Abstract
The formation of several acyl groups and an amide group of Taxol is catalyzed by regioselective CoA thioester-dependent acyltransferases. Several full-length acyltransferase sequences, obtained from a cDNA library constructed from mRNA isolated from Taxus cuspidata cells induced for Taxol production with methyl jasmonate, were individually expressed in Escherichia coli, from which a cDNA clone encoding a 3′-N-debenzoyl- 2′-deoxytaxol N-benzoyltransferase was identified. This recombinant enzyme catalyzes the stereoselective coupling of the surrogate substrate N-debenzoyl-(3′RS)-2′-deoxytaxol with benzoyl-CoA to form predominantly one 3′-epimer of 2′-deoxytaxol. The product 2′-deoxytaxol was confirmed by radio-HPLC,1H-NMR, and chemical ionization-MS. This enzymatic reaction constitutes the final acylation in the Taxol biosynthetic pathway. The full-length cDNA coding for the N-benzoyltransferase has an ORF of 1,323 nucleotides and encodes a 441-residue protein with a calculated molecular weight of 49,040. The recombinant enzyme expressed in E. coli has a pH optimum at 8.0, a kcat ≈ 1.5 ± 0.3 s−1 and Km values of 0.42 mM and 0.40 mM for the N-deacylated taxoid and benzoyl-CoA, respectively. In addition to improving the production yields of Taxol in genetically engineered host systems, this enzyme provides a means of attaching modified aroyl groups to taxoid precursors for the purpose of improving drug efficacy.
Keywords: paclitaxel‖2′-deoxytaxol
The natural product Taxol (generic name paclitaxel, Fig. 1) is a structurally complex diterpenoid comprised of the tricyclic taxane core containing several acyloxy groups, including the 13-O-(N-benzoyl phenylisoserinoyl) side chain, which is a structural requirement for efficacy of the drug against several cancers (1). In particular, the side-chain N-benzoyl function is a necessary structural component for the crucial Taxol bioactivity of binding to tubulin heterodimers and promoting the assembly and stability of microtubules that ultimately disrupts cell division (1).
Figure 1.
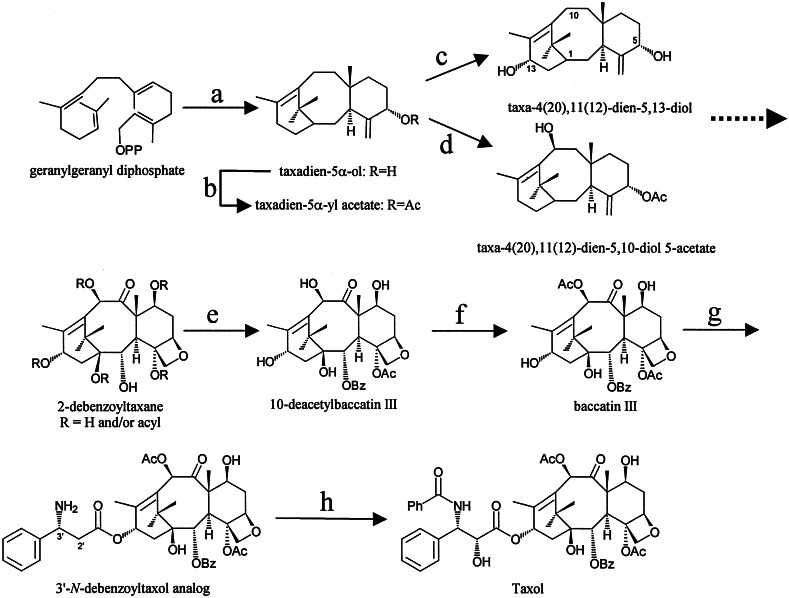
Outline of the Taxol biosynthetic pathway. Illustrated are the cyclization of geranylgeranyl diphosphate to taxadiene by taxadiene synthase and the hydroxylation to taxadien-5α-ol by taxadiene 5α-hydroxylase (a), the acetylation of taxadien-5α-ol by taxadien-5α-ol acetyltransferase (b), the conversion of taxadien-5α-ol to 5,13-diol by a 13α-hydroxylase (c), the hydroxylation of taxadien-5α-yl acetate by a 10β-hydroxylase (d), the formation of a 2-benzoxy taxoid by a taxane 2α-O-benzoyltransferase (e), the conversion of 10-deacetylbaccatin III to baccatin III by a 10-O-acetyltransferase (f), side-chain attachment by the phenylpropanoyltransferase (g), and side-chain benzamidation by DBTNBT to form Taxol (h). The broken arrow indicates multiple convergent steps.
Taxol is presently manufactured semisynthetically by coupling the advanced naturally occurring taxoid 10-deacetylbaccatin III, isolated from the needles of Taxus baccata, to a synthetic side-chain precursor (2, 3). The reliance on the isolation of natural products from Taxus species, or Taxus-derived cell cultures, will continue for the foreseeable future because total synthesis of the drug is not commercially viable. However, isolation from natural sources will become limiting as the clinical applications and resulting demand for Taxol and other advanced, second-generation taxoid drugs increase. Up-regulation of the Taxol biosynthetic pathway, by overexpression of selected genes in Taxus cells or other suitable hosts, can potentially address the supply issue. However, this approach requires, foremost, an understanding of the pathway(s) leading to not only the diterpenoid portion of Taxol but also to the N-benzoyl-phenylisoserinoyl side chain, and the isolation of the responsible genes.
Previous biogenetic studies have indicated that benzamidation of N-debenzoyltaxol is the final step of the Taxol pathway (4) (Fig. 1). To isolate the responsible N-benzoyltransferase gene required the functional screening of a previously acquired family of related acyl/aroyltransferase cDNAs from Taxus (5) by using benzoyl-CoA and a suitable taxoid cosubstrate. Syntheses of N-debenzoylated Taxol derivatives have been described (6, 7), but require multistep synthesis of the side chain (8) or use low-abundance taxoids as starting materials for sequential protection, deacylation, and deprotection to the target substrate. As an alternative, a facile four-step semisynthesis of N-debenzoyl-(3′RS)-2′-deoxytaxol provided a productive surrogate substrate for screening the related (58–69% identity) set of recombinant Taxus acyltransferases (5, 9, 10). Isolation and analysis of the cDNA clone encoding a 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase (DBTNBT) are described herein, and the properties of this recombinant enzyme, which catalyzes the last step of Taxol biosynthesis, are reported.
Materials and Methods
Substrates.
N-debenzoyl-(3′RS)-2′-deoxytaxol and 13-O-[(2S)-α-phenylalanoyl]baccatin III were similarly synthesized as described (11, 12), except that either N-Boc-(3RS)-β-phenylalanine (13) or N-Boc-(2S)-α-phenylalanine (Aldrich) was coupled to 7-TES-baccatin III (11, 12). Deprotection by described methods (7) yielded the desired cosubstrates. [7-14C]Benzoyl-CoA was prepared as described (10). Other CoA thioester salts were purchased from Sigma, and 3RS-β-phenylalanine (3-amino-3-phenylpropionic acid) was purchased from Aldrich. Authentic baccatin III was generously provided by Hauser Chemical Research (Boulder, CO), or was synthesized from 10-deacetylbaccatin III as described (14). Synthesis of (3′RS)-2′-deoxytaxol from the N-debenzoyl (3′RS)-substrate was carried out by the Schotten–Baumann method (7).
Bacterial Expression, N-Benzoyltransferase Assay, and Product Identification.
The isolation of nine Taxus transacylase cDNA clones, that are putatively involved in Taxol biosynthesis, has been described (5). By using an appropriate primer set [pair 1, TX10NDE1F (5′-TGG AGA AGG CAG GTC CAA CAG-3′ and TX10BAM1R (5′-GAT CCT CAC ACT TTA CTT ACA TCC TC-3′); pair 2, TX10NDE2F (5′-TAT GGA GAA GGC AGG CTC ACC AG-3′) and TX10BAM2R (5′-CTC ACA CTT TAC TTA CAT ATT TCT C-3′)] and a sticky-end PCR method (15), these cDNAs were transferred from the previously used pCWori+ vector (5) to pSBET (16). Denaturating and reannealing of the derived amplicon mixtures yielded cohesive-end products for each clone that contained 5′-NdeI and 3′-BamHI terminal overhangs to permit directional ligation into the appropriately digested pSBETa vector. These sequence verified vector constructs were individually transformed into Escherichia coli BL21(DE3) for expression. Each culture harboring a unique transacylase was used to inoculate 100 ml of Luria–Bertani medium supplemented with kanamycin (50 μg/ml) and was grown at 37°C to an A600 = 1.0, and then induced by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.5 mM. The cultures were then incubated at 18°C with shaking (220 rpm) for 16 h, harvested by centrifugation (2,000 × g, 20 min), resuspended in 10 ml of extraction buffer (50 mM Mopso, pH 7.2/5% glycerol/1 mM EDTA/0.5 mM DTT), and then disrupted by sonication for 30 s at 0°C by using a Virsonic 475 (Virtis, Gardiner, NY) with a 1.5-cm probe at maximum power. The resulting homogenates were clarified by centrifugation (45,000 × g, 1 h) to provide the soluble enzyme fraction.
A 1-ml aliquot (5 mg total protein) of the soluble enzyme preparation was incubated with 100 μM of N-debenzoyl-(3′RS)-2′-deoxytaxol and 80 μM (1.5 μCi; 1 Ci = 37 GBq) of [7-14C]benzoyl-CoA for 2 h at 31°C. The reaction mixture was extracted as described for other transferase assays (10), and the organic extract was analyzed by radio-HPLC (Perkin–Elmer HPLC ISS 200 pump coupled to a Perkin–Elmer ABI 785A UV/Visible Detector and a Packard A-100 Radiomatic detector). The samples were separated on a Phenomenex (Torrance, CA) reverse-phase Phenyl-3 column (5 μm, 4.6 × 250 mm) by elution at 1 ml/min with 25:75 (vol/vol) CH3CN/H2O for 5 min, then to 65:35 (vol/vol) CH3CN/H2O with a linear gradient over 40 min, then ramped to 85:15 (vol/vol) CH3CN/H2O and held 5 min, and finally returned to initial conditions. The enzyme preparation from an E. coli transformant bearing the clone designated TAX10 generated a single radioactive product (also detected at 254 nm) that was coincident with authentic (3′RS)-2′-deoxytaxol.
This E. coli transformant that harbored TAX10 was cultured at large scale (4 liter) to generate sufficient enzyme for preparative conversion to product for spectrometric analyses. The resulting product (≈1 mg) of the putative N-benzoyltransferase was purified by preparative TLC [0.5 mm silica gel, 50:50 (vol/vol) EtOAc/hexane], and the material that comigrated with authentic (3′RS)-2′-deoxytaxol (Rf = 0.15) was isolated, dissolved in 0.75 ml of CDCl3 as internal standard, and analyzed by 1H-NMR with a Varian Mercury 300 instrument. Also, a portion of the TLC-purified product (≈30 μg) was dissolved in 5 ml of methanol and, by syringe-pump delivery, was injected at 3 μl/min directly into an atmospheric pressure chemical ionization probe linked to an LCQ (ThermoQuest/Finnigan, San Jose, CA) ion-trap mass detector instrument in the positive-ion mode.
Partial Purification and Characterization of Recombinant N-Benzoyltransferase.
Four liters of E. coli culture that was transformed with the T. cuspidata TAX10 clone grown in 2.8-liter Fernbach flasks, harvested, and extracted as before. The soluble extract was analyzed by SDS/PAGE with Coomassie brilliant blue staining and revealed the presence of a protein of appropriate size (≈50 kDa) that was absent in cultures of control bacteria harboring empty vector. The extract (150 ml, 750 mg of total protein) was loaded onto a diethylaminoethyl-cellulose (Whatman DE-52, Clifton, NJ) column (2.5 × 20 cm; 75-ml bed volume) that was previously equilibrated with 50 mM Mopso (pH 7.2) containing 5% glycerol, 5 mM MgCl2, and 0.5 mM DTT (buffer A). After washing with 3 column volumes of buffer A, elution of the target enzyme was achieved with a linear NaCl gradient (0–500 mM; 500 ml; 10 ml/min). Fractions containing the bulk of the recombinant protein as determined by the described assay (120–200 mM NaCl, ≈80 ml) were pooled and loaded onto a 40-μm type 1 ceramic hydroxyapatite (Bio-Rad) column (2.5 × 20 cm; 50-ml bed volume) that was previously equilibrated with buffer B (buffer A containing 150 mM NaCl). After washing with 3 column volumes of buffer B, proteins were eluted with a linear gradient of 0–100 mM potassium phosphate (pH 7.2; 250 ml; 10 ml/min). Fractions containing the N-benzoyltransferase activity (eluting between 25–50 mM phosphate) were pooled, concentrated to 5 mg/ml (≈10 ml) by using an Amicon Centriprep YM-30 centrifugal concentrator (Millipore), and filtered through a 0.45-μm Acrodisc syringe filter (Pall). The filtrate, containing the partially purified protein (≈50% pure), was flash-frozen (liquid N2) in 100-μl batches and stored at −80°C for subsequent characterization studies.
After defining reaction linearity with respect to protein concentration and time, kinetic constants were determined by using standard assay conditions at reciprocally varied cosubstrate concentrations (0–1 mM) with the remaining reactant at saturation (1 mM). The program kaleidagraph (version 3.08, Synergy Software, Reading, PA) was used for calculations with double-reciprocal plotting of each data set, and the equation for the best fit line (R2 = 0.99) was determined. The data are reported as the mean of duplicate assays with SE less than ±15%. The pH optimum for N-acyltransferase activity was assessed in assays containing 5 μl of partially purified enzyme (≈0.5 μg protein), each diluted with 78-μl buffer comprised of either 25 mM sodium phosphate (at pH 5.5–9.0) or 25 mM 3-[cyclohexylamino]-2-hydroxy-1-propanesulfonic acid (Capso) (at pH 9.5–10).
Results and Discussion
Cloning and Heterologous Expression of a 3′-N-Debenzoyl-2′-Deoxytaxol N-Benzoyltransferase.
Biosynthetic analysis using Taxus brevifolia mature bark tissue as enzyme source demonstrated that N-benzoylation of 3′-debenzoyltaxol is the final step in the Taxol pathway (4). The substantial deduced sequence identity (58–69%) within a family of nine cloned acyltransferases from Taxus (10) [including a taxoid 2-O-benzoyltransferase among the three which have been functionally defined (17)] and between these and a Dianthus anthranilate N-hydroxycinnamoyl/benzoyltransferase (≈50–57%) (18) suggested that one of the six remaining undefined Taxus clones might encode the N-debenzoyltaxol N-benzoyltransferase.
To evaluate the remaining transacylase clones, a suitable taxoid cosubstrate was required. Syntheses of N-debenzoyltaxols have been described (6, 7), but these methods necessitate multiple steps and/or the use of starting materials that are of limited availability. Therefore, a direct and efficient four-step synthesis was implemented to couple 7-TES-baccatin III to the phenylpropanoid N-Boc-(3RS)-β-phenylalanine (11, 12), thus requiring minimal functional group protection. Deprotection (7, 8) of the resulting conjugate afforded N-debenzoyl-(3′RS)-2′-deoxytaxol in high purity as a surrogate substrate for assessing the heterologously expressed acyltransferase clones.
The six full-length, but still unassigned, Taxus cDNA clones, originally in pCWori+ (5), were transferred by a cohesive-end PCR method (15) into pSBETa for expression in E. coli; the latter plasmid is driven by the T7 promoter and also encodes the argU gene for the tRNA for the arginine codons AGA and AGG commonly used by plants. Each pSBET construct was used to individually transform E. coli BL21(DE3), and cultures of each induced bacterium were generated for enzyme preparation and assay under standard conditions (10) with N-debenzoyl-(3′RS)-2′-deoxytaxol and [7-14C]benzoyl-CoA as cosubstrates.
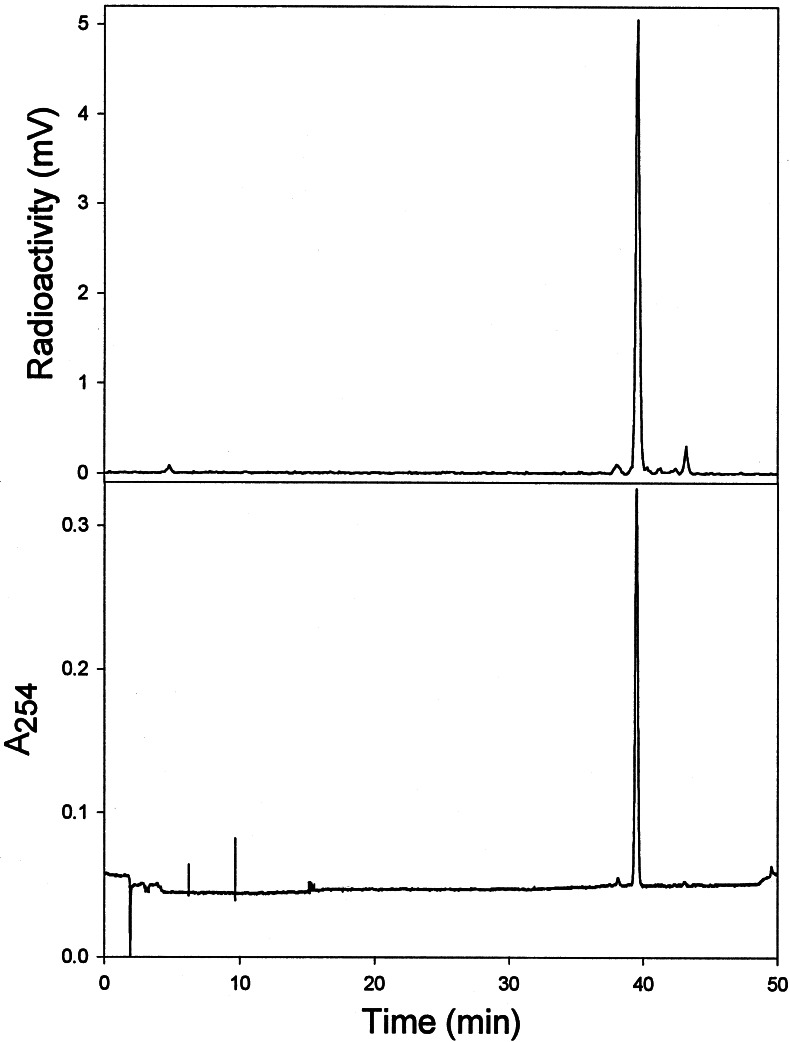
Enzyme expressed from the cDNA designated TAX10 yielded a single biosynthetic product that was revealed by radio-HPLC analysis to possess a retention time of 39.6 ± 0.1 min (with coincident radio and UV traces) corresponding exactly to that of authentic (3′RS)-2′-deoxytaxol (Fig. 2); this analysis does not resolve the diastereoisomers. Control extracts of E. coli host cells identically transformed with empty vector, or with pSBET harboring TAX1, TAX2, or TAX6 of defined function (17), did not yield detectable product when assayed by identical methods. Additionally, no substrate conversion was detected in TAX10 enzyme assays when either cosubstrate was absent or in control assays with boiled protein in the presence of both cosubstrates at saturation.
Figure 2.
Radio-HPLC analysis of the biosynthetic product (Rt = 39.6 ± 0.1 min) generated by the recombinant N-benzoyltransferase using N-debenzoyl-(3′RS)-2′-deoxytaxol and [7-14C]benzoyl-CoA as cosubstrates. The Upper trace shows the radioactivity profile (in mV) and the Lower trace shows the absorbance profile (A254), both of which coincide exactly with the retention time of authentic (3′RS)-2′-deoxytaxol.
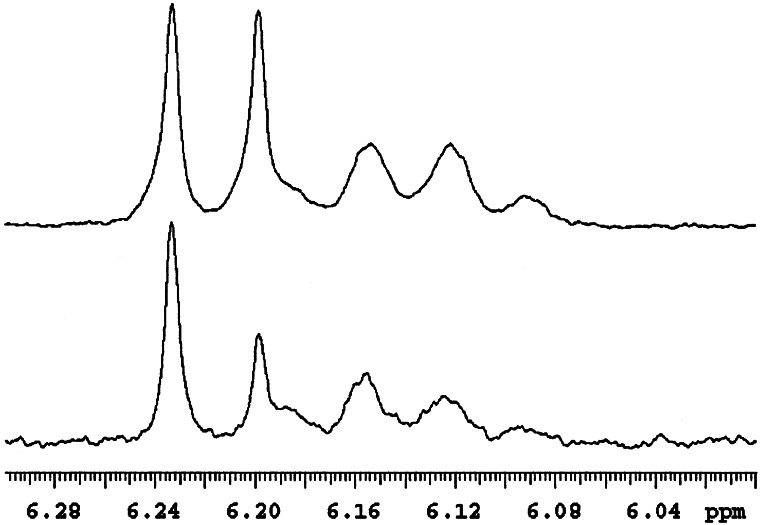
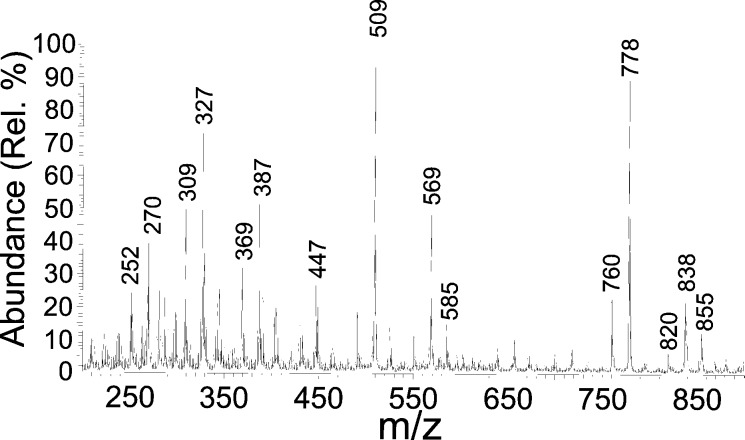
The TAX10 gene overexpressed from pSBET yielded a ≈50-kDa protein (determined by SDS/PAGE), consistent with the size of the encoded enzyme, and this operationally soluble recombinant enzyme from preparative bacterial cultures was partially purified by anion exchange (DE-52) and ceramic hydroxyapatite chromatography. Preparative-scale conversion with this material allowed generation of ≈1 mg of the TLC-purified product (99% pure by HPLC), which was identified by 1H-NMR analysis as 2′-deoxytaxol by comparison to the authentic reference standard. Thus, the TAX10 encoded enzyme was confirmed to be a DBTNBT. 1H-NMR analysis of authentic (3′RS)-2′-deoxytaxol showed the expected diastereoisomeric ratio of 1:1 by comparing the integration of the diagnostic H-10 signal that appears as a singlet (δ 6.196 and 6.232) for each epimer (Fig. 3). A similar detailed analysis of the biosynthetic product revealed that the TAX10 transferase is stereoselective for production of one diastereoisomer in 40% excess (Fig. 3); as yet, the absolute stereochemistry of each product isomer remains to be established. Further analysis by atmospheric pressure chemical ionization mass spectrometry (Fig. 4) revealed that the enzymatic product possesses a mass spectrum identical to that of the authentic standard.
Figure 3.
Partial 1H-NMR spectra (recorded in deuterochloroform) of authentic (3′RS)-2′-deoxyltaxol (Upper) and of the biosynthetic product derived by the recombinant N-benzoyltransferase (TAX10) with N-debenzoyl-(3′RS)-2′-deoxytaxol and benzoyl coenzyme-A as cosubstrates (Lower). The rest of the spectrum of the biosynthetic product was identical to that of the 2′-deoxytaxol standard.
Figure 4.
Atmospheric pressure chemical ionization mass spectrometric analysis of the biosynthetic product generated by the recombinant N-benzoyltransferase with N-debenzoyl-(3′RS)-2′-deoxytaxol and benzoyl-CoA as cosubstrates. Diagnostic ions are at m/z 855 (P+ + H2O), 838 (PH+), 820 (PH+ − H2O), 778 (PH+ − CH3CO2H), 760 (m/z 778 − H2O), 569 (PH+ − PhCH(BzNH)CH2CO2H), 509 (m/z 569 − CH3CO2H), and 270 (PhCH(BzNH)CH2CO2H + H+).
Characterization of the Recombinant N-Benzoyltransferase.
The pH optimum for the recombinant enzyme (designated DBTNBT) was found to be at pH 8.0, with half-maximal velocities near pH 7.0 and 9.0. This pH optimum is similar to those of other defined acyltransferases of the Taxol pathway (17) and to those of other acyltransferases of plant origin (19). Km values of 0.45 mM and 0.41 mM were calculated for 3′-N-debenzoyl-2′-deoxytaxol and benzoyl-CoA, respectively, by double-reciprocal plotting (R2 = 0.99). The kcat value was calculated to be 1.5 ± 0.3 s−1. DBTNBT is apparently regiospecific for acylation at the 3′-amino group of the 13-O-β-phenylalanoyltaxane substrate but not for acylation at the 2′-amino group, as evidenced by the lack of observable amide formation when 13-O-α-phenylalanoylbaccatin III and [7-14C]benzoyl-CoA were used as cosubstrates. The free amino acid β-phenylalanine was also not a productive substrate, indicating that this enzyme requires a 3-amino phenylpropanoyl baccatin ester as cosubstrate for N-acyl group transfer. Additionally, as might be expected, enzymatic amidation of S-α-phenylalanine was not observed.
To evaluate the selectivity of the transferase for the acyl donor, benzoyl-CoA was compared with phenylacetyl-CoA and acetyl-CoA as cosubstrates at saturation (1 mM). Evaluation of Vrel showed that benzoyl-CoA is the superior acyl donor; neither acetyl-CoA nor phenylacetyl-CoA were functional cosubstrates as no product was detected.
Sequence Analysis.
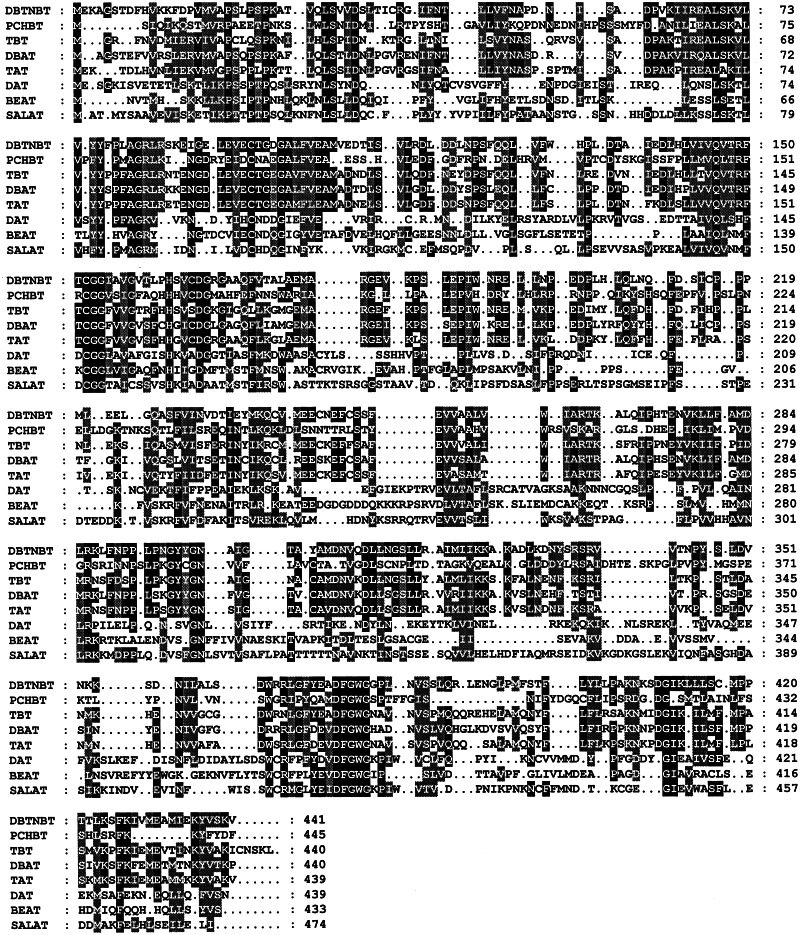
The 1,323-nt coding sequence for DBTNBT (GenBank accession no. AF466397) translates a protein of 441 aa with a calculated molecular weight of 49,040. The deduced amino acid sequence comprises several of the signature features of other Taxus acyltransferases (17) and of other related acyl/aroyltransferases of plant origin (19, 20), including similarities of ≈47–70%, the absence of N-terminal targeting information, a size of ≈50 kDa, and the conserved HXXXDG (H163, D167, and G168) motif (Fig. 5) (19), which may function in acyl transfer catalysis. Deduced sequence comparison of the taxoid N-benzoyltransferase with that of the mechanistically similar N-aroyltransferase anthranilate-N-cinnamoyl/benzoyltransferase from Dianthus revealed significant homology (53% identity, 64% similarity); yet comparison with that of the taxane 2-O-benzoyltransferase of Taxus revealed an even closer relationship (60% identity, 69% similarity) (Fig. 5).
Figure 5.
Deduced sequence alignment of DBTNBT (accession no. AF466397), anthranilate N-hydroxycinnamoyl/benzoyltransferase (PCHCBT, accession no. Z84383), taxane 2α-O-benzoyltransferase (TBT, accession no. AF297618), 10-deacetylbaccatin III 10-O-acetyltransferase (DBAT, accession no. AF193765), taxadien-5α-ol O-acetyltransferase (TAT, accession no. AF190130), deacetylvindoline 4-O-acetyltransferase (DAT, accession no. AF053307), benzyl alcohol acetyltransferase (BEAT, accession no. AF043464), and salutaridinol 7-O-acetyltransferase (SALAT, accession no. AF339913). Columns with residues in black boxes indicate identical amino acids for at least four of the compared sequences, whereas columns with residues in gray boxes indicate similar amino acids for at least four of the compared sequences.
Conclusions
The tricyclic structural core and the aminophenylpropanoyl side chain of the taxoids undergo both O- and N-acyl group transfer reactions, including acetylation, benzoylation, long-chain alkenoyl- and alkanoylation, and cinnamoylation. These diverse enzymatic acylations are, in part, responsible for generating in excess of 350 naturally occurring taxane diterpenoids in Taxus species (21). With the isolation of the DBTNBT cDNA, there are now four acyl/aroyltransferase genes identified that participate in the Taxol biosynthetic pathway (Fig. 1), including the side-chain 3′-N-benzoyltransferase described here, taxane 2α-O-benzoyltransferase, 10-deacetylbaccatin III 10-O-acetyltransferase, and taxadienol 5α-O-acetyltransferase (17). Each of these regioselective transfers contributes to the function of Taxol in promoting tubulin polymerization and stabilization (1). The utilization of these genes can facilitate increased production of Taxol and may allow the biosynthetic generation of Taxol analogs with modified acyl groups and substitution patterns to improve water solubility and/or potency.
There are now eight defined and functionally expressed cDNA clones from Taxus that encode catalysts for nearly half of the 20-step Taxol biosynthetic pathway. These include genes for the branch-point enzyme geranylgeranyl diphosphate synthase (22), the committed step catalyst taxadiene synthase (23), two taxane core hydroxylases (24, 25), and four acyl/aroyltransferases (Fig. 1). There remain to be defined at least six hydroxylases, an apparent oxidase, an epoxidase, an oxirane-to-oxetane oxomutase, an aminomutase, and the side-chain propanoyl-CoA transferase. The latter transferase may soon be acquired by the productive cloning strategy described here.
Acknowledgments
We thank Greg Helms (NMR Center) and Steven Halls (M. J. Murdock Biological Mass Spectrometry Laboratory) of Washington State University for technical assistance. This investigation was supported in part by National Institutes of Health Grant CA-55254, eXegenics Incorporated (Dallas), and McIntire–Stennis Project 0967 from Washington State Agricultural Research Center.
Abbreviation
- DBTNBT
N-debenzoyl-2′-deoxytaxol N-benzoyltransferase
Footnotes
Data deposition: The sequence reported in the paper has been deposited in the GenBank database (accession no. AF466397).
References
- 1.Kingston D G I. Chem Commun. 2001;10:867–880. [Google Scholar]
- 2.Guénard D, Guéritte-Voegelein F, Potier P. Acc Chem Res. 1993;26:160–167. [Google Scholar]
- 3.Georg G I, Ali S, Zygmut J, Jayasinghe L R. Exp Opin Ther Patents. 1994;4:109–120. [Google Scholar]
- 4.Floss H G, Mocek U. In: Taxol: Science and Applications. Suffness M, editor. Boca Raton, FL: CRC Press; 1995. pp. 191–208. [Google Scholar]
- 5.Walker K, Schoendorf A, Croteau R. Arch Biochem Biophys. 2000;374:371–380. doi: 10.1006/abbi.1999.1609. [DOI] [PubMed] [Google Scholar]
- 6.Gunatilaka A A L, Chordia M D, Kingston D G I. J Org Chem. 1997;62:3775–3778. [Google Scholar]
- 7.Georg G I, Boge T C, Cheruvallath Z S, Harriman G C B, Hepperle M, Park H, Himes R H. Bioorg Med Chem Lett. 1994;4:335–338. [Google Scholar]
- 8.Kingston D G I. J Nat Prod. 2000;63:726–734. doi: 10.1021/np000064n. [DOI] [PubMed] [Google Scholar]
- 9.Walker K, Croteau R. Proc Natl Acad Sci USA. 2000;97:583–587. doi: 10.1073/pnas.97.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker K, Croteau R. Proc Natl Acad Sci USA. 2000;97:13591–13596. doi: 10.1073/pnas.250491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiina I, Suenaga Y, Nakano M, Mukaiyama T. Bull Chem Soc Jpn. 2000;73:2811–2818. [Google Scholar]
- 12.Saitoh K, Shiina I, Mukaiyama T. Chem Lett. 1998;7:679–680. [Google Scholar]
- 13.Tarbell D S, Yamamoto Y, Pope B M. Proc Natl Acad Sci USA. 1972;69:730–732. doi: 10.1073/pnas.69.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cravallee C, Didier E, Pecquet P. Tetrahedron Lett. 1998;39:4263–4266. [Google Scholar]
- 15.Zeng G. BioTechniques. 1998;25:206–208. doi: 10.2144/98252bm05. [DOI] [PubMed] [Google Scholar]
- 16.Schenk P M, Baumann S, Mattes R, Steinbiss H. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 17.Walker K, Croteau R. Phytochemistry. 2001;58:1–7. doi: 10.1016/s0031-9422(01)00160-1. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, Reinhard K, Schiltz E, Matern U. Plant Mol Biol. 1997;35:777–789. doi: 10.1023/a:1005878622437. [DOI] [PubMed] [Google Scholar]
- 19.St-Pierre B, De Luca V. Recent Adv Phytochem. 2000;34:285–315. [Google Scholar]
- 20.Grothe T, Lenz R, Kutchan T M. J Biol Chem. 2001;276:30717–30723. doi: 10.1074/jbc.M102688200. [DOI] [PubMed] [Google Scholar]
- 21.Baloglu E, Kingston D G I. J Nat Prod. 1999;62:1448–1472. doi: 10.1021/np990176i. [DOI] [PubMed] [Google Scholar]
- 22.Hefner J, Ketchum R E B, Croteau R. Arch Biochem Biophys. 1998;360:62–74. doi: 10.1006/abbi.1998.0926. [DOI] [PubMed] [Google Scholar]
- 23.Wildung M R, Croteau R. J Biol Chem. 1996;271:9201–9204. doi: 10.1074/jbc.271.16.9201. [DOI] [PubMed] [Google Scholar]
- 24.Schoendorf A, Rithner C D, Williams R M, Croteau R B. Proc Natl Acad Sci USA. 2001;98:1501–1506. doi: 10.1073/pnas.98.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jennewein S, Rithner C D, Williams R M, Croteau R B. Proc Natl Acad Sci USA. 2001;98:13595–13600. doi: 10.1073/pnas.251539398. [DOI] [PMC free article] [PubMed] [Google Scholar]