Abstract
Ste11 is the mitogen-activated protein kinase (MAPK) kinase kinase in the MAPK cascades that mediate mating, high osmolarity glycerol, and filamentous growth responses in Saccharomyces cerevisiae. We show stimulation of the mating pathway by pheromone promotes an accelerated turnover of Ste11 through a MAPK feedback and ubiquitin-dependent mechanism. This degradation is pathway specific, because Ste11 is stable during activation of the high osmolarity glycerol pathway. Because the steady-state amount of Ste11 does not change significantly during pheromone induction, we infer that maintenance of MAPK activation involves repeated cycles in which naïve Ste11 is activated and then targeted for degradation. This model predicts that elimination of active Ste11 would rapidly curtail MAPK activation upon attenuation of the upstream signal. This prediction is confirmed by the finding that blocking ubiquitin-dependent Ste11 degradation during pheromone induction abolishes the characteristic attenuation profile for MAPK activation.
A prevalent mechanism for intracellular signal transmission involves three sequentially acting enzymes that function in what is known as a mitogen-activated protein kinase (MAPK) cascade. The prototype MAPK kinase kinase (MEKK) is the yeast enzyme Ste11, which was identified for its role in pheromone-induced mating differentiation (1, 2). In the mating MAPK cascade, Ste11 phosphorylates and activates the MAPK kinase (MEK), Ste7, which in turn phosphorylates and activates two MAPKs, Fus3 and Kss1 (3–7). Although Fus3 and Kss1 kinase activity is induced to an equivalent extent in pheromone-treated cells, Fus3, but not Kss1, has specificity for critical substrates that are dedicated to the mating differentiation response (7). Ste11 also serves as the MEKK in two other pathways, one that mediates responses to nutritional changes and another that mediates responses to hyperosmotic stress. Nitrogen deprivation in diploid cells and glucose depletion in haploid cells promote transitions to filamentous forms characteristic of pseudohyphal and invasive growth in the respective cell types (8, 9). The cascade that mediates the transition to the filamentous form in both cell types is made up of Ste11, Ste7, and Kss1 (10–13). The pathway that mediates the high osmolarity glycerol (HOG) response has two branches. One branch responds to activation of the Sln1-Ypd1-Skk1 two-component osmotic sensor, whereas the other responds to activation of the Src homology 3-containing Sho1 membrane protein (14–16). Ssk2 and Ssk22 are redundant MEKKs in the Sln1 branch, and Ste11 is the MEKK in the Sho1 branch. Ssk2/22 and Ste11 activate the MEK Pbs2, which in turn activates the MAPK Hog1 (16, 17).
Even though Ste11 is a component of three separate pathways (mating, filamentation, and osmotic stress) the stimulus of one pathway elicits the appropriate response without spuriously activating other Ste11-dependent outputs. For example, hyperosmolarity stimulates the HOG response without inducing mating differentiation (16). This biological specificity is caused, at least in part, by the physical association of Ste11 with components of the different pathways (16, 18). In the mating pathway, Ste11, Ste7, Fus3, and indirectly Kss1 are in a complex with the scaffold Ste5 (7, 19, 20). In the HOG pathway, the MAPK kinase Pbs2 functions as a scaffold through its interactions with Ste11, Hog1, and the membrane protein Sho1 (16). These different assemblies are thought to poise Ste11 for activation by the stimulus of a given pathway and therefore restrict the input signal to activation of a single MAPK cascade. We reasoned that different assemblies might also impose different modes of regulation on Ste11 in each of its pathways. This view prompted us to examine more closely the regulation of Ste11 during pheromone-induced signaling and to test whether the same regulatory mechanisms were common to its function in the HOG response pathway.
Materials and Methods
Yeast Genetic Procedures and Strains.
Unless otherwise specified, yeast growth media and genetic manipulations were used as described by Sherman et al. (21). Yeast strains were transformed by the method of Ito et al. (22). For the construction of strains with various deletions, we used the replacement method of Rothstein (23). Gene replacements were confirmed either by Southern blot or PCR analysis.
Yeast strains C699–1 (ste11Δ∷URA3) and C699–5 (bar1Δ∷HisG) were derived from strain C699 (MATa ade2–1 can1–100 his3–12, 16 leu2–3, 112 trp1–1 ura3–1) by gene replacement (strain C699 is isogenic to strain W303–1A). An XbaI fragment from plasmid pNC202 carries the ste11Δ∷URA3 allele that was used for replacement at the STE11 locus (2). An EcoRI–SalI fragment from plasmid C1329 (provided by K. Nasmyth, Institute for Molecular Pathology, Vienna) carries the bar1Δ∷HisG-URA3-HisG allele that was used for replacement at the BAR1 locus. The bar1Δ∷HisG allele was generated from the resulting strain by selection on 5-fluororotic acid medium (24). This medium provides a positive selection for isolates in which the URA3 marker is excised by recombination within the direct HisG repeats (25). Strain C699–15 (bar1Δ∷HisG FUS1-lacZ∷LEU2) was derived from strain C699–5 by integration of pNC296 at the LEU2 locus (6). C699–75 (bar1Δ∷HisG FUS1-lacZ∷LEU2 fus3Δ∷ADE2, kss1Δ∷URA3) was derived from strain C699–15 by replacement of the FUS3 locus with a BamHI–SalI fragment from pSL2223 carrying the fus3Δ∷ADE2 allele and the KSS1 locus using an EcoRI–SphI fragment from pBC65 carrying the kss1Δ∷URA3 allele (20, 26). Strain E929–6C (MATa cyc1 CYC7-H2 can1leu2–3, 112 trp1Δ 1 ura3–52) and strain FP50 (MATa ssk2Δ∷LEU2 ssk22Δ∷LEU2 ste11∷HIS3 ura3 leu2 his3), which retains the function of only the Sho1- and Ste11-dependent branch of the osmotic response pathway, have been described (6, 16).
Recombinant DNA Procedures and Plasmids.
Bacterial transformations, bacterial DNA preparations, plasmid constructions, and DNA restriction enzyme digestions were performed by standard methods (27). pNC245 (GAL-STE11M CEN3 TRP1) and pNC734 (GAL-STE11M CEN4 URA3) are plasmids that allow galactose-dependent expression of a Ste11 derivative with a C-terminal fusion to an 11-aa myc epitope. Construction of pNC245 has been described (2). pNC734 carries the same GAL-STE11M allele in the YCp50 (CEN4 URA3) vector (28). YEpdoa4C571S (URA3) is a 2-μm-based plasmid allowing expression of the dominant negative doa4C571S allele (29). pGA1903 (TPI1-FUS3M CEN3 TRP1) was used for constitutive expression of myc epitope-tagged Fus3 (Fus3M) (6).
Ste11 Protein Stability Determinations.
Cultures were grown to early log phase (1–2 × 107 cells/ml) in the appropriate selective medium containing 2% galactose for expression of GAL-STE11M. Cells were harvested, washed, and switched to medium containing 2% dextrose to inhibit further GAL-STE11M transcription. After removal of initial samples for protein extract preparation (time 0), incubation of the cultures continued without or with pheromone (50 nM α-factor) for induction of the mating pathway activation cascade or with 0.4 M NaCl for induction of the HOG cascade. Whole-cell protein extracts were prepared for immune blot analysis either by the method of Mattison et al. (30) or by a modified Pfiefer et al. (31) procedure in which the ammonium sulfate fractionation step was omitted.
Immune Blot Analysis.
Whole-cell protein extracts (100 μg) were fractionated by using 10% SDS/PAGE. The 9E10 anti-myc mAbs were used at 10 μg/ml to detect Ste11M or Fus3M (2, 32). Antitubulin mAbs were used at 0.17 μg/ml to detect the constitutively expressed Tub1 reference protein (Accurate Chemicals). Goat anti-mouse IgG conjugated to alkaline phosphatase (0.17 μg/ml) was used as the secondary antibody for colorimetric detection with the Promega Protoblot immuno blot system. Quantification of protein signals on immune blot images was accomplished by using Scion IMAGE 1.60c software.
Immune Precipitation Kinase Assay.
The Fus3M Doa4 (E929–6C pGA1903) and Fus3M Doa4C571S (E929–6C pGA1903 YEpdoa4C571S) strains were grown in appropriate selective medium to a density of ≈1 × 107 cells/ml. Mating pheromone α-factor was added to a final concentration of 3 μM. Extracts from samples were prepared at the indicated times for immune complex kinase assays as described by Errede et al. (3). The same extract preparations were used for Fus3M immune blot analysis (see above). Immune complexes were isolated under conditions where activities vs. immune complex (beads) are in the linear range: 200 μg of extract using 16 μg anti-myc 9E10 antibodies, 25 μl protein A Sepharose beads (Amersham Pharmacia). Kinase assays with γ-32ATP (DuPont-NEN) and myelin basic protein (MBP) as substrate were performed directly on the washed immune complexes as described (3). Proteins in the immune complex kinase reaction were fractionated by SDS/PAGE (12.5%), and the amount of 32P incorporated into MBP was determined by PhosphorImage analysis (Molecular Dynamics) of the dried gel.
Results
Pheromone Stimulation Decreases Ste11 Protein Stability.
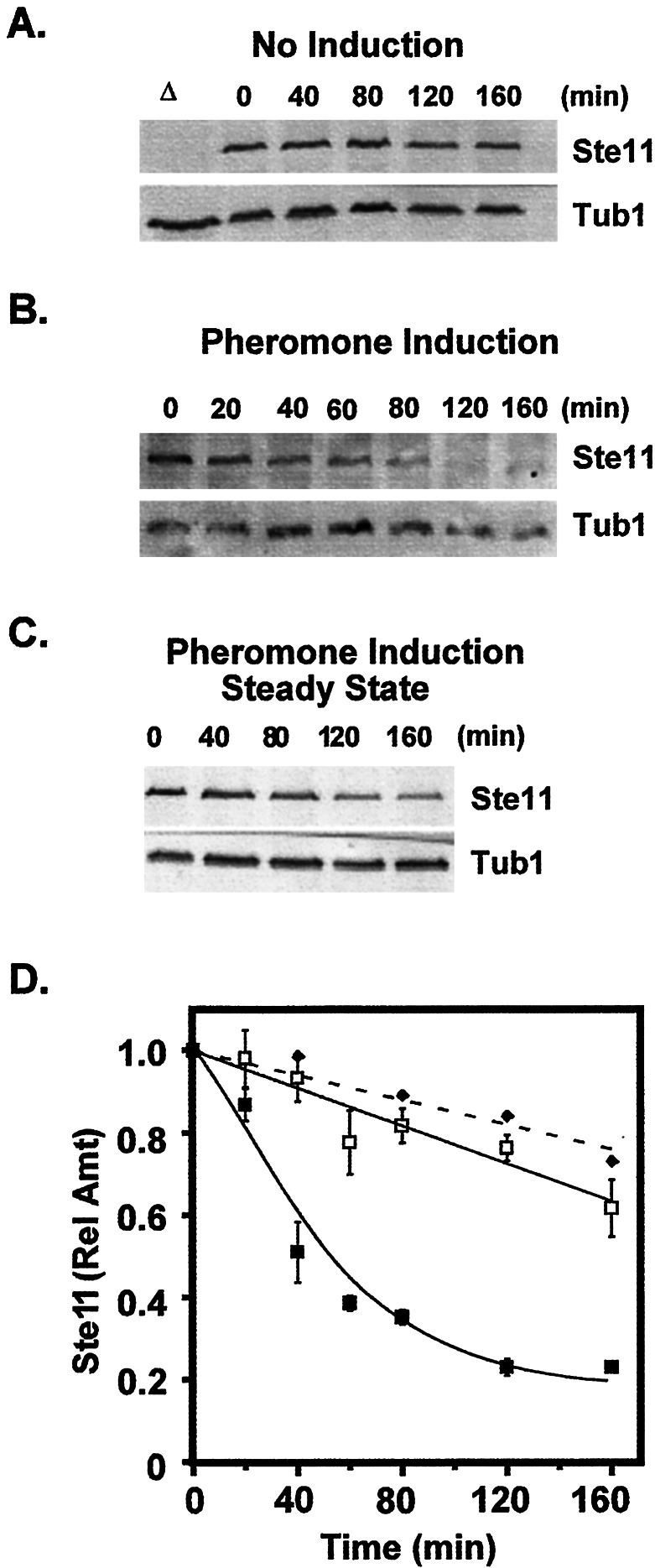
We compared the half-life of Ste11 protein in unstimulated and pheromone-stimulated cells. To make this comparison, we used a plasmid that expresses a myc epitope-tagged version of Ste11 (Ste11M) under control of the regulated GAL1,10 promoter (2). A yeast strain with this allele was grown to early log phase under conditions (galactose medium) that allowed transcription of Ste11M. The culture was switched to medium (glucose medium) that blocks further transcription of Ste11M. Concurrent with the change in medium, one portion of the culture was induced with mating pheromone and the other was maintained as a no induction reference. Whole-cell protein extracts were prepared from aliquots of the cultures at the indicated times after the shift, and the amount of Ste11M protein that persisted in the samples was assessed by immune blot analysis using anti-myc antibodies (Fig. 1). The amount of Ste11M protein in each sample is reported as the ratio of the signal for Ste11M to that for an internal reference (Tub1). Because GAL-STE11 mRNA may persist and continue to be translated, the protein half-lives measured by this approach may be longer than what would be measured for endogenous Ste11 in the absence of protein synthesis. Nevertheless, the relative difference in the half-life in unstimulated vs. stimulated cultures provides a reasonable indicator of whether protein stability is being affected. This analysis reveals that the Ste11 protein has a significantly shorter half-life (t1/2 = 50 min) under pheromone-inducing conditions compared with noninducing conditions (t1/2 200 min) (Fig. 1 A, B, and D). For reference, we examined the amount of Ste11M in cultures undergoing pheromone induction with galactose in the medium so that transcription of GAL-STE11M continued (Fig. 1 C and D). Because the steady-state amount of Ste11M shows little decrease during exposure to pheromone, we infer that new protein synthesis offsets the loss caused by accelerated protein turnover (see below).
Figure 1.
Pheromone induction accelerates degradation of Ste11. Representative immune blots showing the time-dependent amount of Ste11M (A–C Upper) from extracts of a wild-type strain (C699–15 pNC734) without induction (A) and with pheromone induction (B and C). Extracts from cultures switched to dextrose show the persistence of pre-existing Ste11M (A and B) whereas those from cultures maintained in galactose show the steady-state amount of Ste11M (C). The steady-state amount of tubulin (Tub1) in each sample serves as an internal reference (A–C Lower). (D) Plots of the relative amount of Ste11M (Ste11M/Tub1) vs. time from no induction dextrose (□), pheromone induction dextrose (■), and pheromone induction galactose (⧫) cultures. The amount of Ste11M at each time is the average density of the Ste11M signal divided by the average density of the Tub1 signal. Relative amounts at different times are normalized to the amount of Ste11M at t = 0 for the corresponding time course. Values shown on the plots are the average from three or more independent experiments. Bars show the average deviation for each point.
A Feedback and Ubiquitin-Dependent Process Mediates the Pheromone-Induced Change in Ste11 Protein Half-Life.
To confirm that the apparent depletion of Ste11M in the absence of transcription was caused by protein turnover and to test whether turnover was mediated by an ubiquitin-dependent pathway, we used the doa4-C571S active site mutation (29). Doa4 is a deubiquitinating enzyme that recycles ubiquitin from both proteosome-bound ubiquitinated intermediates and membrane proteins destined for destruction in the vacuole (33–35). Expression of the doa4-C571S mutation from a high-copy plasmid in wild-type cells inhibits degradation of ubiquitin-dependent substrates, presumably by competition with the normal Doa4 for binding to component(s) of the proteolytic machinery (29).
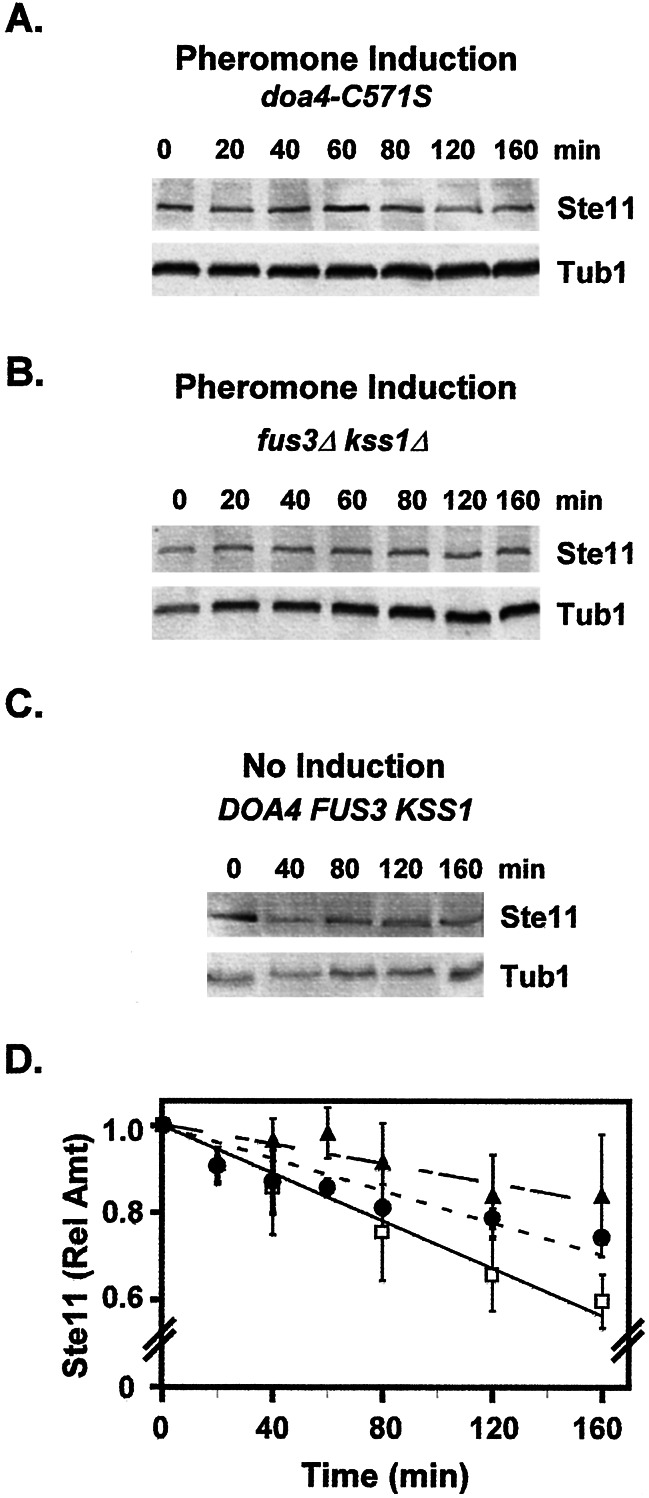
In contrast to cultures undergoing pheromone induction in the DOA4 reference strain, the half-life of Ste11M in the doa4-C571S overexpressing strain was found to be >200 min, which is the same or greater than that observed for Ste11M in the culture without pheromone induction (Fig. 2 A, C, and D). This result establishes that pheromone induction accelerates Ste11 degradation through an ubiquitin-dependent mechanism. We have not been successful at detecting Ste11-ubiquitin or any other ubiquitin-conjugated proteins in yeast and therefore have not been able to ascertain whether Ste11 is directly ubiquitinated.
Figure 2.
Pheromone-induced degradation of Ste11 occurs through a MAPK feedback and ubiquitin-dependent mechanism. Representative immune blots showing the time-dependent decrease in pre-existing Ste11M (A–C Upper) from extracts of a wild-type strain (C699–15 pNC245) without pheromone induction (C), the same strain overexpressing the dominant negative doa4-C571S allele (C699–15 pNC245 YEpdoa4C571S) during pheromone induction (A) and an isogenic fus3Δkss1Δ strain (C699–75 pNC245) during pheromone induction (B). The steady-state amount of tubulin (Tub1) in each sample serves as an internal reference (A–C Lower). (D) Plots of the relative amount of Ste11M (Ste11M/Tub1) vs. time from wild type no pheromone (□), doa4-C571S with pheromone (▴), and fus3Δkss1Δ with pheromone (●) strains. Determination of the relative amount of Ste11 at each time point is as described in Fig. 1. Values are the average from three independent experiments. Bars show the average deviation for each point.
The diminished stability of Ste11 under pheromone-inducing conditions could be either a direct or indirect consequence of its activation. One indirect mechanism might involve a feedback loop that depends on Fus3 and Kss1. To test this possibility, the above protocol was applied to determine the half-life of Ste11M under pheromone-inducing conditions in an isogenic fus3Δkss1Δ strain. The Ste11 protein has the same or greater half-life in the fus3Δkss1Δ strain undergoing pheromone induction as in the wild-type strain without pheromone induction (Fig. 2 B–D). As it was previously established that Ste11 is activated by pheromone in this strain background (6), the finding here eliminates the possibility that degradation of Ste11 is caused merely by its conversion to an active form. Instead, Ste11 degradation depends on a feedback mechanism involving the MAPKs of this pathway.
Ste11 Protein Stability Is Not Affected by Hyperosmotic Stress.
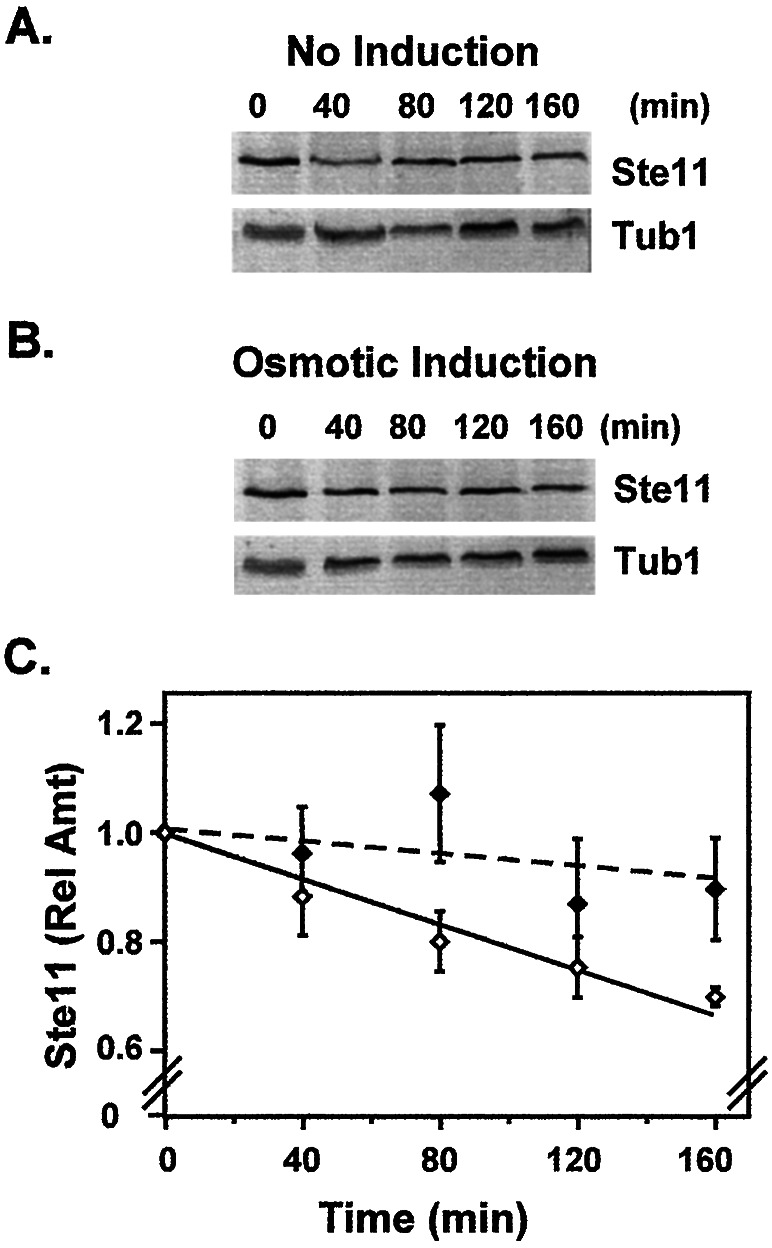
To test whether Ste11 is similarly regulated in the HOG pathway, we used the above protocol to measure the half-life of Ste11M in cultures undergoing high osmolarity (0.4 M NaCl) induction. In contrast to pheromone stimulation, Ste11M half-life is as long or longer than that observed in the absence of stimulation (Fig. 3). O'Rourke and Herskowitz (36) reported a Hog1-dependent feedback mechanism that normally down-regulates the Sho1 branch of the HOG pathway. Our result shows that target selection and/or the mode of inhibition associated with the Hog1-dependent mechanism differs from that of the Fus3/Kss1-dependent mechanism. Further, this outcome establishes that Ste11 is subject to different modes of regulation in the two pathways.
Figure 3.
Osmotic induction does not accelerate Ste11 degradation. Representative immune blots showing the time-dependent decrease in pre-existing Ste11M (A and B Upper) from extracts of a ssk2Δ ssk22Δ strain (FP50 pNC734) without (A) or with osmotic induction (B). The steady-state amount of Tub1 in each sample serves as an internal reference (A and B). (C) Plots of the relative amount of Ste11M (Ste11M/Tub1) vs. time from the no induction (open symbols) and 0.4 M NaCl induction (closed symbols) time courses. Determination of the relative amount of Ste11 at each time point is as described in Fig. 1. Values are the average from three independent experiments. Bars show the average deviation for each point.
Ubiquitin-Dependent Degradation Contributes to Attenuation of MAPK Activation in Response to Pheromone.
The responsiveness of cells to persistent pheromone induction diminishes with time. This phenomenon occurs through several desensitization or adaptation mechanisms that impinge on different components of the signaling pathway (37–45). The feedback and ubiquitin-dependent degradation of Ste11 could be an additional mechanism that contributes to desensitization. If this is the case, the absence of ubiquitin-dependent degradation should compromise normal signal attenuation and might even increase the signal intensity because the active pool of Ste11 is not eliminated.
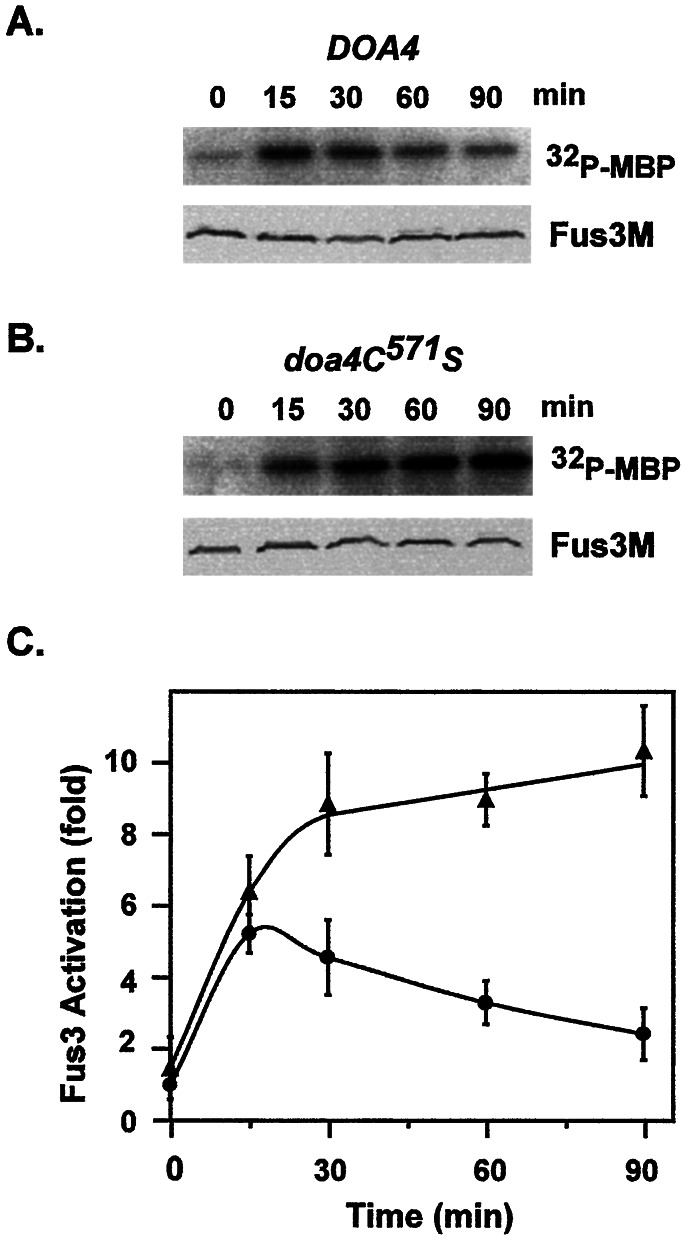
To test this prediction, we compared the Fus3 activation profile for DOA4 cells where Ste11 turnover occurs to that for doa4-C571S cells, where Ste11 is stable. To allow measurement of Fus3 kinase activity by immune complex assays, the DOA4 and doa4C571S strains expressed a myc epitope-tagged version of Fus3 (Fus3M) from the constitutive TPI1 promoter (6). Protein extracts were prepared from samples before pheromone addition (t = 0) and at the indicated times during pheromone induction. The amount of Fus3M present in each extract was assessed by immune blot analysis using anti-myc antibodies and was found to remain constant throughout the time course in both DOA4 and doa4C571S strain backgrounds (Fig. 4 A and B Lower). Immune complex kinase assays were performed with each extract by using MBP as substrate. Fus3 activity during pheromone induction in the DOA4 background showed the typical time-course profile (Fig. 4 A Upper and C). During the first 15–30 min of exposure to pheromone, Fus3 activity increases ≈5-fold relative to the unstimulated culture. This increase in activity then declines during adaptation to the persistent stimulus. By contrast, more active Fus3 accumulated in the doa4-C571S background (7- to 10-fold) during the initial phase of the induction profile and, as predicted, the active pool, which persists in this background, delayed or prevented the adaptation phase of Fus3 kinase activation (Fig. 4 B Upper and C).
Figure 4.
Ubiquitin-dependent degradation is required for down-regulation of MAPK activity during the adaptive response to pheromone. Representative PhosphorImage of 32P incorporation into MBP (A and B Upper) from Fus3M immune complex kinase assays and immune blots of Fus3M (A and B Lower) by using extracts from (A) a DOA4 strain (E929–6C pGA1903) and (B) a doa4C571S (E929–6C pGA1903 YEpdoa4C571S) strain during a pheromone induction time course. (C) Plots of pheromone-induced Fus3 activation in DOA4 (●) and doa4C571S (▴) strains. Activities are determined from PhosphorImage quantification of 32P incorporation into MBP. Fus3 activation is in arbitrary units relative to the average value obtained for Fus3 immune complex assays using extracts isolated from the respective DOA4 and doa4C571S strain extracts at t = 0 min. Plot shows the average from four assays using extracts from three independent time courses. Bars show the average deviation for each point.
Discussion
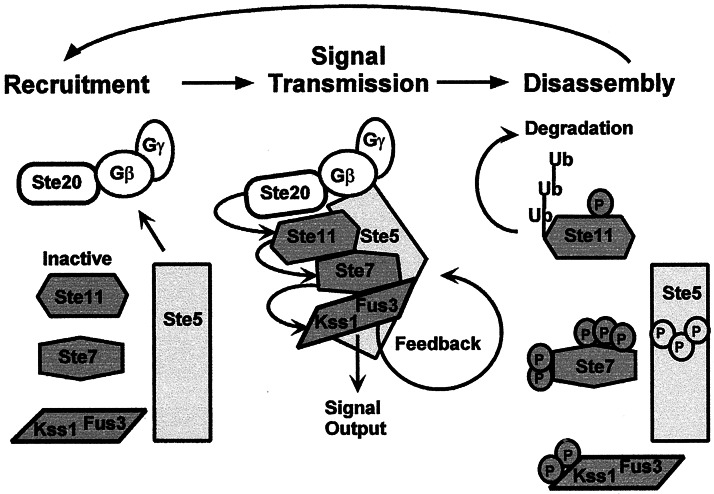
The feedback-dependent regulation of Ste11 turnover during pheromone induction suggests that the active pool of Ste11 is preferentially targeted for ubiquitin-mediated degradation. The question then arises as to how signal output is sustained during the time typically required for mating differentiation and cell fusion (≈90 min). One mechanism to meet this requirement could involve a cycle involving activation of Ste11 from a “naïve” pool that is constantly replenished with newly synthesized protein (Fig. 5). In this cycle, Ste5 binds inactive Ste11 and takes it to the plasma membrane for phosphorylation and activation by Ste20 (46, 47). The active Ste11 continues the phosphorylation cascade, leading to MAPK activation (3, 4). Once activated, Fus3/Kss1 then directly or indirectly promotes the rapid ubiquitin-dependent degradation of active Ste11. This model is consistent with the failure of van Drogen et al. (48) to detect a functional Ste11–green fluorescent protein fusion at the tip of mating projections where other components of the activation cascade were observed to accumulate during pheromone induction. Because protein phosphorylation is often a prerequisite for ubiquitination, it is plausible that Fus3/Kss1 directly phosphorylate Ste11 and that this modification within the complex at the plasma membrane targets Ste11 for degradation. The possibility of direct feedback phosphorylation seems reasonable because Fus3 (or Kss1) feedback phosphorylates other components of the cascade (3, 49, 50). Although seemingly wasteful, the cycle of synthesis and degradation would impart high signal-to-noise characteristics to the cascade and possibly contribute to the specificity of signal output.
Figure 5.
Model for the cycle of Ste11 synthesis, activation, and degradation during pheromone-induced stimulation of the mating pathway MAPK activation cascade. See text for explanation.
Another aspect of the above model is that ubiquitin-dependent Ste11 degradation provides an inherent brake to the pathway that can stop the activation cascade immediately upon attenuation of the upstream signal. Our comparison of Fus3 activation profiles in the wild-type and doa4C571S strains establishes that ubiquitin-dependent degradation is necessary for attenuation (Fig. 4). The accumulation of an active pool of Ste11 in the doa4C571S background would counter the inactivation of Fus3 by the Msg5 dual-specificity phosphatase and explain abolishment of the characteristic attenuation profile (44). The only other components upstream of Fus3 in the pathway that are known to undergo ubiquitin-dependent degradation are the receptor and the Gα subunit (41, 51). Although the doa4C571S background used here would also stabilize these two upstream components, their stabilization would not abolish adaptation. G protein activation is down-regulated by receptor phosphorylation, a mechanism that is independent of ubiquitin-mediated degradation (40). Therefore, desensitization to pheromone still occurs with respect to G protein activation. Further, a lysine to arginine substitution mutation in Gα-Gpa1 that prevents its ubiquitin-dependent degradation causes an enhanced adaptation response (52). It remains possible that other components of the pathway upstream from Fus3 might be subject to ubiquitin-dependent degradation and would also contribute to attenuation of Fus3 activation. We have previously shown that Ste7 protein persists during pheromone induction in the absence of protein synthesis (6) and thus is unlikely to contribute to attenuation by this mechanism. Further, the observations of van Drogen et al. (48) concerning Ste5, Ste11, Ste7, and Fus3 localization and persistence during pheromone induction suggest that Ste11 indeed may be the component of the activation cascade that is most acutely affected by ubiquitin-dependent degradation. This view argues that persistence of Ste11 is likely to be primarily responsible for abrogation of the MAPK attenuation profile that we observed in our analysis.
Acknowledgments
This research was supported by Public Health Grant GM-39582 from the National Institutes of Health.
Abbreviations
- MAPK
mitogen-activated protein kinase
- HOG
high osmolarity glycerol
- MBP
myelin basic protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hartwell L H. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhodes N, Connell L, Errede B. Genes Dev. 1990;4:1862–1874. doi: 10.1101/gad.4.11.1862. [DOI] [PubMed] [Google Scholar]
- 3.Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G. Nature (London) 1993;362:261–264. doi: 10.1038/362261a0. [DOI] [PubMed] [Google Scholar]
- 4.Neiman A M, Herskowitz I. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevenson B J, Rhodes N, Errede B, Sprague G F., Jr Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z, Gartner A, Cade R, Ammerer G, Errede B. Mol Cell Biol. 1993;13:2069–2080. doi: 10.1128/mcb.13.4.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkreutz A, Boucher L, Tyers M. Curr Biol. 2001;11:1266–1271. doi: 10.1016/s0960-9822(01)00370-0. [DOI] [PubMed] [Google Scholar]
- 8.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 9.Cullen P J, Sprague G F., Jr Proc Natl Acad Sci USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook J G, Bardwell L, Thorner J. Nature (London) 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Styles C A, Fink G R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 12.Madhani H D, Styles C A, Fink G R. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 13.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 14.Maeda T, Takekawa M, Saito H. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 15.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 16.Posas F, Saito H. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 17.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 18.Yashar B, Irie K, Printen J A, Stevenson B J, Sprague G F, Jr, Matsumoto K, Errede B. Mol Cell Biol. 1995;15:6545–6553. doi: 10.1128/mcb.15.12.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi K Y, Satterberg B, Lyons D M, Elion E A. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 20.Printen J A, Sprague G F., Jr Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 22.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothstein R J. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 24.Boeke J D, LaCroute F, Fink G R. Mol Gen Genet. 1994;197:354–356. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 25.Alani E, Cao L, Kleckner N. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Courchesne W E, Kunisawa R, Thorner J. Cell. 1989;58:1107–1119. doi: 10.1016/0092-8674(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Johnston M, Davis R W. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papa F R, Hochstrasser M. Nature (London) 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- 30.Mattison C P, Spencer S S, Kresge K A, Lee J, Ota I M. Mol Cell Biol. 1999;19:7651–7660. doi: 10.1128/mcb.19.11.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeifer K, Arcangioli B, Guarente L. Cell. 1987;49:9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- 32.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papa F R, Amerik A Y, Hochstrasser M. Mol Biol Cell. 1999;10:741–756. doi: 10.1091/mbc.10.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swaminathan S, Amerik A Y, Hochstrasser M. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amerik A Y, Nowak J, Swaminathan S, Hochstrasser M. Mol Biol Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Rourke S M, Herskowitz I. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Y, Davis N G. Mol Cell Biol. 2000;20:563–574. doi: 10.1128/mcb.20.2.563-574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenness D D, Spatrick P. Cell. 1986;46:345–353. doi: 10.1016/0092-8674(86)90655-0. [DOI] [PubMed] [Google Scholar]
- 39.Reneke J E, Blumer K J, Courchesne W E, Thorner J. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, Konopka J B. Mol Cell Biol. 1996;16:247–257. doi: 10.1128/mcb.16.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hicke L, Riezman H. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 42.Dohlman H G, Song J, Ma D, Courchesne W E, Thorner J. Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apanovitch D M, Slep K C, Sigler P B, Dohlman H G. Biochemistry. 1998;37:4815–4822. doi: 10.1021/bi9729965. [DOI] [PubMed] [Google Scholar]
- 44.Doi K, Gartner A, Ammerer G, Errede B, Shinkawa H, Sugimoto K, Matsumoto K. EMBO J. 1994;13:61–70. doi: 10.1002/j.1460-2075.1994.tb06235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhan X L, Deschenes R J, Guan K L. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- 46.Sette C, Inouye C J, Stroschein S L, Iaquinta P J, Thorner J. Mol Biol Cell. 2000;11:4033–4049. doi: 10.1091/mbc.11.11.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drogen F, O'Rourke S M, Stucke V M, Jaquenoud M, Neiman A M, Peter M. Curr Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- 48.van Drogen F, Stucke V M, Jorritsma G, Peter M. Nat Cell Biol. 2001;3:1051–1059. doi: 10.1038/ncb1201-1051. [DOI] [PubMed] [Google Scholar]
- 49.Errede B, Ge Q Y. Philos Trans R Soc London B. 1996;351:143–148. doi: 10.1098/rstb.1996.0010. ; discussion 148–149. [DOI] [PubMed] [Google Scholar]
- 50.Kranz J E, Satterberg B, Elion E A. Genes Dev. 1994;8:313–327. doi: 10.1101/gad.8.3.313. [DOI] [PubMed] [Google Scholar]
- 51.Madura K, Varshavsky A. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 52.Marotti L A, Jr, Newitt R, Wang Y, Aebersold R, Dohlman H G. Biochemistry. 2002;41:5067–5074. doi: 10.1021/bi015940q. [DOI] [PubMed] [Google Scholar]