Abstract
Legionella pneumophila is a motile intracellular pathogen of macrophages and amoebae. When nutrients become scarce, the bacterium induces expression of transmission traits, some of which are dependent on the flagellar sigma factor FliA (σ28). To test how particular components of the L. pneumophila flagellar regulon contribute to virulence, we compared a fliA mutant with strains whose flagellar construction is disrupted at various stages. We find that L. pneumophila requires FliA to avoid lysosomal degradation in murine bone marrow-derived macrophages (BMM), to regulate production of a melanin-like pigment, and to regulate binding to the dye crystal violet, whereas motility, flagellar secretion, and external flagella or flagellin are dispensable for these activities. Thus, in addition to flagellar genes, the FliA sigma factor regulates an effector(s) or regulator(s) that contributes to other transmissive traits, notably inhibition of phagosome maturation. Whether or not the microbes produced flagellin, all nonmotile L. pneumophila mutants bound BMM less efficiently than the wild type, resulting in poor infectivity and a loss of contact-dependent death of BMM. Therefore, bacterial motility increases contact with host cells during infection, but flagellin is not an adhesin. When BMM contact by each nonmotile strain was promoted by centrifugation, all the mutants bound BMM similarly, but only those microbes that synthesized flagellin induced BMM death. Thus, the flagellar regulon equips the aquatic pathogen L. pneumophila to coordinate motility with multiple traits vital to virulence.
Legionella pneumophila is a ubiquitous gram-negative motile bacterium that replicates inside protozoa in aquatic environments (27, 77, 86). When aerosols from contaminated water sources are inhaled by humans, L. pneumophila can also replicate within phagocytic alveolar macrophages (43), causing the life-threatening pneumonia Legionnaires' disease (60). Like many intracellular pathogens, L. pneumophila alternates between two distinct life cycle phases: an intracellular replicative phase and an infectious transmissive phase (reviewed in references 65 and 76). Transmissive microbes express a number of virulence-associated traits absent in replicative microbes, prominent among them flagellar production and subsequent motility.
In addition to motility, two other transmissive-phase virulence traits are the ability to (i) isolate the microbial vacuole from the endosomal network of phagocytes to avoid lysosomal degradation (lysosome avoidance) and (ii) induce rapid, replication-independent macrophage death (cytotoxicity). One L. pneumophila structure essential for both traits is the type IV secretion system encoded by the dot/icm loci (11, 21, 51, 78, 82, 93). In addition, a link between flagellation and virulence-associated traits is well established (15, 17, 38, 72), and regulators that control flagellation also affect both lysosome avoidance and cytotoxicity (7, 36, 66; also see below). However, the specific mechanisms that link flagellation and virulence are not known and are the subject of this report.
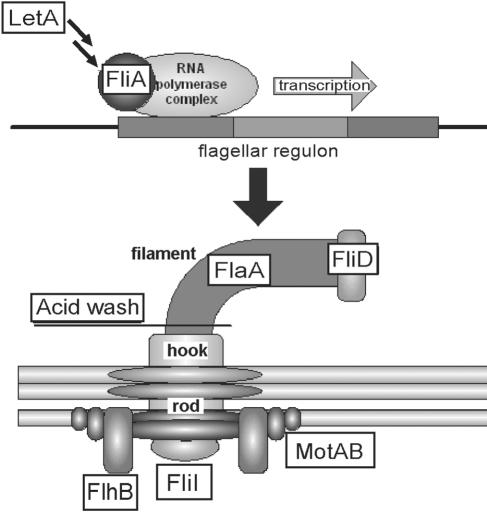
Motility is conferred by the flagellum, a large protein complex encoded by flagellar regulon genes whose temporal expression pattern is highly coordinated (Fig. 1) (reviewed in references 2 and 59). The ancestor of type III secretion systems, the flagellar secretion apparatus, exports the majority of flagellar components to the bacterial periplasm or external milieu, including rod, hook, flagellin, and capping proteins. The basal body stabilizes the flagellum, and the rotational forces generated by the Mot ion channels are transmitted from the basal body through the hook to the external filament, causing filament rotation. In addition to the structural genes necessary to form the flagellum, a number of regulators are dedicated to L. pneumophila flagellin (flaA) expression, and several of these also govern virulence (reviewed in references 40 and 65). These include the flagellar sigma factor FliA (36, 38, 39) and the LetA/S two-component system (29, 36, 58). LetA/S indirectly induces motility and a host of other transmission traits by inhibiting the global regulator CsrA, resulting in the expression of fliA and flaA (26, 65, 66). As in Pseudomonas and Vibrio sp. flagellar regulation (23, 71), the L. pneumophila master flagellar regulators are RpoN and the coactivator FleQ (48), whereas FliA (σ28) may independently govern several flagellar elements, including flaA expression.
FIG. 1.
Flagellar schematic, indicating components analyzed in this study. LetA is a two-component response regulator that governs a broad array of phenotypes, including flagellation and motility. FliA, a flagellar sigma factor (σ28), transcribes several vital genes of the flagellar regulon, including flaA. FlaA, flagellin, is the protein that comprises the external flagellar filament. MotAB is the ion channel that drives flagellar filament rotation. FliD (Hap2) caps the filament and allows for flagellin polymerization into a function flagellum. FlhB is an integral component of the flagellar basal body, required for flagellar secretion. FliI, flagellar ATPase, is required for flagellar secretion. An acid wash treatment was used to dissociate external proteins, including flagellar filaments.
To establish a replication niche, many virulent bacteria utilize flagella at multiple stages of infection. Flagella not only mediate chemotaxis toward favorable environments but also contribute to various virulence traits, including microbial adhesion or invasion (3, 33, 47, 52, 80, 90) and toxin delivery (31, 54, 84, 100). Further, flagellar production is often coregulated with other virulence systems (reviewed in references 40 and 70). L. pneumophila coordinately regulates flagellation and virulence-associated traits, including intracellular replication. Flagella per se are dispensable for replication in macrophages or amoebae (24, 36, 48, 61). However, L. pneumophila requires the regulator FliA for motility, for intracellular growth in the social amoeba Dictyostelium discoideum (38), and for efficient infection and cytotoxicity in murine bone marrow-derived macrophages (BMM) (36, 38, 66). Accordingly, the flagellar regulon may directly influence lysosome avoidance and/or cytotoxicity, two early events during macrophage infection that are not easily detected solely by determining bacterial CFU. Although the flaA locus is one target of FliA induction (36, 38, 39), other targets remain unknown but likely include genes within the flagellar regulon such as flhB and fliD (reviewed in reference 40). Because fliA mutants have multiple defects, FliA may also regulate other genes outside the flagellar system. To test which flagellar components contribute to the L. pneumophila transmission phenotype, including lysosome avoidance and cytotoxicity, we systematically constructed and analyzed a group of mutants defective at various stages of flagellar development (Fig. 1).
MATERIALS AND METHODS
Bacterial strains, media, and reagents.
L. pneumophila Lp02 (thyA hsdR rpsL; MB110), a virulent thymine auxotroph derived from the serogroup 1 clinical isolate Philadelphia 1 (10), was the parent for all mutant strains analyzed (Table 1). All mutants are indicated by both strain number and the lpg locus designation from the L. pneumophila Philadelphia 1 genome (20). L. pneumophila was cultured at 37°C in N-(2-acetamido)-2-aminoethanesulfonic acid (ACES; Sigma)-buffered yeast extract (AYE) broth in 5-ml aliquots on a rotating wheel or on ACES-buffered charcoal yeast extract (CYE) agar, both supplemented with 100 μg ml−1 thymidine (AYET and CYET, respectively). Bacteria from frozen stocks were streaked onto CYET plates with appropriate antibiotics, and <2-week-old colonies were cultured in broth overnight to mid-exponential phase, then subcultured in fresh AYET for an additional 16 to 24 h prior to experimentation. Exponential (E)-phase cultures were defined as having an optical density at 600 nm (OD600) of 0.4 to 2.0, and post-exponential (PE) cultures as having an OD600 of 3.5 to 4.5 and exhibiting full motility and a compact, coccoid shape. We noted that a minority of nonmotile wild-type (WT) E-phase microbes are at least partially flagellate (see Fig. 2), are cytotoxic to BMM (see Fig. 5), and avoid degradation in BMM (see Fig. 3C). Although plate-grown microbes are cultured in broth for >24 h to obtain synchronous E-phase cultures for analysis, a variable minority of WT E-phase microbes do maintain a subset of PE-phase transmission traits, including flagellation, perhaps from either their prior growth on agar or the lag phase in broth. Indeed, variability was observed for the cytotoxicity of WT E microbes, in part dependent on the optical density of the culture (Fig. 5B). Antibiotics were not used in broth culture, unless noted. To ascertain CFU, serial dilutions of bacteria were incubated on duplicate CYET plates for 4 days and resultant colonies counted. The proton ionophore carbonyl cyanide m-chlorophenylhydrazone (10 μM; Sigma) or the sodium-channel-blocking agent phenamil (0.8 mM, Sigma) was used to block motility by coincubation with microbes for 10 min.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype/phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F−endA1 hsdR17 (r− m+) supE44 thi-l recA1 gyrA (Nal1) relA1 Δ(lacZYA-argF−)U169 φ80dlacZΔM15 λpirRK6 | Laboratory collection |
| L. pneumophila | ||
| MB110 | Lp02 wild type; thyA hsdR rpsL | 10 |
| MB413 | Lp02 letA 22-3::kan mutant (lpg2646) | 36 |
| MB416 | Lp02 letS 36::kan mutant (lpg1912) | 36 |
| MB410 | Lp02 fliA 35::kan mutant (lpg1782) | 36 |
| MB510 | Lp02 fliA 35::kan mutant pMMBGent-FliA | This work |
| MB550 | Lp02 fliA 35::kan mutant preg-FliA | This work |
| MB462 | Lp02 fliA 35::kan mutant pMMBGent-Δmob vector control | This work |
| MB460 | Lp02 dotA::gent mutant (lpg2646) | 66 |
| MB379 | Lp02 rpoS::kan mutant (lpg1284) | 8 |
| MB473 | Lp02 pMMBGentΔmob; vector control | 35 |
| MB552 | Lp02 fliD::kan mutant (lpg1338) | This work |
| MB553 | Lp02 fliD::gent mutant | This work |
| MB554 | Lp02 flhB::gent “A” mutant (lpg1786) | This work |
| MB555 | Lp02 flhB::gent “C” mutant | This work |
| MB556 | Lp02 flhB::kan mutant | This work |
| MB534 | Lp02 flaA::kan mutant (lpg1340) | This work |
| MB557 | Lp02 flaA::kan mutant pMMBGent-flaA | This work |
| MB558 | Lp02 flaA::kan mutant pMMBGentΔmob, vector control | This work |
| MB532 | Lp02 flaA::gent mutant | This work |
| MB560 | Lp02 motAB::gent “A” mutant (lpg1780-81) | This work |
| MB561 | Lp02 motAB::gent “C” mutant | This work |
| MB562 | Lp02 motAB::gent “A” pMMB206-motAB | This work |
| MB563 | Lp02 motAB::gent “A” pMMB206; vector control | This work |
| MB564 | Lp02 fliI::cam mutant (lpg1757) | 61 |
| MB565 | Lp02 flI::cam fliD::gent “2A” double mutant | This work |
| MB566 | Lp02 flI::cam fliD::gent “2B” double mutant | This work |
| MB567 | Lp02 flaA::gent fliD::kan “A” double mutant | This work |
| MB568 | Lp02 flaA::gent fliD::kan “B” double mutant | This work |
| MB573 | Lp02 flhB::gent “A” fliA 35::kan double mutant | This work |
| Plasmids | ||
| pMMBGent-Δmob | pMMB67EH derivative; Δmob lacIq Ptac Gentr; broad-host-range vector | 35 |
| pMMB206-Δmob | pMMB66EH derivative; Δmob lacIq PtaclacUV5 Camr | 66 |
| pUC4K | Source of 1.3-kb kanamycin resistance GenBlock | Pharmacia |
| pGEMT-Easy | MCS within coding region of β-lactamase α fragment linearized with single-T overhangs, 3 kb; Ampr | Promega |
| pGEMT-Gent | pGEMT-Easy with 1.7-kb Gentr cassette from BH038 cloned MCS, source of Gent cassette; Ampr Gentr | This work |
| pGEMT | MCS within coding region of β-lactamase α fragment, lacking EcoRI sites of T-Easy; Ampr | Promega |
| pGEM-flaA | pGEMT-Easy with 1.876-kb PCR-amplified flaA chromosomal region, ligated into T overhangs; Ampr | This work |
| pGEM-ΔflaA-gent | pGem-flaA with 1.7-kb Gentr cassette disrupting internal EcoRI sites at 393-455 bases of 1,428-base FlaA ORF | This work |
| pGEM-ΔflaA-kan | pGem-flaA with 1.3-kb Kanr cassette disrupting internal EcoRI sites at 393-455 bases of 1,428-base FlaA ORF | This work |
| pMMBGent-FlaA | pMMBGent-Δmob with ∼3.0 kb of FlaA genomic region with native promoter and ORF ligated into MCS Complementing plasmid, Gentr | This work |
| pGEM-fliA | pGEMT-Easy with 0.94-kb PCR-amplified flaA chromosomal region, ligated into T overhangs; Ampr | This work |
| pMMBGent-FliA | 940-pp FliA genomic region from pGEM-fliA cut with EcoRI, isolated, and ligated into EcoRI site of pMMBGent-Δmob, noninducible, complementing plasmid, Gentr | This work |
| pregFliA | pMMB206-Δmob with ∼750-bp FliA RBS and ORF PCR product cloned into PtaclacUV5 site, IPTG-inducible FliA expression; Camr | This work |
| pGEM-fliD | pGEMT (not T-Easy) with 3.457-kb PCR-amplified fliD chromosomal region, ligated into T overhangs; Ampr | This work |
| pGEM-ΔfliD-kan | pGem-fliD with 1.3-kb Kanr cassette disrupting internal EcoRI site at base 641 of 1,626-base FliD ORF; Ampr Kanr | This work |
| pGEM-ΔfliD-gent | pGem-fliD with 1.7-kb Gentr cassette disrupting internal EcoRI site at base 641 of 1,626-base FliD ORF; Ampr Gentr | This work |
| pGEM-motAB | pGEMT-Easy with 1,600-bp PCR-amplified motAB chromosomal region, ligated into T overhangs; Ampr | This work |
| pGEM-ΔmotAB-gent1 (A) | pGEM-motAB with 1.9-kb Gentr cassette inserted between StuI and ClaI sites in opposite orientation to motAB, losing ∼130 bp of motAB region; Ampr Gentr | This work |
| pGEM-ΔmotAB-gent2 (C) | pGEM-motAB with 1.9-kb Gentr cassette inserted between StuI and ClaI sites in same orientation as motAB, losing ∼130 bp of motAB region; Ampr Gentr | This work |
| pMMB206-motAB | 1.7-kb motAB ORF region amplified with BamH1-MotA5 and SalI-MotB3 primers, digested, and ligated into pMMB206-Δmob at the PtaclacUV5 site, IPTG-inducible MotAB expression; Camr | This work |
| pGEM-flhB | pGEMT-Easy with 3.4-kb PCR-amplified flhB chromosomal region ligated into T overhangs; Ampr | This work |
| pGEM-ΔflhB-EcoRV | pGem-flhB with addition of internal N-terminal EcoRV site at base 137 of the 1,140-bp FlhB ORF and loss of ∼770 bp of the ORF | This work |
| pGEM-ΔflhB-gent | pGem-flhB-EcoR V with 1.7-kb Gentr cassette inserted at EcoRV site disrupting the truncated FlhB; Ampr Gentr | This work |
MCS, multicloning site; RBS, ribosome binding site.
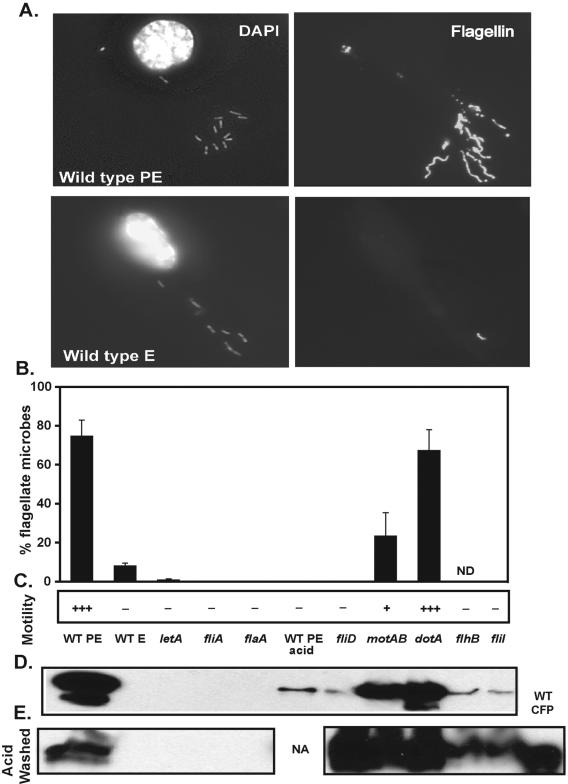
FIG. 2.
Flagellation and flagellin levels of flagellar regulon mutants. (A) Microbial and BMM DNAs (DAPI) (left column) and flagellin (polyclonal anti-flagellum antibody) (right column) were visualized by IF microscopy. Representative images of infected BMM with 10 to 15 motile WT PE or nonmotile WT E microbes are shown. (B) The mean percentage of flagellate microbes ± SD was calculated from observed punctate or filamentous anti-flagellin staining of >100 DAPI-positive microbes in duplicate coverslips from two to four independent experiments. Note that many strains had 0% flagellation and therefore lack data bars. (C) Motility of broth cultures was assessed qualitatively as follows: +++, 50 to 100% motility; ++, 5 to 50% motility; +, 1 to 5% motility; −, <1% motility. (D and E) Flagellin was quantified by Western blot analysis of untreated whole microbes (D) or acid-washed microbes (E) normalized by cell density (expressed as OD600), with WT CFPs as a positive control. One representative image from two (E) or three (D) independent Western blot analyses is depicted. Note that microbes were fourfold more concentrated for acid washing followed by Western blot analysis (E). ND, not determined; NA, not applicable.
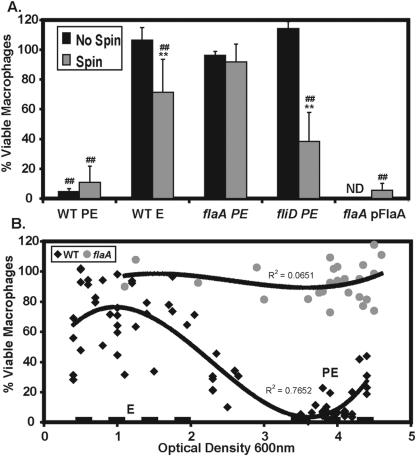
FIG. 5.
Motility and flagellin promote BMM cell death, and levels of BMM death depend on culture density. To test whether motility and flagellin contribute to Legionella-induced cell death, we examined BMM death by using flagellar regulon mutants at an MOI of 30 to 60. (A) BMM were incubated with microbes for 1 h with or without an initial mild centrifugation; then BMM viability was quantified by reduction of Alamar Blue. Mean percent viable BMM ± SD is shown for three or more experiments; data points from each experiment that fell within an MOI range of 30 to 60 were pooled. Differences that were statistically significant by a two-tailed Student t test (P < 0.01) are indicated by double asterisks for centrifugation versus no centrifugation and by double number signs for flaA PE microbes versus the strain indicated. (B) To correlate levels of BMM killing with growth phase and flagellin expression, we tested BMM viability after an initial centrifugation and 1-h infection with microbes as described for panel A. The values for WT and flaA mutant cytotoxicity obtained in multiple individual experiments are displayed on a scatterplot with macrophage viability (y axis) versus OD600 of the culture (x axis), including only data from WT and flaA microbes at MOIs between 30 and 60. A best-fit line was constructed in Excel (Microsoft) using a third-order polynomial equation for both strains, with the correlation coefficient (R) indicated. Dashed lines indicate the OD values used for E-phase (OD600, 0.4 to 2.0) and PE-phase (OD600, 3.5 to 4.5) cultures.
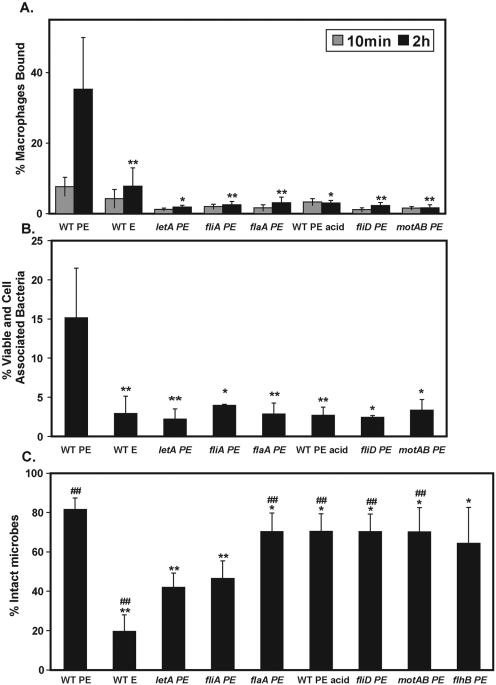
FIG. 3.
Motility but not flagellin promotes BMM infection, whereas FliA is required for lysosome avoidance. (A) Percent macrophages bound. To test if motility promotes efficient BMM infection, monolayers were incubated with the strains indicated at an MOI of ∼1 for either 10 min or 2 h after a mild centrifugation and were then fixed. The mean percentage of BMM bound ± SD indicates anti-Legionella staining that colocalized with BMM and was calculated from duplicate coverslips in three or more independent experiments. (B) Percent viable and cell-associated microbes (infectivity). To test the contributions of motility and the flagellar regulon to the sum of contact, entry, and survival in BMM, macrophage monolayers were incubated for 2 h after a mild centrifugation with each strain at an MOI of ∼1. Cell-associated viable microbes were enumerated as CFU and expressed as the mean percentage of the initial microbial inoculum recovered ± SD, calculated from two to four independent experiments. (C) Lysosome avoidance. To quantify the ability of L. pneumophila strains to avoid degradation, BMM were infected at an MOI of 1 to 5 for 10 min after a mild centrifugation. BMM were washed extensively, the incubation was continued for 2 h, and the microbes were then examined by IF microscopy. Degraded versus intact microbes were determined by the shape of the bacterium and expressed as the mean percentage of microbes that were intact ± SD. Asterisks indicate statistically significant differences (**, P < 0.01 by a two-tailed Student t test; *, P < 0.05) in comparison with WT PE microbes at 2 h postinfection; ##, significant differences (P < 0.01) between fliA PE bacteria and the strain indicated. Note that in panel C, flhB PE microbes were significantly different from letA PE microbes (P < 0.05) and the difference from fliA PE microbes was borderline significant (P = 0.07).
Mutant and plasmid construction and complementation.
All strains and plasmids (Tables 1 and 2) were constructed using standard techniques, as described elsewhere (66). All cloning was done in the Escherichia coli strain DH5α. At least two independent isolates of each L. pneumophila mutant were analyzed in all assays, but results for only one isolate are presented.
TABLE 2.
Primers
| Primer | Sequencea | Wild-type amplicon size (kb) |
|---|---|---|
| fliA-UPPER | 5′-TAG CTG AAT CGG TGA GTA AT-3′ | |
| fliA-LOWER | 5′-TCC TAT TAA AGT CAA AGT ATC CAT-3′ | 0.9 |
| fliAUP2-EcoR1 | 5′-ACG GGA ATT CAT AAC TAC AAT CTG CTG TTA TTG-3′ | |
| fliADOWN-BamH1 | 5′-ACG GGA TCC TTA TTA TTC CGG TAA TCT TGA TCT A-3′ | 0.75 |
| dotAUpper2165L | 5′-CGC ATT GGT ACT AGC CTT TCG TTA-3′ | |
| dotALower2166L | 5′-ACC ATC CTC ATA TTC CAC TTC CTT-3′ | 3.4 |
| fliD_UP | 5′-TGG CGC AAT AGG TGG AAC TG-3′ | |
| fliD_DOWN | 5′-TGG CGG CAA ATA ATG GAA AAA A-3′ | 3.4 |
| flhB_UP | 5′-TGG CCC ATG GTG CGT ATT-3′ | |
| flhB_DOWN | 5′-AGG GGA ACT TAT GGC CAA CTT ATC-3′ | 3.4 |
| flhB-Int_Down-EcoRV | 5′-ACG GAT ATC CCA AGT CCG GTA AAT AAT AAA ATG-3′ | |
| FlhB-Int-UP | 5′-ACG ATG ATG AGC AAT CAC TAG TGG GGT AAT-3′ | NAb |
| flaAUP | 5′-CTC CAG CCC AGC AGC CAC TTC-3′ | |
| flaADOWN | 5′-GAG CAC CCA TAT TAG CCC GAT TAG-3′ | 1.9 |
| flaA-Sacl | 5′-CGA GCT CTC TCA AGC GTG CGG AAA TG-3′ | |
| flaA-Kpn1 | 5′-CGG GGT ACC GCC CTG GCT ATC TTG AAC TAT GC-3′ | 3.0 |
| motA8510 | 5′-CAA GAT TAC CGG AAT AAA AAA TAA G-3′ | |
| motB8511 | 5′-TTT AAT AAA TAA TTA CCC AAG AG-3′ | 1.6 |
| BamHI-MotA5 | 5′-GCG CGG ATC CGC GAG CAG GAT TTG AAT TTG AAG G-3′ | |
| SalI-MotB3 | 5′-GCA CGT CGA CGC CTT TTA ATA AAT AAT TAC CCA AG-3′ | 1.7 |
| fliI_UP | 5′-TTA ATT CAA AAG CCC GTT CA-3′ | |
| fliI_DOWN | 3′-CCA CCT CCC CTT TAT ATT GTT TAG-3′ | 2.3 |
Underlined nucleotides indicate restriction sites.
NA, not applicable.
To construct a flaA flagellin mutant (fliC, lpg1340), the flaA ∼1,900-bp genomic locus was amplified from Lp02 genomic DNA using primers flaAUP and flaADOWN, then ligated to create pGEM-flaA. A 65-bp N-terminal region of the flaA open reading frame (ORF) was removed by digestion with HindIII, and the remaining pGEM-flaA was blunted with Klenow fragment and dephosphorylated with calf intestinal phosphatase (New England Biolabs). The 1.3-kb kanamycin resistance cassette from pUC4K or the 1.7-kb gentamicin resistance cassette from pGEMT-Gent was purified by EcoRI digestion, blunted with Klenow fragment, and ligated into the digested pGEM-flaA plasmid to create pGEM-ΔflaA-kan or pGEM-ΔflaA-gent. The disrupted flaA locus was amplified by PCR (Roche; High Fidelity), transferred to L. pneumophila Lp02 by natural competence as described previously (8, 87), and selected for by antibiotic resistance. Multiple putative flaA::kan and flaA::gent mutants were screened by PCR for mutant-sized loci, and two independent isolates (MB534, MB532) were tested and found similar in all phenotypic assays; MB534 data are shown. To construct a flaA complementing plasmid that expressed FlaA from its native promoter, a ∼3.0-kb genomic region including flaA was amplified with primers flaA-Sac1 and flaA-Kpn1, by adding SacI and KpnI sites to the respective ends, and ligated into the multicloning site of digested pMMBGent-Δmob, creating pMMBGent-flaA. This plasmid was electroporated into flaA::kan (MB557) cells, then selected by gentamicin resistance.
To construct a motAB motility ion channel mutant (lpg1780 to lpg1781), a 1.6-kb motAB genomic region, immediately 3′ to the FliA ORF, was amplified with primers motA8510 and motB8511 and ligated to create pGEM-motAB. pGEM-motAB was digested with StuI and ClaI, dropping out a 129-bp fragment including the 3′ terminus of motA and the 5′ terminus of motB, blunted, and the 1.7-kb Gentr fragment was digested with EcoRI, blunted, and ligated to create pGEM-ΔmotAB-gent. Two independent isolates of this plasmid were obtained, with the Gentr fragment in opposite orientations. Each plasmid was used as a template for PCR, and the resulting PCR product was transferred to Lp02 by natural competence. Multiple motAB::gent mutants were isolated and verified as described above. Two independent motAB::gent isolates (MB560, MB561), one from each PCR product, were tested and found similar in all assays; MB560 data are shown. To complement the mutant, the 1.7-kb motAB ORF amplified with primers BamHI-motA5 and SalI-motB3 was ligated into the pMMB206Δmob plasmid to create the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible plasmid pMMB206-motAB, and the resultant plasmid was electroporated into the motAB::gent mutant to create MB562.
To create a fliD flagellar cap mutant (Hap2, lpg1338), a 3.4-kb fliD genomic region was amplified by PCR with primers FliD_UP and FliD_DOWN and ligated to create pGEM-fliD. pGEM-fliD was digested with EcoRI to disrupt the fliD ORF; then either the Kanr or the Gentr cassette was ligated to create pGEM-ΔfliD-kan or pGEM-ΔfliD-gent. Multiple independent fliD mutants in Lp02 were isolated by natural competence and verified as described above, and two isolates (MB552, MB553) were tested in all assays and found similar; MB552 data are shown.
To create the flhB flagellar secretion-defective mutant (lpg1786), a 3.4-kb flhB genomic region was amplified by PCR with primers flhB_UP and flhB_DOWN and ligated to create pGEM-flhB. To add an EcoRV site while simultaneously deleting a ∼800-bp portion of the ORF, we performed PCR from plasmid pGEM-flhB outward from the ORF with primers flhB-Int-Down-EcoRV and flhB-Int-Up, digested the PCR product with EcoRV, and religated to create pGEM-ΔflhB-EcoRV. pGEM-ΔflhB-EcoRV was digested with EcoRV, and either a blunted Kanr or Gentr cassette was ligated to create pGEM-ΔflhB-gent or pGEM-ΔflhB-kan. Multiple independent flhB mutants in Lp02 were isolated by natural competence and verified as described above, and a majority of assays were performed with two independent isolates (MB554, MB555); MB554 data are shown. It is possible that downstream ORFs were also disrupted in this strain, including the flagellar secretion gene flhA.
A fliA (σ28) transposon mutant was previously isolated and characterized (36). To complement this mutant, we followed two approaches. First, a 940-bp EcoRI fragment from pGEM-fliA containing the FliA ORF and putative promoter was ligated to create the noninducible complementing vector pMMBGent-FliA, which was electroporated into the fliA mutant to create MB510. Second, a 750-bp region with the fliA ORF and ribosomal binding site was amplified with primers fliAUP2-EcoRI and fliADOWN-BamHI and ligated to create the IPTG-inducible vector pregFliA, which was then electroporated into the fliA mutant to create MB550.
Macrophage cultures.
BMM were isolated from the femur exudates of female A/J mice (Jackson Laboratory, Bar Harbor, Maine) and cultured in L-cell supernatant-conditioned medium for 1 week before replating onto 24- or 96 well plates as described elsewhere (66, 89). Macrophages were cultured in RPMI medium plus 10% heat-inactivated fetal bovine serum (FBS), a medium that does not support extracellular L. pneumophila growth.
Quantification of pigment production, CV binding, sodium sensitivity, and heat resistance.
The soluble melanin-like pigment produced during late-PE phase was measured by spectrophotometry (97). Broth cultures maintained in the PE phase for 2 to 4 days were centrifuged at 16,000 × g for 10 min; then the OD550 of culture supernatants was determined. Crystal violet (CV; Sigma) binding was quantified by taking aliquots of broth cultures, adjusting to an OD600 of 1.0 in 1-ml volumes, then incubating with 5 μg/ml of CV for 10 min at 37°C. After centrifugation for 5 min at 16,000 × g, supernatants were measured at OD600 and results expressed as the percentage of total CV remaining in the supernatant (not bound by microbes). PE cultures used for CV binding had an OD600 of >4.0 (mid- to late-PE phase), since inconsistent results were obtained with lower culture densities. Sodium sensitivity was calculated by plating 10-fold serial dilutions of broth cultures in phosphate-buffered saline (PBS) onto CYET agar with or without 100 mM NaCl, then enumerating CFU after a 5- to 6-day incubation, as described elsewhere (17, 66). Heat resistance was determined by incubating microbes for 20 min at 57°C and plating for CFU as described elsewhere (36).
CFPs.
To isolate a crude flagellar preparation (CFP) (81), 400 ml of a PE-phase broth culture of L. pneumophila was gently pelleted at 6,000 × g for 20 min at 4°C. Bacterial pellets were resuspended in 40 ml of PBS and vortexed vigorously for 5 min to dislodge surface structures, including flagella. Loss of motility was confirmed by microscopy where appropriate. Bacteria were centrifuged again at 6,000 × g for 20 min at 4°C, and the PBS supernatant containing flagellar fragments was passed through 0.45-μm filters and collected by centrifugation at 100,000 × g for 3 h at 4°C. CFPs were suspended in 500 μl of PBS, aliquoted, and stored at −20°C until use.
Acid washing of microbes to remove flagella.
To dissociate external flagella, we employed a brief acid wash as described previously (68). Microbes were collected by gentle centrifugation at 5,000 × g for 5 min, resuspended in PBS, pH 2.0, for 5 min at 37°C, and then washed twice with sterile PBS, pH 7.4. Complete loss of motility was confirmed by microscopy where appropriate, and experiments determined no loss of viability for every PE-phase L. pneumophila strain tested, as noted previously (66). Control experiments verified that PBS washes without acid did not affect motility or other virulence traits of WT PE-phase microbes (data not shown).
Percentage of intact L. pneumophila organisms in BMM.
The percentage of intact microbes after a 2-h incubation in macrophages was determined by immunofluorescence (IF) microscopy, as described previously, with minor modifications (8, 66). In brief, BMM were infected with microbes at a multiplicity of infection (MOI) of ∼1, incubated 10 min, washed three times to remove extracellular microbes, and then incubated for an additional 2 h. Fixation, antibody staining, and mounting were performed as described previously (66), and duplicate coverslips were scored for intact versus degraded L. pneumophila, as gauged by anti-Legionella staining of “intact” rod shapes versus dispersed or rounded “degraded” particles (66). Results from four or more experiments were expressed as means ± standard deviations (SD), except for flhB, which was analyzed twice. To detect microbial delivery to lysosomes, we prelabeled the lysosomal network with Texas Red-ovalbumin (TRov; 50 μg/ml; Molecular Probes); infection, fixation, and antibody straining were performed as described above, and colocalization between L. pneumophila staining and TRov was scored (49). To detect colocalization with the endosomal/lysosomal protein LAMP-1, fixed monolayers were incubated with rat monoclonal antibody 1D4B against LAMP-1 at 1:1,000 and subsequently with an Oregon Green-conjugated goat anti-rat antibody and visualized by IF microscopy as described elsewhere (49).
Percentage of BMM bound.
To determine the ability of microbes to contact and adhere to BMM over time, we determined the fraction bound (percent infected BMM). Infection of BMM in 24-well plates was initiated by centrifugation at 400 × g for 10 min; then cultures were incubated for an additional 10 min or 2 h. At the indicated time postinfection, coverslips were washed, fixed, and microbes enumerated with an anti-Legionella antibody by microscopy as described previously (66). Duplicate coverslips were scored by counting total BMM in five or more independent fields and the total anti-Legionella-stained particles in those fields, including both microbes that were intact and those that were degraded. The actual MOI, calculated by determining CFU of inoculates, varied from 0.5 to 5. To pool data from multiple experiments, results were expressed as mean percent BMM bound ± SD, and all data were normalized to an MOI of 1.
Infectivity and intracellular growth.
Infectivity is a gauge of the ability of L. pneumophila to bind, enter, and survive inside BMM during a 2-h incubation, as previously described (17, 66). In brief, BMM were infected with strains at an MOI of ∼1, with or without an initial centrifugation at 400 × g for 10 min, as indicated. Cells were incubated for 2 h at 37°C, washed three times with fresh RPMI, and lysed into 1 ml of PBS; then intracellular CFU were enumerated. Results are expressed as the percentage of the inoculated CFU recovered from lysed BMM at 2 h postinfection. To gauge intracellular growth, the pooled BMM supernatant and lysate were plated for CFU at various times postinfection, as described elsewhere (66).
Quantifying flagellation and motility.
In preliminary experiments, we determined that the fragile flagella are best preserved when microbes are internalized in BMM phagosomes. To determine flagellation of microbial strains, BMM on coverslips were briefly infected with microbes at an MOI of ∼10 by an initial centrifugation of 5 min at 400 × g and were fixed immediately thereafter. The primary mouse monoclonal antibody 2A5 against Legionella flagella (a gift of N. C. Engleberg, Ann Arbor, MI) was used at 1:100, and a secondary Oregon Green-conjugated anti-mouse antibody (Molecular Probes) was used at 1:1,000, as described elsewhere (17). Microbes were counted by 4′,6′-diamidino-2-phenylindole (DAPI) staining, and colocalization with punctate, filamentous anti-flagellin staining was scored. Flagellin levels of crude flagellar preparations or whole microbes were also gauged by Western blot analysis, essentially as described elsewhere (48), using a 1:100 dilution of the antiflagellar monoclonal antibody 2A5. Whole microbes were normalized by optical density before Western blot analysis. We were unable to detect flagellin in concentrated supernatants from WT or fliD microbes, perhaps due to high flagellin levels required for detection by antibody 2A5. To gauge flagellation by transmission electron microscopy (TEM), broth-grown PE-phase microbes were fixed in 1% phosphotungstic acid and applied to grids for examination. To gauge motility qualitatively, microbes cultured to the PE phase in broth were examined by inverted microscopy as wet mounts, with relative motility levels based on observations of rapid, directed motion of many microbes.
Macrophage cell death (cytotoxicity): Alamar Blue reduction.
To quantify BMM viability after L. pneumophila infection, microbes were coincubated with BMM in RPMI-10% FBS in 96-well plates, with or without an initial centrifugation at 400 × g for 10 min as indicated. Duplicate wells were coincubated for an additional 1 h with twofold dilutions of microbes spanning a range of MOIs; then the medium was removed, RPMI-FBS plus 10% Alamar Blue (AccuMed) was added for 6 to 12 h, and viability was determined by spectrophotometer, as described previously (17, 35). The actual MOI of the inoculum was determined by enumerating CFU. Because L. pneumophila thymine auxotrophs were analyzed, the potential contribution of intracellular replication to cytotoxicity was eliminated by omitting exogenous thymidine.
RESULTS
Flagellar regulon mutants, flagellation, and motility.
We first determined the levels of flagellation and motility for our panel of flagellar regulon mutants. As the transmissive- and replicative-phase prototypes in each assay, we used broth-grown WT PE and WT E L. pneumophila, respectively (17, 65, 88). WT PE microbes were highly motile and ∼75% had a single, monopolar flagellum, whereas WT E microbes were completely nonmotile and ∼10% flagellate (Fig. 2B), with minimal flagellin protein detected by Western blot analysis (Fig. 2D) (17). As an additional reference strain, we tested dotA mutants (MB460) that lack an integral component of the Dot/Icm type IV secretion system. dotA mutants were as motile and flagellate as WT PE organisms (Fig. 2). Both WT PE and dotA PE whole-microbe lysates and crude flagellar preparations possessed an abundant ∼48-kDa protein that was identified as flagellin by Western blot analysis (Fig. 2D and data not shown).
To eliminate production of flagellin, we disrupted flaA, the gene that encodes the major component of the external flagellar filament (24, 37). As expected, the flaA mutant (MB534) was nonmotile, and its motility defect was fully complemented by the pMMBGent-FlaA vector. Further, flaA PE crude flagellar preparations and whole-microbe lysates lacked flagellin, and TEM revealed microbes with no external flagella (Fig. 2D and data not shown). As an alternate means of removing the external flagella, microbes were briefly washed with PBS (pH 2.0), a treatment that dissociates the external flagella but does not kill PE-phase microbes (data not shown) (68). Like flaA PE bacteria, WT PE acid-washed microbes were nonmotile and completely lacked external flagella (Fig. 2B), although Western blot analysis of whole-microbe lysates revealed detectable but reduced levels of total flagellin protein (Fig. 2D), which presumably represent an intracellular pool.
To disrupt motility but not flagellin production or secretion, we constructed a mutant lacking FliD (Hap2), the flagellar capping protein required for assembly of secreted flagellin into functional flagella (45, 46, 53, 55, 83). fliD mutants (MB552) were completely nonmotile and also lacked intact external flagella (Fig. 2B and C), although moderate amounts of flagellin were detected in whole-microbe lysates (Fig. 2D). Further, fliD PE microbes produced flagellin at levels comparable to those of WT PE microbes; Western blot analysis of both acid-washed strains revealed similar flagellin levels (Fig. 2E). Rare fliD microbes whose surfaces stained diffusely with an anti-flagellin antibody were observed by microscopy, a pattern not observed for any other strain examined. In other bacterial species, removal of the flagellar cap does not disrupt flagellin production or secretion but instead perturbs its polymerization (46, 83), likely accounting for the continued production, secretion, and subsequent adhesion of flagellin to the surfaces of L. pneumophila fliD mutants.
To block flagellar secretion, we disrupted the integral component of the flagellar secretion apparatus, FlhB (62, 63), or the required flagellar ATPase, FliI (61, 91). Because flagellar secretion is required for building the rod, hook, and flagellar filament (Fig. 1) (reviewed in reference 13), flhB (MB554) and fliI (MB564) mutants were nonmotile and nonflagellate, validating the likely impairment of flagellar secretion (Fig. 2 and data not shown), as described previously for the L. pneumophila fliI mutant (61). Nevertheless, flagellar secretion-deficient strains of L. pneumophila accumulated flagellin protein, since the flaA promoter is still highly active in mutants that lack FliI or FlhB (B. Hammer and A. Molofsky, unpublished), and Western blot analysis detected flagellin associated with whole-microbe lysates (Fig. 2D). When the secretion mutants were first acid washed to determine intracellular protein levels, their flagellin levels were only modestly decreased relative to those of WT or fliD microbes (Fig. 2E). Thus, fliI and flhB mutants contain flagellin that is neither secreted by flagellar secretion nor assembled into a filament.
In an attempt to disrupt motility without eliminating external flagella, we mutated motAB (MB560), the locus that encodes the ion channels required to drive flagellar motion (25, 102). L. pneumophila has two sets of nonhomologous motAB genes, similar to the arrangement in Pseudomonas aeruginosa (25); we disrupted the lpg1780-to-lpg1781 operon adjacent to fliA. As expected, only 1 to 5% of motAB microbes displayed directed motion, and expression of exogenous motA and motB from the pMMB206-motAB plasmid restored motility to wild-type levels. However, only ∼25% of the mutant cells bore intact flagella, and whole-microbe flagellin levels were modestly decreased compared with those of WT PE or dotA PE microbes (Fig. 2D). Likewise, both motility and flagellation were significantly decreased when L. pneumophila was treated with either of two drugs reported to block flagellar motion but not assembly by other bacteria, namely, the proton ionophore carbonyl cyanide m-chlorophenylhydrazone or the sodium channel-blocking agent phenamil (5, 34, 41, 67). Therefore, both genetic and pharmacological data indicate that nonmotile flagella are not as stable as motile flagella on L. pneumophila under the conditions tested. Here the motAB mutant is a tool to examine the impact of an intermediate level of flagellation coupled with a very low level of motility; since a significant percentage of the mutant microbes retain intact but immobile flagella, the strain allows the contributions of motility and flagella to other traits to be separated.
To discern the functions of the two primary flagellar regulators previously implicated in virulence, we analyzed mutants that lack the two-component response regulator LetA or the flagellar sigma factor FliA (29, 36, 37, 58). Both letA (MB413) and fliA (MB410) mutants lack flaA promoter activity and flagellin protein and are completely nonflagellate and nonmotile (Fig. 2) (36, 39), defects that are partially complemented by the exogenous expression of pMMBGent-LetA or pMMBGent-FliA, respectively (36) (data not shown). Using this panel of flagellar regulon mutants (Fig. 1), we examined the contributions of external flagella, flagellin protein, motility, flagellar secretion, and flagellar regulators to L. pneumophila virulence-associated traits.
Motility, but not flagellin protein, contributes prominently to infectivity toward BMM.
An intracellular pathogen infects a host cell in several steps: (i) the host cell is contacted, (ii) the microbe adheres intimately, (iii) the microbe is engulfed, and (iv) the microbe survives intracellularly. As a sum of these steps, we measure the infectivity of L. pneumophila by incubating BMM with microbes at an MOI of ∼1 for 2 h, then enumerating the CFU of intracellular microbes as a percentage of the initial inoculum. A correlation exists between the transmissive-phase traits of motility and infectivity (17, 36, 66). Here we specifically tested the contributions of particular flagellar regulon genes to each step of BMM infection.
We first examined binding to BMM microscopically, an assay that measures only (i) microbial contact and (ii) intimate adherence. At 10 min postinfection, motile WT PE microbes associated with BMM slightly better than all nonmotile strains (Fig. 3A). In contrast, by 2 h, motile WT PE microbes bound BMM ∼6-fold more often than all nonmotile strains (Fig. 3A). At either a low or a high MOI, no differences in BMM binding or total infectivity were detected for nonmotile strains, whether the cells lacked external flagellin protein (flaA PE and acid-washed WT PE microbes), failed to polymerize flagellin (fliD), or formed a nonfunctional flagellum (motAB) (Fig. 3A; also data not shown). If polymerized flagellin promoted BMM binding, we would likely detect increased binding by the motAB mutants, since ∼25% of the microbes remain flagellate (Fig. 2). Thus, we conclude, in agreement with the results of others (24), that flagellin does not contribute significantly to BMM adhesion. Instead, motility likely promotes numerous BMM contacts during the 2-h infection period.
A similar pattern was observed when infectivity was measured by enumerating cell-associated CFU after a 2-h incubation. Even when cell contact was promoted by initiating the assay with a mild centrifugation, nonmotile microbes did not infect as efficiently as the WT (Fig. 3B) (data not shown). Consistent with previous results (17), motile WT PE microbes were ∼15% infectious, a proportion similar to that of motile dotA PE microbes (data not shown), whereas all nonmotile strains tested were <5% infectious. Although nonmotile strains (acid-washed WT PE microbes, flaA, fliI, motAB, and fliD) infected BMM poorly, the microbes that did enter and survive the initial infection replicated efficiently, as did several nonmotile, nonflagellate strains described previously: WT E, letA PE, fliI PE, and fliA PE organisms (36, 61, 66; A. Molofsky, unpublished). Thus, motility but not flagellin likely promotes contacts with BMM and increases infectivity, but an intact flagellum, flagellar secretion, or motility is not required for subsequent microbial replication.
FliA promotes lysosome avoidance in BMM independently of the flagellar regulon.
L. pneumophila avoids degradation in macrophage lysosomes, a trait integral to its survival and pathogenesis in humans (42, 88). Therefore, we examined how efficiently each flagellar regulon mutant avoided lysosomal degradation. By scoring the morphology of each intracellular microbe by IF microscopy, trafficking was analyzed independently of BMM binding efficiency (see Materials and Methods) (66). After a 2-h incubation, approximately 80% of intracellular WT PE microbes were intact, as judged by their rod shape, whereas only 20% of WT E microbes were intact (Fig. 3C), confirming the effect of growth phase on lysosome avoidance (17). Similarly to letA mutants (36), fliA PE mutants were degraded twice as frequently as WT PE controls (Fig. 3C), a phenotype largely complemented by expression of pMMBGent-FliA (data not shown). fliA mutants also colocalized with lysosomal TRov (49) five times more frequently than did WT PE microbes (data not shown). Thus, the flagellar sigma factor FliA induces expression of factors that protect L. pneumophila from degradation by macrophages.
To ascertain how FliA contributes to lysosome avoidance, we tested other flagellar regulon mutants, including the known or likely targets of FliA regulation: flaA, fliD, and flhB (40). Compared to the fliA or letA mutants, significantly more of each of the other intracellular nonmotile microbes were intact, including acid-washed WT PE, flaA, fliD, flhB, and motAB bacteria, although each nonmotile strain was degraded slightly more frequently than WT PE microbes (Fig. 3C). Further, flaA and fliD PE mutants avoid acquisition of the late endosomal/lysosomal protein LAMP-1 with an efficiency similar to that of WT PE microbes (data not shown). Thus, intact flagella, flagellin protein, and flagellar secretion are not required for lysosome avoidance; motility may play a minor role. In contrast, the flagellar sigma factor FliA (σ28) is required for proper trafficking. Therefore, L. pneumophila has coopted FliA to regulate genes beyond the flagellar regulon, namely, virulence factors that promote lysosome avoidance.
FliA regulates melanin-like pigment production and crystal violet binding independently of the flagellar regulon.
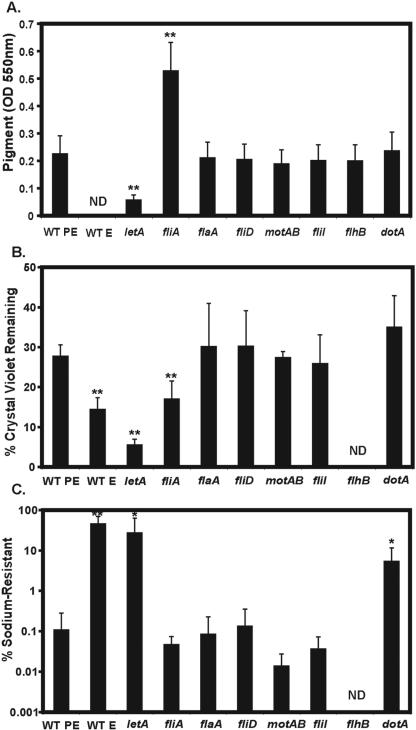
To investigate whether additional transmission traits are regulated by FliA, we next assayed pigment production. As it reaches the transmissive PE phase, L. pneumophila produces a soluble, melanin-like pigment that is believed to protect the microbe from UV damage in nature but is dispensable for efficient infection and replication in eukaryotic cells (95, 97, 98). There is a strong correlation between expression of transmissive-phase virulence traits and pigment production: the letA, letS, letE, and relA transmission mutants are all defective for pigmentation (7; reviewed in references 65, 66, and 103). In contrast, fliA (σ28) mutants are hyperpigmented (Fig. 4A) (66), a phenotype significantly suppressed when fliA expression is restored by the pregFliA plasmid (data not shown). To discern if the effect of FliA on pigmentation is dependent on its induction of other flagellar genes, we quantified the soluble pigment produced by the panel of strains. External flagella/flagellin, flagellar secretion, motility, and type IV secretion did not influence pigmentation: flaA, fliD, flhB, fliI, motAB, and dotA single mutants and flaA fliD (MB567) and fliI fliD (MB565) double mutants pigmented similarly to WT PE microbes (Fig. 4A; also data not shown). Further, flagellar secretion did not contribute to hyperpigmentation by the fliA mutant; a fliA flhB (MB573) double mutant maintained excess pigmentation (data not shown).
FIG. 4.
Pigment production, crystal violet binding, and sodium sensitivity of flagellar regulon mutants. (A) Of the flagellum-specific components, only FliA affects pigment production. Soluble pigment was quantified as the OD550 of supernatants obtained from cultures 2 to 4 days after the strains entered PE phase. (B) Of the flagellum-specific components, only FliA regulates crystal violet binding. Regulated changes to the microbial surface were detected by bacterial binding to the dye crystal violet and are expressed as the percentage of CV remaining in the culture supernatant. (C) The flagellar secretion system does not confer sensitivity to sodium. Percent sodium resistance was calculated by enumerating colony formation on CYET medium with or without 100 mM NaCl. All results in all panels are pooled means ± SD from three or more independent experiments. Asterisks indicate statistically significant differences in comparison to WT PE microbes (**, P < 0.01; *, P < 0.05) determined by a two-tailed Student t test. ND, not done.
To test if FliA regulates other nonflagellar properties, we measured the propensity of broth-grown L. pneumophila to bind the dye crystal violet (14), a surface-dependent property that is elevated in E-phase relative to PE-phase microbes (E. Fernandez-Moreira, unpublished) (Fig. 4B). letA and fliA PE-phase mutants bind excess CV compared with WT PE microbes, although to different degrees (Fig. 4B). Like the pigmentation phenotype, CV binding was not dependent on external flagella/flagellin, flagellar secretion, motility, or type IV secretion: PE-phase flaA, fliD, fliI, motAB, and dotA mutants bound CV similarly to WT PE microbes (Fig. 4B). Further, removal of flagella and other surface proteins by acid washing of WT PE microbes did not significantly alter microbial CV binding (data not shown). Since pigment production/secretion, crystal violet binding, and lysosomal avoidance are not dependent on the flagellar regulon, FliA must regulate these traits independently of its control of flagellation and motility.
The flagellar regulon does not influence heat resistance or type IV secretion-dependent sodium sensitivity.
As additional transmissive-phase phenotypes, we tested if FliA and the flagellar regulon impacted L. pneumophila resistance to heat or sensitivity to sodium chloride. Heat resistance and salt sensitivity are dependent on both the PE phase and regulators of transmission, including LetA and LetE (7, 66), whereas salt sensitivity also requires the Dot/Icm type IV secretion system (17, 79, 94). The flagellar regulon was not required for heat tolerance; fliA, flaA, and fliI mutants were as heat resistant as WT PE microbes (data not shown). Since the flagellar pore secretes proteins, we reasoned it may also be a conduit for small ions, thereby conferring sodium sensitivity. As reported previously, WT E microbes or PE-phase mutants lacking the type IV secretion gene dotA, the response regulator letA, or the alternate sigma factor rpoS (MB379) were defective for sodium sensitivity, efficiently forming colonies on agar that contained 100 mM sodium (Fig. 4C; also data not shown) (8, 36). Like WT PE microbes, all PE flagellar regulon mutants tested were sensitive to sodium, including fliA, flaA, fliD, fliI, and motAB. Thus, unlike the type IV Dot/Icm secretion machinery, the type III flagellar secretion apparatus does not confer sodium sensitivity. Furthermore, defects in the flagellar regulon do not appear to impact type IV secretion, as judged by sodium sensitivity, endosomal avoidance, and intracellular growth (Fig. 4C and data not shown).
Legionella induction of BMM cell death requires motility and flagellin.
As L. pneumophila becomes motile, it expresses cytolytic activity against red blood cells and macrophages. Since cytotoxicity is contact dependent (44, 51), we determined whether the poor BMM binding of the nonmotile strains (Fig. 3A) affected their cytotoxicity. As predicted, the WT E, flaA PE, fliD PE, and fliA PE nonmotile strains were completely noncytotoxic at all MOIs tested, whereas motile WT PE microbes were highly cytotoxic to BMM (Fig. 5A; supplemental Fig. 1C; data not shown). When contact of the nonmotile strains with BMM was promoted by mild centrifugation, both WT E and fliD PE microbes killed significantly more BMM; nevertheless, flaA microbes remained noncytotoxic at all culture densities tested (Fig. 5A and B), a defect fully complemented by exogenous expression of pMMBGent-FlaA (Fig. 5A). Additional evidence indicated a requirement for flagellin in BMM cell death: all nonmotile strains tested that lacked flagellin were similarly defective for BMM cytotoxicity, including PE-phase letA, fliA, flaA, and acid-washed WT microbes (A. Molofsky, unpublished). The nonmotile, flagellin-producing motAB, flhB, and fliI strains displayed intermediate levels of BMM killing, slightly greater than that of WT E microbes (A. Molofsky, unpublished). Further, the degree of BMM killing correlated with growth phase-dependent flagellin expression by L. pneumophila cultured in broth (Fig. 5B) (66). Therefore, BMM cytotoxicity was absolutely dependent not only on motility to initiate host-cell contacts, but also on the production of flagellin protein.
DISCUSSION
In nature, L. pneumophila is prevalent in fresh-water sources, where it replicates after infecting single-celled protozoa, a lifestyle likely dependent on motility. Indeed, a vast majority of Legionella species express flagellin (FlaA) and are motile (15, 37, 69), indicating that this trait confers fitness in aquatic environments. Flagellate L. pneumophila organisms are also observed in lung tissue of patients with Legionnaires' disease (18), where the microbes can infect and replicate inside alveolar macrophages. L. pneumophila is not flagellate during intracellular replication but becomes so during the transmissive phase, when the microbes lyse their spent host and search for a new replication niche (17, 65, 76). Here we report that components of the flagellar regulon also contribute to several virulence-associated transmissive traits, namely, macrophage binding, lysosome avoidance, pigment production, surface modifications, and cytotoxicity.
The flagellar sigma factor FliA (σ28) and the response regulator LetA promote efficient lysosome avoidance in macrophages, the hallmark of L. pneumophila pathogenesis. LetA likely contributes to lysosome avoidance by relieving CsrA repression of FliA expression (6, 66). Motility appears to play a minor role in lysosome avoidance (Fig. 3C), perhaps by orienting a polar type IV secretion apparatus toward the BMM (9, 75) or by promoting contact and subsequent binding of microbes with active transmission programs. However, FliA also induces lysosome avoidance independently of its targets in the flagellar regulon, as evidenced by the fact that fliA mutants are degraded more frequently than other nonmotile strains (Fig. 3C). Indeed, FliA contributes to at least two other transmission traits that are independent of motility, namely, pigmentation (Fig. 4A) and crystal violet binding (Fig. 4B). Likewise, FliA regulates microbial growth in the social amoeba D. discoideum independently of the flaA locus (38). Accordingly, the classic definition of FliA as a flagellum-specific sigma factor is not accurate for L. pneumophila. FliA must cooperate with other regulators to govern the cohort of transmission genes required for efficient lysosome avoidance, since both letA and fliA mutants evade degradation more frequently than replicative-phase WT E microbes do (Fig. 3C). Identification of the targets of FliA regulation, including those outside the flagellar regulon, will yield insight into how this pathogen avoids destruction in macrophages, sentinels of innate immunity.
FliA apparently induces lysosome avoidance by a mechanism that is coordinated with, but distinct from, the Dot/Icm effectors that immediately isolate L. pneumophila from the endosomal network. First, all flagellar regulon mutants (fliA, flaA, fliD, and fliI) have a functional Dot/Icm secretion system, as gauged by their replication in BMM (data not shown) and sodium sensitivity (Fig. 4C). Second, >50% of fliA PE mutants are degraded in BMM, whereas dotA mutants remain intact in a late endosomal vacuole (data not shown) (49). Finally, unlike dotA mutants, fliA and letA cells have altered crystal violet binding, an assay that reflects surface properties (14) and correlates to evasion of lysosomes (E. Fernandez Moreira and M. Swanson, unpublished). Therefore, by a mechanism that requires FliA but is independent of type IV secretion, L. pneumophila modifies or exports to its surface virulence factors that delay or alter its delivery to degradative lysosomes (49). One potential target of FliA regulation is either synthesis or modification of the pathogen's unusually hydrophobic lipopolysaccharide (32, 57, 101), a process known to be coregulated with flagellation in E. coli (56).
The flagellar regulon also promotes binding to host cells (Fig. 3A), a trait vital to both infection and cytotoxicity. Flagellin itself does not significantly mediate BMM adherence; all nonmotile strains bind BMM similarly to WT PE microbes at 10 min postinfection, whether or not they produce or polymerize flagellin (Fig. 2D and 3A). Instead, the occurrence of fewer contacts with BMM during the 2-h infection period likely accounts for the poor binding (Fig. 3A) and infectivity (Fig. 3B) of nonmotile bacteria. Once L. pneumophila has bound to macrophages, its motility, flagellin, and flagellar secretion apparatus are largely dispensable for either survival (Fig. 3C) or replication (data not shown). Thus, our study corroborates and extends the previous report that L. pneumophila flaA mutants are defective for early stages of infection of amoebae and macrophages (24).
A hallmark of Legionnaires' disease is extensive inflammation with lysis of inflammatory cells and necrosis of the alveolar epithelium, a pathology that has been linked in mice to a bacterial cytotoxin(s) (4, 12, 16, 50). The selective advantage for L. pneumophila cytotoxicity was thought to be escape from amoebae (30, 64), though egress may also occur by vacuolar exocytosis mediated by two Dot/Icm substrates, LepA and LepB (19). In vitro, lysis of red blood cells and certain eukaryotic hosts, including amoebae and macrophages, is mediated by the contact- and type IV secretion system-dependent pore-forming activity of L. pneumophila (4, 30, 44, 51). Several other L. pneumophila factors (ProA, Lly, or PlaB) can promote hemolysis, but mutations in each cause little or no defect in macrophage cytotoxicity (28, 73, 99). In contrast, the RtxA protein is partially required for macrophage cytotoxicity, by promoting either the pore-forming activity or macrophage adhesion (22). Our analysis expands the limited list of L. pneumophila factors required for cytotoxicity against macrophages to include flagellin.
There are several plausible models by which flagellin could act in concert with type IV secretion to promote BMM cell death. Flagellin does not contribute directly to pore formation, as flagellin is dispensable for lysis of red blood cells (A. Molofsky and C. Madigan, unpublished). Instead, flagellin protein could promote cytotoxicity by orienting the pathogen's type IV secretion system toward the host cell, as both the type IV secretion system and flagellum are likely located at its poles (9). However, this model seems unlikely, since unlike dot/icm mutants, flagellin-deficient strains traffic and replicate in BMM as efficiently as WT microbes (Fig. 3C and data not shown). A number of observations are consistent with the idea that flagellin could target L. pneumophila to regions of the BMM surface that are susceptible to either delivery or activity of its cytotoxin(s): Cohorts of toxins are cholesterol dependent and active in “lipid rafts,” microdomains rich in glycosphingolipids, GM1 gangliosides, and cholesterol (reviewed in reference 92); transmissive L. pneumophila enters macrophages at lipid-raft-rich regions (96); and Pseudomonas aeruginosa flagellin binds asialo-GM1 ganglioside (1), a component of lipid rafts. It is also possible that flagellin itself triggers BMM death, since Pseudomonas syringae flagellin elicits a hypersensitive reaction that kills tomato cells, a death that is enhanced with a fliD mutant that hypersecretes flagellin (83). However, since CFPs were not cytotoxic to BMM, and purified flagellin failed to complement the defective cytotoxicity of nonflagellate mutants (data not shown), we hypothesize a requirement for type IV secretion to function either directly, to deliver flagellin to the host cell cytoplasm, or indirectly, to create a vacuolar pore by which excess surface flagellin might leak intracellularly. Understanding of the interplay between microbial virulence factors and macrophage defense mechanisms can be obtained by testing these models directly.
In addition to knowledge of how components of the L. pneumophila flagellar regulon contribute to virulence, we also gained insight into their coordinate regulation. Since inactive flagella (motAB PE) are somewhat unstable on L. pneumophila, either nonmotile flagella are more susceptible to physical stress or a feedback loop may promote the shedding of nonfunctional flagella. Secondly, flagellar secretion is not absolutely required for FliA function; flhB or fliI secretion mutants are not defective for pigmentation, crystal violet binding, or lysosome avoidance (Fig. 3C and 4), a situation that differs from flagellar regulation by enteric bacteria (reviewed in reference 2). Secretion mutants (flhB, fliI) as well as the capping mutant (fliD) do contain less total flagellin than whole WT PE microbes (Fig. 2D), but this likely reflects the large amount of external polymerized flagellin of WT PE microbes. In support of this hypothesis, when microbes were acid washed to remove external flagellin (Fig. 2D and E), WT PE microbes had levels of flagellin similar to those of both the capping mutant (fliD) and the motility mutant (motAB), and only slightly greater than those of the secretion mutants (flhB, fliI).
L. pneumophila produces a pigment in the transmissive PE phase (65), a trait sensitive to FliA activity. Unlike the letA regulatory mutant, which is defective for expression of all transmission traits tested, fliA mutants are as sodium sensitive (Fig. 4C) and heat resistant as the WT, but they are hyperpigmented (Fig. 4A). Strikingly, excess pigmentation was also observed when wild-type or mutant strains were cultured either in a stringent chemically defined medium (74), or with gentamicin for plasmid maintenance, or in the absence of csrA expression (data not shown) (66). We hypothesize that a variety of metabolic stresses induce pigment production, perhaps due to activation of gluconeogenic pathways that synthesize aromatic amino acids, the precursors of the melanin-like pigment (85). Since fliA mutants grow similarly to the WT in broth culture, most likely some accessory metabolic pathway, such as aromatic amino acid synthesis, accounts for hyperpigmentation.
In summary, our analysis demonstrates that the L. pneumophila flagellar regulon genes impact multiple virulence traits in different manners. Motility promotes contact with host cells, the prelude to subsequent pathogen-phagocyte interactions, including lysosome avoidance or host cell death (cytotoxicity). In addition to its role in motility, the flagellar sigma factor FliA is required for lysosome avoidance, pigmentation, and crystal violet binding by mechanisms that are independent of flagellin production, secretion, or assembly. Finally, flagellin protein itself directly or indirectly promotes the death of macrophages. Thus, flagellar regulation has evolved to promote not only motility but also key elements of L. pneumophila pathogenesis, thereby providing the intracellular pathogen with a mechanism to coordinate expression of transmission traits.
Acknowledgments
We thank J. D. Sauer for scientific discussions, M. O'Riordan for insight and guidance, especially during the sabbatical of M.S.S., and C. A. Madigan for excellent experimental assistance and discussions. Also thanks to David Friedman and his lab for sharing equipment and reagents.
This project was supported by NIH grant AI 44212-01. A.B.M. was supported by NIH National Research Service Award 56-T32-GM07544 from the National Institute of General Medical Sciences and a University of Michigan Frederick G. Novy Fellowship.
Editor: D. L. Burns
REFERENCES
- 1.Adamo, R., S. Sokol, G. Soong, M. I. Gomez, and A. Prince. 2004. Pseudomonas aeruginosa flagella activate airway epithelial cells through asialoGM1 and toll-like receptor 2 as well as toll-like receptor 5. Am. J. Respir. Cell Mol. Biol. 30:627-634. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 3.Allen-Vercoe, E., and M. J. Woodward. 1999. The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype Enteritidis to chick gut explant. J. Med. Microbiol. 48:771-780. [DOI] [PubMed] [Google Scholar]
- 4.Alli, O. A. T., L.-Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68:6431-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atsumi, T., S. Sugiyama, E. J. Cragoe, Jr., and Y. Imae. 1990. Specific inhibition of the Na+-driven flagellar motors of alkalophilic Bacillus strains by the amiloride analog phenamil. J. Bacteriol. 172:1634-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachman, M. A., and M. S. Swanson. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 72:2468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachman, M. A., and M. S. Swanson. 2004. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect. Immun. 72:3284-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 40:1201-1214. [DOI] [PubMed] [Google Scholar]
- 9.Bardill, J. P., J. L. Miller, and J. P. Vogel. 2005. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol. Microbiol. 56:90-103. [DOI] [PubMed] [Google Scholar]
- 10.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 11.Berger, K. H., J. J. Merriam, and R. I. Isberg. 1994. Altered intracellular targeting properties associated with mutations in the Legionella pneumophila dotA gene. Mol. Microbiol. 14:809-822. [DOI] [PubMed] [Google Scholar]
- 12.Blackmon, J. A., M. D. Hicklin, and F. W. Chandler. 1978. Legionnaires' disease. Pathological and historical aspects of a ‘new’ disease. Arch. Pathol. Lab. Med. 102:337-343. [PubMed] [Google Scholar]
- 13.Blocker, A., K. Komoriya, and S. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohach, G. A., and I. S. Snyder. 1983. Characterization of surfaces involved in adherence of Legionella pneumophila to Fischerella species. Infect. Immun. 42:318-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosshardt, S. C., R. F. Benson, and B. S. Fields. 1997. Flagella are a positive predictor for virulence in Legionella. Microb. Pathog. 23:107-112. [DOI] [PubMed] [Google Scholar]
- 16.Brieland, J., P. Freeman, R. Kunkel, C. Chrisp, M. Hurley, J. Fantone, and C. Engleberg. 1994. Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. Am. J. Pathol. 145:1537-1546. [PMC free article] [PubMed] [Google Scholar]
- 17.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 66:3029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler, F. W., B. M. Thomason, and G. A. Hebert. 1980. Flagella on Legionnaires' disease bacteria in the human lung. Ann. Intern. Med. 93:715-716. [DOI] [PubMed] [Google Scholar]
- 19.Chen, J., K. S. de Felipe, M. Clarke, H. Lu, O. R. Anderson, G. Segal, and H. A. Shuman. 2004. Legionella effectors that promote nonlytic release from protozoa. Science 303:1358-1361. [DOI] [PubMed] [Google Scholar]
- 20.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 21.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle, T. B., A. C. Hawkins, and L. L. McCarter. 2004. The complex flagellar torque generator of Pseudomonas aeruginosa. J. Bacteriol. 186:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fettes, P. S., V. Forsbach-Birk, D. Lynch, and R. Marre. 2001. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int. J. Med. Microbiol. 291:353-360. [DOI] [PubMed] [Google Scholar]
- 27.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 28.Flieger, A., K. Rydzewski, S. Banerji, M. Broich, and K. Heuner. 2004. Cloning and characterization of the gene encoding the major cell-associated phospholipase A of Legionella pneumophila, plaB, exhibiting hemolytic activity. Infect. Immun. 72:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34:187-194. [DOI] [PubMed] [Google Scholar]
- 30.Gao, L. Y., and Y. A. Kwaik. 2000. The mechanism of killing and exiting the protozoan host Acanthamoeba polyphaga by Legionella pneumophila. Environ. Microbiol. 2:79-90. [DOI] [PubMed] [Google Scholar]
- 31.Ghelardi, E., F. Celandroni, S. Salvetti, D. J. Beecher, M. Gominet, D. Lereclus, A. C. Wong, and S. Senesi. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girard, R., T. Pedron, S. Uematsu, V. Balloy, M. Chignard, S. Akira, and R. Chaby. 2003. Lipopolysaccharides from Legionella and Rhizobium stimulate mouse bone marrow granulocytes via Toll-like receptor 2. J. Cell Sci. 116:293-302. [DOI] [PubMed] [Google Scholar]
- 33.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 34.Gosink, K. K., and C. C. Hase. 2000. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 182:4234-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33:721-731. [DOI] [PubMed] [Google Scholar]
- 36.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44:107-118. [DOI] [PubMed] [Google Scholar]
- 37.Heuner, K., L. Bender-Beck, B. C. Brand, P. C. Luck, K. H. Mann, R. Marre, M. Ott, and J. Hacker. 1995. Cloning and genetic characterization of the flagellum subunit gene (flaA) of Legionella pneumophila serogroup 1. Infect. Immun. 63:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heuner, K., C. Dietrich, C. Skriwan, M. Steinert, and J. Hacker. 2002. Influence of the alternative σ28 factor on virulence and flagellum expression of Legionella pneumophila. Infect. Immun. 70:1604-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heuner, K., J. Hacker, and B. C. Brand. 1997. The alternative sigma factor σ28 of Legionella pneumophila restores flagellation and motility to an Escherichia coli fliA mutant. J. Bacteriol. 179:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heuner, K., and M. Steinert. 2003. The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int. J. Med. Microbiol. 293:133-143. [DOI] [PubMed] [Google Scholar]
- 41.Heytler, P. G., and W. W. Prichard. 1962. A new class of uncoupling agents—carbonyl cyanide phenylhydrazones. Biochem. Biophys. Res. Commun. 7:272-275. [DOI] [PubMed] [Google Scholar]
- 42.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiplies intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Husmann, L. K., and W. Johnson. 1994. Cytotoxicity of extracellular Legionella pneumophila. Infect. Immun. 62:2111-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeda, T., K. Oosawa, and H. Hotani. 1996. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J. Mol. Biol. 259:679-686. [DOI] [PubMed] [Google Scholar]
- 46.Ikeda, T., S. Yamaguchi, and H. Hotani. 1993. Flagellar growth in a filament-less Salmonella fliD mutant supplemented with purified hook-associated protein 2. J. Biochem. (Tokyo) 114:39-44. [DOI] [PubMed] [Google Scholar]
- 47.Inglis, T. J., T. Robertson, D. E. Woods, N. Dutton, and B. J. Chang. 2003. Flagellum-mediated adhesion by Burkholderia pseudomallei precedes invasion of Acanthamoeba astronyxis. Infect. Immun. 71:2280-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacobi, S., R. Schade, and K. Heuner. 2004. Characterization of the alternative sigma factor σ54 and the transcriptional regulator FleQ of Legionella pneumophila, which are both involved in the regulation cascade of flagellar gene expression. J. Bacteriol. 186:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joshi, A. D., S. Sturgill-Koszycki, and M. S. Swanson. 2001. Evidence that Dot-dependent and -independent factors isolate the Legionella pneumophila phagosome from the endocytic network in mouse macrophages. Cell Microbiol. 3:99-114. [DOI] [PubMed] [Google Scholar]
- 50.Katz, S. M., and S. Hahemi. 1982. Electron microscopic examination of the inflammatory response to Legionella pneumophila in guinea pigs. Lab. Investig. 46:24-32. [PubMed] [Google Scholar]
- 51.Kirby, J. E., J. P. Vogel, H. L. Andrews, and R. R. Isberg. 1998. Evidence of pore-forming ability by Legionella pneumophila. Mol. Microbiol. 27:323-336. [DOI] [PubMed] [Google Scholar]
- 52.Kirov, S. M., M. Castrisios, and J. G. Shaw. 2004. Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72:1939-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komoriya, K., N. Shibano, T. Higano, N. Azuma, S. Yamaguchi, and S. I. Aizawa. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767-779. [DOI] [PubMed] [Google Scholar]
- 54.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landini, P., and A. J. Zehnder. 2002. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J. Bacteriol. 184:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luneberg, E., N. Zetzmann, D. Alber, Y. A. Knirel, O. Kooistra, U. Zahringer, and M. Frosch. 2000. Cloning and functional characterization of a 30 kb gene locus required for lipopolysaccharide biosynthesis in Legionella pneumophila. Int. J. Med. Microbiol. 290:37-49. [DOI] [PubMed] [Google Scholar]
- 58.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219:241-248. [DOI] [PubMed] [Google Scholar]
- 59.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 60.McDade, J. E., C. C. Shepard, D. W. Fraser, T. R. Tsai, M. A. Redus, and W. R. Dowdle. 1977. Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory diseases. N. Engl. J. Med. 297:1197-1203. [DOI] [PubMed] [Google Scholar]
- 61.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minamino, T., T. Iino, and K. Kutuskake. 1994. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J. Bacteriol. 176:7630-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Molmeret, M., O. A. Alli, M. Radulic, M. Susa, M. Doric, and Y. A. Kwaik. 2002. The C-terminus of IcmT is essential for pore formation and for intracellular trafficking of Legionella pneumophila within Acanthamoeba polyphaga. Mol. Microbiol. 43:1139-1150. [DOI] [PubMed] [Google Scholar]
- 65.Molofsky, A. B., and M. S. Swanson. 2004. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol. Microbiol. 53:29-40. [DOI] [PubMed] [Google Scholar]
- 66.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50:445-461. [DOI] [PubMed] [Google Scholar]
- 67.Nishihara, T., and E. Freese. 1975. Motility of Bacillus subtilis during growth and sporulation. J. Bacteriol. 123:366-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okuda, S., R. Igarashi, Y. Kusui, Y. Kasahara, and H. Morisaki. 2003. Electrophoretic mobility of Bacillus subtilis knockout mutants with and without flagella. J. Bacteriol. 185:3711-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ott, M., P. Messner, J. Heesemann, R. Marre, and J. Hacker. 1991. Temperature-dependent expression of flagella in Legionella. J. Gen. Microbiol. 137:1955-1961. [DOI] [PubMed] [Google Scholar]
- 70.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 71.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 72.Pruckler, J. M., R. F. Benson, M. Moyenuddin, W. T. Martin, and B. S. Fields. 1995. Association of flagellum expression and intracellular growth of Legionella pneumophila. Infect. Immun. 63:4928-4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quinn, F. D., and L. S. Tompkins. 1989. Analysis of a cloned sequence of Legionella pneumophila encoding a 38 kD metalloprotease possessing haemolytic and cytotoxic activities. Mol. Microbiol. 3:797-805. [DOI] [PubMed] [Google Scholar]
- 74.Ristroph, J. D., K. W. Hedlund, and S. Gowda. 1981. Chemically defined medium for Legionella pneumophila growth. J. Clin. Microbiol. 13:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rohde, M., J. Puls, R. Buhrdorf, W. Fischer, and R. Haas. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49:219-234. [DOI] [PubMed] [Google Scholar]
- 76.Rowbotham, T. J. 1986. Current views on the relationships between amoebae, legionellae and man. Isr. J. Med. Sci. 22:678-689. [PubMed] [Google Scholar]
- 77.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roy, C. R., and R. R. Isberg. 1997. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect. Immun. 65:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scharfman, A., S. K. Arora, P. Delmotte, E. Van Brussel, J. Mazurier, R. Ramphal, and P. Roussel. 2001. Recognition of Lewis x derivatives present on mucins by flagellar components of Pseudomonas aeruginosa. Infect. Immun. 69:5243-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scherer, D. C., I. DeBuron-Connors, and M. F. Minnick. 1993. Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of human erythrocytes. Infect. Immun. 61:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Segal, G., M. Purchell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shimizu, R., F. Taguchi, M. Marutani, T. Mukaihara, Y. Inagaki, K. Toyoda, T. Shiraishi, and Y. Ichinose. 2003. The ΔfliD mutant of Pseudomonas syringae pv. tabaci, which secretes flagellin monomers, induces a strong hypersensitive reaction (HR) in non-host tomato cells. Mol. Genet. Genomics 269:21-30. [DOI] [PubMed] [Google Scholar]
- 84.Song, Y. C., S. Jin, H. Louie, D. Ng, R. Lau, Y. Zhang, R. Weerasekera, S. Al Rashid, L. A. Ward, S. D. Der, and V. L. Chan. 2004. FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC 43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol. Microbiol. 53:541-553. [DOI] [PubMed] [Google Scholar]
- 85.Steinert, M., M. Flugel, M. Schuppler, J. H. Helbig, A. Supriyono, P. Proksch, and P. C. Luck. 2001. The Lly protein is essential for p-hydroxyphenylpyruvate dioxygenase activity in Legionella pneumophila. FEMS Microbiol. Lett. 203:41-47. [DOI] [PubMed] [Google Scholar]
- 86.Steinert, M., U. Hentschel, and J. Hacker. 2002. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol. Rev. 26:149-162. [DOI] [PubMed] [Google Scholar]
- 87.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 89.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tasteyre, A., M. C. Barc, A. Collignon, H. Boureau, and T. Karjalainen. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thomas, J., G. P. Stafford, and C. Hughes. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. USA 101:3945-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tweten, R. K., M. W. Parker, and A. E. Johnson. 2001. The cholesterol-dependent cytolysins. Curr. Top. Microbiol. Immunol. 257:15-33. [DOI] [PubMed] [Google Scholar]
- 93.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 94.Vogel, J. P., C. Roy, and R. R. Isberg. 1996. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann. N. Y. Acad. Sci. 797:271-272. [DOI] [PubMed] [Google Scholar]
- 95.Warren, W. J., and R. D. Miller. 1979. Growth of Legionnaires' disease bacterium (Legionella pneumophila) in chemically defined medium. J. Clin. Microbiol. 10:50-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watarai, M., I. Derre, J. Kirby, J. D. Growney, W. F. Dietrich, and R. R. Isberg. 2001. Legionella pneumophila is internalized by a macropinocytotic uptake pathway controlled by the Dot/Icm system and the mouse Lgn1 locus. J. Exp. Med. 194:1081-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wiater, L. A., A. B. Sadosky, and H. A. Shuman. 1994. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol. Microbiol. 11:641-653. [DOI] [PubMed] [Google Scholar]
- 98.Wintermeyer, E., M. Flugel, M. Ott, M. Steinert, U. Rdest, K. H. Mann, and J. Hacker. 1994. Sequence determination and mutational analysis of the lly locus of Legionella pneumophila. Infect. Immun. 62:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wintermeyer, E., U. Rdest, B. Ludwig, A. Debes, and J. Hacker. 1991. Characterization of legiolysin (lly), responsible for hemolytic activity, colour production, and fluorescence of Legionella pneumophila. Mol. Microbiol. 5:1135-1143. [DOI] [PubMed] [Google Scholar]
- 100.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zahringer, U., Y. A. Knirel, B. Lindner, J. H. Helbig, A. Sonesson, R. Marre, and E. T. Rietschel. 1995. The lipopolysaccharide of Legionella pneumophila serogroup 1 (strain Philadelphia 1): chemical structure and biological significance. Prog. Clin. Biol. Res. 392:113-139. [PubMed] [Google Scholar]
- 102.Zuberi, A. R., C. W. Ying, H. M. Parker, and G. W. Ordal. 1990. Transposon Tn917lacZ mutagenesis of Bacillus subtilis: identification of two new loci required for motility and chemotaxis. J. Bacteriol. 172:6841-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]