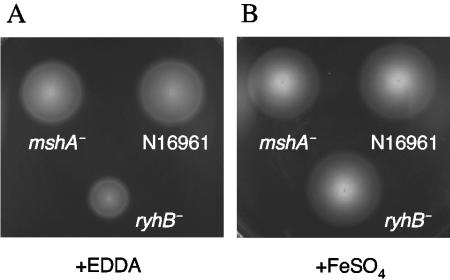
Flagellar rotation and motility are closely linked to biofilm formation in V. cholerae (21, 56). In the early stages of biofilm formation, motility is necessary for approaching a surface and for spreading across that surface to initiate microcolony formation (56). In addition, nonmotile mutants with defects in the sodium-driven flagellar motor exhibit decreased EPS expression and subsequently do not form wild-type three-dimensional biofilms (21). In a chemotaxis assay, the N16961 ryhB mutant ARM711 consistently showed an approximately 50% decrease in swarm zone in low-iron conditions compared with its parental strain, while the mshA mutant was unaffected (Fig. 7A). This suggests that RyhB is needed for optimal chemotactic motility. Thus, flagellum-dependent microcolony formation or the flagellum-dependent signaling pathway leading to EPS production could be compromised in the ryhB mutant strain. These results are consistent with the decrease in flagellar and chemotaxis gene expression observed in the microarray analysis of the ryhB mutant (Table 5). As was noted in the biofilm assays, the defect was suppressed by adding iron to the medium (Fig. 7B). Interestingly, added succinate significantly increased the swarm zones of all the strains tested, including the wild-type and mshA mutant strains (data not shown), suggesting that V. cholerae may have a strong chemotactic response to succinate.
FIG. 7.
The ryhB mutant has decreased motility and/or chemotaxis under conditions of iron starvation. The wild-type strain (N16961), the mshA mutant (SAC101), and the ryhB mutant (ARM711) were inoculated into chemotaxis agar containing 20 μg/ml EDDA (A) or 40 μM FeSO4 (B) and incubated at 37°C for 8 to 10 h, after which the swarm zone of each strain was measured.
RyhB is not required for V. cholerae virulence.
It has been shown that V. cholerae Hfq is required for the virulence of V. cholerae in infant mice (5). Because RyhB likely acts in concert with Hfq, it is possible that RyhB is also required for some aspect of virulence. To test this, the infant mouse model of V. cholerae intestinal colonization was used. In this assay, equal numbers of the ryhB mutant and its parental strain were inoculated intragastrically into 5-day-old infant mice, and the ability of the mutant strain to compete with the wild-type strain was assessed by determining the ratio of viable mutant cells to wild-type cells recovered after 24 h. Because the classical strain O395 and the El Tor strain N16961 display differences in virulence gene expression and in biofilm formation, we tested both the 0395 ryhB mutant ARM572 and the N16961 ryhB mutant ARM711. The 0395 mutant showed no defect in colonization (Table 7). The competitive index (the output ratio normalized to the input ratio) of the ryhB mutant was slightly above 1.0, suggesting that the ryhB mutant may have a small competitive advantage; however, since it is less that twofold, this result may not be significant.
TABLE 7.
In vivo competition of V. cholerae ryhB mutants with their wild-type parental strains
| Strain | Competitive indexa
|
No. of mice | |
|---|---|---|---|
| Mean (SD) | Range | ||
| ARM572b | 1.67 (0.38) | 1.28-2.08 | 6 |
| ARM711c | 2.51 (1.82) | 0.67-8.1 | 7 |
| ARM711c,d | 1.11 (0.31) | 0.82-1.56 | 7 |
The competitive index of the specified strain relative to its wild-type parental strain was determined by normalizing the output ratio of the two competing strains to the input ratio, as described in Materials and Methods.
ARM572 competed against O395.
ARM711 competed against N16961.
Ten-fold-lower inoculum.
The N16961 ryhB mutant also exhibited no defect in colonization compared with its parental strain (Table 7). In fact, ARM711 appears to have an even greater competitive advantage over its parental strain than that exhibited by the O395 ryhB mutant ARM572 over its parent. This effect was not observed when a 10-fold-smaller inoculum size was administered.
DISCUSSION
In this study, we provide the initial characterization of V. cholerae RyhB. This small RNA is a functional homolog of E. coli RyhB, and like the E. coli RNA, it represses the synthesis of SodB and several TCA cycle enzymes. V. cholerae and E. coli RyhB share significant overall sequence similarity; the central portions of V. cholerae RyhB and E. coli RyhB are identical at 31 of 34 nucleotides. The region of E. coli RyhB involved in base pairing with the sodB message in E. coli is contained within this highly conserved stretch. In addition, a 22-nucleotide stretch with nearly complete complementarity (20- out of 22-nucleotide complementary) to V. cholerae RyhB was found upstream of sodB in the N16961 genome (16) (Fig. 1D), suggesting a base-pairing mechanism of regulation by RyhB similar to the one described in E. coli. Consistent with these findings, it was clear from Northern analyses (Fig. 3), microarrays (Table 3), and real-time PCR (data not shown) that sodB is one of the most highly regulated targets of RyhB in strains representing both the classical and the El Tor biotypes of V. cholerae. Among other predicted targets, based on RyhB function in E. coli and P. aeruginosa, were the succinate dehydrogenase operon, aconitase, and fumarase, all of which appear to be RyhB regulated in V. cholerae as well, as determined by microarray analysis. Many other genes involved in cellular metabolism, including aerobic- and anaerobic-respiration and electron transport genes, were identified in microarrays as possible RyhB targets in V. cholerae. Several of these are down-regulated in the ryhB mutant, suggesting either repression of an upstream negative regulator by RyhB in the well-documented manner or direct positive regulation of these targets by RyhB, possibly through stabilization of the mRNA. This mode of direct positive regulation by a small RNA was recently described for the GadY sRNA, which induces the acid response regulator GadX by stabilizing its mRNA (38); however, no such activity has been reported for RyhB.
Although V. cholerae RyhB functions similarly to E. coli RyhB, there are distinctive features of V. cholerae RyhB, perhaps the most intriguing of which is its greater length. Whereas E. coli RyhB is 90 nucleotides long, V. cholerae RyhB is expressed as an RNA molecule of >200 nucleotides. It does not appear that the V. cholerae RyhB transcript is further processed, at least not under the conditions tested, since no lower-molecular-weight species were detectable in any significant amounts in Northern analyses. In addition, only one transcriptional start site for ryhB was detected by 5′ RACE. Thus, the unique 5′ and 3′ regions of the RyhB transcript are likely part of the mature expressed sRNA and may contain sites for interactions with additional targets in V. cholerae. Among the putative targets identified through a BLAST search of the V. cholerae genome with the extended regions of V. cholerae RyhB, none showed up in our arrays as being RyhB regulated. However, in the absence of information about the folding of the RNA and the locations of bases available for interactions with targets, it is not possible to rule out a direct role of these sequences in regulation of target genes. The 5′ region may also be important for the structure or stability of the sRNA molecule in V. cholerae. The RyhBΔ5′ sRNA still regulated sodB expression, but to a lesser extent than the full-length RyhB produced from the same vector (data not shown). In addition, RyhBΔ5′ was expressed at a lower level than wild-type RyhB (data not shown).
At least one of the genes regulated by RyhB in E. coli (the ferritin gene bfr) is not regulated by RyhB in V. cholerae. In E. coli, the bfd and bfr genes, although apparently linked (2), are oppositely regulated in response to iron and Fur. While bfd is induced in low iron, consistent with classical Fur-mediated regulation, bfr is repressed (J. M. Grogan, unpublished data, cited in reference 42). This repression is mediated through RyhB, but the mechanism is not known (26). In V. cholerae, both bfd and bfr are negatively regulated by iron and Fur (Mey et al., unpublished), and there is no evidence that the bfd-bfr transcript is a direct target of V. cholerae RyhB (Fig. 3 and data not shown). Interestingly, significant homology to V. cholerae RyhB was found upstream of bfr in N16961 (data not shown), leaving open the possibility that a level of regulation exists that was not detectable under the conditions used in our studies. However, the identity is with the coding strand, not the template strand, which would entail a mechanism for RyhB regulation different than base pairing with the mRNA. It has been suggested that a Fur-regulated antisense RNA may be encoded within E. coli bfd-bfr (2); such an antisense RNA within the V. cholerae bfd-bfr locus could be a potential target for RyhB.
Some of the altered regulation seen in the V. cholerae ryhB mutant may reflect alterations in iron metabolism. Clues that the V. cholerae ryhB mutant may be somewhat iron stressed were gained from the microarray analyses. A number of the genes whose expression was altered in the absence of RyhB or in the presence of RyhB overproduction encode proteins involved in iron transport or ones that require heme or iron for their functions. The iron- and Fur-regulated hemolysin gene hlyA is consistently derepressed in the ryhB mutant. The rfn-related cluster, which includes a putative iron-sulfur cluster protein-encoding gene, rfnB, is also repressed by RyhB. The rnf operon from Rhodobacter capsulatus was shown to be repressed in low iron (18), consistent with iron regulation via a mechanism similar to the V. cholerae RyhB circuit. However, it is not the case that the entire iron regulon is affected, i.e., there is not a global switching on of all the iron-regulated genes, which would be the expected result under iron starvation conditions. In fact, the fhu, feo, and ent iron transport systems, which are negatively regulated by iron and Fur in V. cholerae (45; Mey et al., unpublished), were repressed in a ryhB mutant. Further, there was no effect of the ryhB mutation on growth of the cells under the conditions used for microarray analysis of RNA levels (data not shown). Rather, these results point to a more subtle effect on iron homeostasis.
The ability of V. cholerae to form biofilms in aquatic environments is critical for its persistence in these environments and for efficient spread to the human host (13). Interestingly, iron appears to play an integral role in the regulation of biofilm formation in the El Tor strain N16961. Adding iron chelators to the assay medium at levels that did not inhibit planktonic growth caused a concentration-dependent decrease in biofilm production in the wild-type strain. Further, rugose switching was suppressed in the wild-type strain in the presence of iron chelator, even though the density of the culture reached the critical point at which rugose switching begins to occur in iron-replete media (data not shown). This suggests that the effect of iron stress on EPS production and biofilm formation is not simply a result of reduced growth in low-iron conditions. Moderate (subbacteriostatic) iron limitation has been linked to poor biofilm formation in several other bacterial species, including P. aeruginosa and Staphylococcus aureus (58), suggesting that sensitivity of biofilms to small shifts in iron homeostasis may be a common theme among bacterial pathogens. In V. cholerae, this effect could be mediated via RyhB. The N16961 ryhB mutant, although able to maintain wild-type planktonic growth in the biofilm medium, failed to form biofilms unless excess iron was added, signaling that the ryhB mutant probably does experience subtle iron stress within the biofilm. In addition, the N16961 ryhB mutant exhibited decreased chemotaxis in low-iron, but not high-iron, medium and was more sensitive to iron chelators than its wild-type parental strain (data not shown). These data are consistent with the expression profile of a ryhB mutant, indicating that RyhB is necessary for regulating normal cellular iron metabolism.
There are several direct mechanisms by which RyhB could influence biofilm formation in response to iron limitation. A constitutively expressed target of RyhB, which would normally be repressed by RyhB in iron-depleted medium, since RyhB is maximally expressed under those conditions, might be inappropriately turned on. Such a target could be the succinate dehydrogenase operon, which is highly up-regulated in the RyhB mutant. The net effect of this would likely be significantly lower cellular succinate levels. It has been proposed that succinate may be a component of the EPS produced by V. cholerae growing in biofilms (62). Interestingly, adding exogenous succinate to the growth medium restored biofilm formation in the ryhB mutant, suggesting that succinate bypasses the RyhB requirement. Since high levels of iron in the medium do not repress the expression of sdh in the ryhB mutant (data not shown), it is not completely clear why adding iron would overcome the succinate depletion potentially caused by increased succinate dehydrogenase activity in the ryhB mutant. Iron-replete cells may simply contain higher levels of all TCA cycle intermediates due to the increased growth rate and respiration under those conditions. It is also possible that the formation of iron-sulfur clusters, which are an integral part of the succinate dehydrogenase complex, is somewhat decreased in high iron, since one set of genes involved in their assembly (suf) is negatively controlled by iron and Fur (27, 41). Another gene whose overexpression in the ryhB mutant could negatively influence biofilm formation is ompU. In Vibrio anguillarum, eliminating ompU caused a 10-fold increase in biofilm growth, suggesting that OmpU has an inhibitory effect on biofilm formation (54).
The effects of RyhB on biofilm formation may also reflect the reduced expression of flagellar genes in the ryhB mutant. The microarray analysis indicated that the ryhB mutant had decreased expression of a number of genes in the motility and chemotaxis pathways, suggesting that the proper assembly and function of flagella may be affected by RyhB. The microarray results are consistent with the observed decrease in motility of the N16961 ryhB mutant ARM711 under low-iron conditions (Fig. 7). Reduced motility may directly inhibit biofilm formation or may affect biofilm formation by interfering with the flagellum-dependent signaling pathway that regulates EPS production (21). These hypotheses are under investigation.
Biofilm formation in V. cholerae is a complex process. In strains that possess a functional HapR, the relationship between quorum sensing and biofilm formation has been clearly established. When these strains are at low cell density, two known quorum-sensing pathways and at least one unknown pathway converge to phosphorylate the central regulator LuxO (31). LuxO in its phosphorylated state acts via Hfq and four sRNAs to repress the expression of HapR (22, 51, 65). Since HapR is a negative regulator of the vps cluster that controls synthesis of EPS, the net result is EPS production and biofilm growth (14). However, many toxigenic, virulent strains of V. cholerae, including O395 and N16961, contain defective hapR alleles, and thus, it is not clear if there is a link between the LuxO-mediated pathways and biofilm formation in these strains. In strains possessing a functional HapR, mutating HapR is associated with the rugose phenotype, consistent with the role of HapR in repressing the vps genes. Thus, it might be expected that V. cholerae strains that do not encode a functional HapR are by default rugose; however, this is clearly not the case, since N16961 and O395 are both smooth under laboratory conditions. Because N16961 is capable of rugose switching, and because the N16961 rugose variant produces higher levels of EPS and more robust biofilms than the smooth cells (1), it follows that a negative regulatory circuit separate from the HapR pathway exists in this strain. The recent discovery that an Hfq-dependent sRNA pathway regulates the integration of quorum-sensing signals with biofilm formation in a HapR+ strain (22) suggests the possibility that a separate sRNA pathway could be involved in the negative regulation of EPS production and biofilm formation in HapR− strains. While our studies show that RyhB is required for wild-type biofilm growth, consistent with repression by RyhB of a negative regulator of biofilm formation, overproduction of RyhB in N16961 did not result in a rugose phenotype or in increased biofilms (data not shown), indicating that RyhB probably does not directly regulate the phase-variable rugose-switching pathway in N16961.
Many small RNAs require Hfq, and V. cholerae has a functional Hfq protein (5, 22). It was shown recently that Hfq is required for V. cholerae virulence in both a HapR+ (20) and a HapR− (5) strain background. In the HapR+ strain, Hfq and multiple small RNAs were required for expression of the toxin-coregulated pilus (TCP) (22), a key virulence determinant of V. cholerae, whereas in the HapR− strain, no such effect on TCP expression was seen (5). This emphasizes that the sRNA pathways leading to virulence gene expression and biofilm formation in HapR+ and HapR− strains are fundamentally different. We investigated whether V. cholerae RyhB might be involved in Hfq-mediated regulation of virulence in the hapR mutant strains N16961 and O395. This is probably not the case, since a ryhB mutant in either O395 or N16961 exhibited no defects in the ability to compete with the parental strain during intragastric growth.
The ability to form biofilms or to properly regulate biofilm formation has been shown to be important for V. cholerae virulence. Excessive biofilm formation in V. cholerae impedes colonization of infant mice (43, 57, 64), while eliminating biofilm growth altogether may negatively impact growth in vivo, as has been reported for strains with mutations in galU (35), vpsR (43), and motX (21). However, there are examples of mutations that eliminate biofilm formation but do not affect virulence, including mutations in the structural gene for the MSHA pilus, mshA (50); in galE (35); and, as reported here, in ryhB, indicating that biofilm growth per se may not be required for colonization of the mammalian host. Rather, the link between absent or inappropriate biofilm formation and colonization defects could be explained by the coordinate regulation of biofilm formation and virulence gene expression in many V. cholerae strains (14, 21, 31, 57, 64, 65). Nevertheless, by contributing to the environmental persistence and infectivity of V. cholerae (13), biofilms should be considered an important aspect of the pathogenic potential of this organism.
Acknowledgments
We gratefully acknowledge Elizabeth E. Wyckoff for helpful discussions and critical reading of the manuscript; Amanda Oglesby, Vanamala Kanukurthy, and Erin Murphy for expert technical assistance with the microarrays; and Carolyn Fisher and Mary Lozano for help with the mouse experiments. We thank Vishy Iyer and his laboratory for providing equipment and invaluable assistance during the printing of the microarrays and for advice and assistance in performing the array analyses. We are indebted to Bob McLean for his advice in setting up the biofilm assays and to Susan Gottesman for sharing unpublished information on the V. cholerae ryhB sequence.
This work was supported by a grant from the Foundation for Research and by grant AI50669 from the National Institutes of Health. V. cholerae microarray slides were also provided through the Pathogen Functional Genomics Resource Center of NIAID.
Editor: F. C. Fang
REFERENCES
- 1.Ali, A., M. H. Rashid, and D. K. Karaolis. 2002. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl. Environ. Microbiol. 68:5773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., P. M. Harrison, and J. R. Guest. 1989. Cloning, sequencing, and mapping of the bacterioferritin gene (bfr) of Escherichia coli K-12. J. Bacteriol. 171:3940-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butterton, J. R., M. H. Choi, P. I. Watnick, P. A. Carroll, and S. B. Calderwood. 2000. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J. Bacteriol. 182:1731-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma E expression. Mol. Microbiol. 53:345-354. [DOI] [PubMed] [Google Scholar]
- 6.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elkins, M. F., and C. F. Earhart. 1989. Nucleotide sequence and regulation of the Escherichia coli gene for ferrienterobactin transport protein FepB. J. Bacteriol. 171:5443-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8998-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissmann, T. A., and D. Touati. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 13.Hall-Stoodley, L., and P. Stoodley. 2005. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 13:7-10. [DOI] [PubMed] [Google Scholar]
- 14.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 15.Haralalka, S., S. Nandi, and R. K. Bhadra. 2003. Mutation in the relA gene of Vibrio cholerae affects in vitro and in vivo expression of virulence factors. J. Bacteriol. 185:4672-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, D. P., and S. M. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jouanneau, Y., H. S. Jeong, N. Hugo, C. Meyer, and J. C. Willison. 1998. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus—characterization of two membrane-bound iron-sulfur proteins. Eur. J. Biochem. 251:54-64. [DOI] [PubMed] [Google Scholar]
- 19.Killion, P. J., G. Sherlock, and V. R. Iyer. 2003. The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD). BMC Bioinformatics 4:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauriano, C. M., C. Ghosh, N. E. Correa, and K. E. Klose. 2004. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 186:4864-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 23.Litwin, C. M., and S. B. Calderwood. 1994. Analysis of the complexity of gene regulation by Fur in Vibrio cholerae. J. Bacteriol. 176:240-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 25.Masse, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 28.Mekalanos, J. J., D. J. Swartz, G. D. Pearson, N. Harford, F. Groyne, and M. de Wilde. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551-557. [DOI] [PubMed] [Google Scholar]
- 29.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 32.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moll, I., T. Afonyushkin, O. Vytvytska, V. R. Kaberdin, and U. Blasi. 2003. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9:1308-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niederhoffer, E. C., C. M. Naranjo, K. L. Bradley, and J. A. Fee. 1990. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J. Bacteriol. 172:1930-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 38.Opdyke, J. A., J. G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 40.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quail, M. A., P. Jordan, J. M. Grogan, J. N. Butt, M. Lutz, A. J. Thomson, S. C. Andrews, and J. R. Guest. 1996. Spectroscopic and voltammetric characterisation of the bacterioferritin-associated ferredoxin of Escherichia coli. Biochem. Biophys. Res. Commun. 229:635-642. [DOI] [PubMed] [Google Scholar]
- 43.Rashid, M. H., C. Rajanna, D. Zhang, V. Pasquale, L. S. Magder, A. Ali, S. Dumontet, and D. K. Karaolis. 2004. Role of exopolysaccharide, the rugose phenotype and VpsR in the pathogenesis of epidemic Vibrio cholerae. FEMS Microbiol. Lett. 230:105-113. [DOI] [PubMed] [Google Scholar]
- 44.Rogers, H. J. 1973. Iron-binding catechols and virulence in Escherichia coli. Infect. Immun. 7:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers, M. B., J. A. Sexton, G. J. DeCastro, and S. B. Calderwood. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182:2350-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Runyen-Janecky, L. J., M. Hong, and S. M. Payne. 1999. Virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect. Immun. 67:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmehl, M., A. Jahn, A. Meyer zu Vilsendorf, S. Hennecke, B. Masepohl, M. Schuppler, M. Marxer, J. Oelze, and W. Klipp. 1993. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol. Gen. Genet. 241:602-615. [DOI] [PubMed] [Google Scholar]
- 48.Stoebner, J. A., and S. M. Payne. 1988. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 56:2891-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vance, R. E., J. Zhu, and J. J. Mekalanos. 2003. A constitutively active variant of the quorum-sensing regulator LuxO affects protease production and biofilm formation in Vibrio cholerae. Infect. Immun. 71:2571-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 53.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 54.Wang, S. Y., J. Lauritz, J. Jass, and D. L. Milton. 2003. Role for the major outer-membrane protein from Vibrio anguillarum in bile resistance and biofilm formation. Microbiology 149:1061-1071. [DOI] [PubMed] [Google Scholar]
- 55.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weinberg, E. D. 2004. Suppression of bacterial biofilm formation by iron limitation. Med. Hypotheses 63:863-865. [DOI] [PubMed] [Google Scholar]
- 59.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyckoff, E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyckoff, E. E., A.-M. Valle, S. L. Smith, and S. M. Payne. 1999. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J. Bacteriol. 181:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 63.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]