Abstract
A histone-like nucleoid structure (H-NS) is a major component of the bacterial nucleoid and plays a crucial role in the global gene regulation of enteric bacteria. Here, we cloned and characterized the gene for the H-NS-like protein VpaH in Vibrio parahaemolyticus. vpaH encodes a protein of 134 amino acids that shows approximately 55%, 54%, and 41% identities with VicH in Vibrio cholerae, H-NS in V. parahaemolyticus, and H-NS in Escherichia coli, respectively. The vpaH gene was found in only trh-positive V. parahaemolyticus strains and not in Kanagawa-positive or in trh-negative environmental strains. Moreover, the G+C content of the vpaH gene was 38.6%, which is lower than the average G+C content of the whole genome of this bacterium (45.4%). These data suggest that vpaH was transmitted to trh-possessing V. parahaemolyticus strains by lateral transfer. The vpaH gene was located about 2.6 kb downstream of the trh gene, in the convergent direction of the trh transcription. An in-frame deletion mutant of vpaH lacked motility on semisolid motility assay plates. Western blot analysis and electron microscopy observations revealed that the mutant was deficient in lateral flagella biogenesis, whereas there was no defect in the expression of polar flagella. Additionally, the vpaH mutant showed a decreased adherence to HeLa cells and a decrease in biofilm formation compared with the wild-type strain. Introduction of the vpaH gene in the vpaH-negative strain increased the expression of lateral flagella compared with the wild-type strain. In conclusion, our findings suggest that VpaH affects lateral flagellum biogenesis in trh-positive V. parahaemolyticus strain TH3996.
Vibrio parahaemolyticus is a gram-negative halophilic bacterium that causes food-borne gastroenteritis most frequently associated with the consumption of raw or undercooked seafood (5, 42). Consumption of sufficiently high numbers of the organism can cause gastroenteritis in humans (20). In a few cases, infection results in primary septicemia and wound infection, similar to Vibrio vulnificus infections (6, 13). Most of the clinical isolates of V. parahaemolyticus, from patients with diarrhea, show hemolysis on a special blood agar (Wagatsuma agar) (36). This hemolysis is called the Kanagawa phenomenon (KP), which is induced by the thermostable direct hemolysin (TDH) (18, 40). In 1988, Honda et al. reported that KP-negative V. parahaemolyticus causes gastroenteritis in humans and identified a new hemolysin in clinical isolates of KP-negative V. parahaemolyticus strains. This new hemolysin was referred to as TDH-related hemolysin (TRH), which has biological activities similar to TDH and shares approximately 67% identity in amino acid sequence with TDH (19). The pathogenic mechanisms of V. parahaemolyticus have not been well elucidated, but the TDH and TRH produced by this bacterium are considered major virulence factors involved in gastrointestinal disorders (18, 40).
A histone-like nucleoid structure (H-NS) is a small, abundant nucleoid-associated protein that mediates chromosomal DNA condensation (21, 45). H-NS has a homodimer structure and binds preferentially to curved DNA sequences (47), although there is no apparent sequence specificity for H-NS binding to DNA. Characterization of dominant-negative mutants and mutational analysis reveal that the C-terminal region is required for DNA binding, whereas the N-terminal region is implicated in protein-protein interactions (44, 48). No information, however, is available concerning the structure of H-NS, except for the organization of its C-terminal domain that has been resolved by nuclear magnetic resonance (38). H-NS affects the expression of many unrelated genes, including proVWX, bgl, appY, ompC, and fimB in Escherichia coli (2, 8, 16), and the expression of some virulence genes in Salmonella enterica serovar Typhimurium, Shigella spp., and Vibrio cholerae (14, 25, 31, 32). The majority of genes are negatively regulated by H-NS, although some of them, such as the flagella regulon, are positively regulated (4, 17). Despite these numerous observations, the role of H-NS in bacterial physiology has not been completely elucidated.
Recent genome sequencing of the V. parahaemolyticus strain RIMD2210633 (Kanagawa phenomenon-positive, serotype O3:K6) revealed that an H-NS gene (hns) was located on the large chromosome of the organism (24). In the present study, we identified an additional H-NS homologue gene, vpaH, in the trh-positive V. parahaemolyticus strain TH3996. We then constructed a vpaH deletion mutant from this TH3996 strain and analyzed the phenotype.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
V. parahaemolyticus and E. coli strains and plasmids used in this study are listed in Table 1. V. parahaemolyticus TH3996 (34) was used for the cloning and deletion of vpaH and lafTU. All V. parahaemolyticus strains used in this study were obtained from the Laboratory for Culture Collection, Research Institute for Microbial Diseases, Osaka University. E. coli DH5α and SM10λpir (29) strains were used for the general manipulation of plasmids and the mobilization of plasmid into V. parahaemolyticus, respectively. The bacteria were cultured at 37°C with shaking in Luria-Bertani (LB) medium (for E. coli) and LB medium supplemented with 3% NaCl (for V. parahaemolyticus). Thiosulfate-citrate-bile-sucrose agar (Nissui Seiyaku, Tokyo, Japan) was used to screen mutant strains. Antibiotics were used at the following concentrations: 100 μg/ml for ampicillin and 5 μg/ml for chloramphenicol.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | hsdR recA lacZYAπ80 lacZΔM15 | Laboratory collection |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Muλpir R6K | 29 |
| V. parahaemolyticus | ||
| TH3996 | Clinical isolate, trh+ure+ | 34 |
| 3996-M1 | TH3996, vpaH disrupted | This study |
| 3996-M2 | TH3996, lafTU disrupted | This study |
| 3996-C1 | 3996-M1, vpaH complemented | This study |
| RIMD2212189 | Clinical isolate, KP positive, tdh+vpaH deficient | RIMDa |
| RIMD2212189-1 | RIMD2212189 with the vpaH gene introduced | This study |
| Plasmid | ||
| pUC119 | Cloning vector, Apr | 49 |
| pT7Blue T-vector | Multicopy (ColE1 ori) TA cloning vector, Apr | Novagen, Inc. |
| pYAK1 | Suicide vector, R6Kori, sacB, Cmr | 22 |
| pSA19CP | Plasmid vector derived from V. parahaemolyticus, Cmr | 30 |
| pKS-1 | 1.8-kb SpeI fragment cloned into the XbaI site on pUC119 | This study |
| pKS-2 | pSA19CP introduced with the PCR-amplified TH3996 vpaH gene | This study |
RIMD, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan.
DNA manipulations.
DNA manipulations were performed as previously described (37).
DNA sequencing and analysis.
Nucleotide sequences were determined with a Li-Cor model 4000L automated DNA sequencer (Lincoln, Nebr.) using universal M13 IRD41 infrared-dye-labeled primers. Computer analysis of DNA sequences was performed with the DNASIS program (Hitachi Software, Tokyo, Japan). Homology searches against representative sequences were done through the National Center for Biotechnology Information using the BLAST network service. The amino acid sequence alignment and phylogenetic analysis were carried out using the computer program CLUSTAL X (version 1.8) (43).
Dot hybridization analysis.
To determine the distribution of the vpaH gene among V. parahaemolyticus strains, dot blot hybridization was carried out as previously described (34). Probe for dot blot hybridization analysis was prepared by PCR using oligonucleotide primers that were synthesized based on the sequence of the vpaH gene: vpaH-F, 5′-ATGAGCGAATTCGAAAAAACG-3′, and vpaH-R, 5′-AGAGCAAAATCTAAAAGTTT-3′. The probe (size, 401 bp) was labeled using a random-primer extension method with the PCR-digoxigenin probe synthesis kit (Boehringer Mannheim). DNA was fixed to the membrane by UV cross-linking using a GS Gene Linker UV chamber (Bio-Rad). Hybridization was carried out at 42°C under high stringency conditions (50% concentration of formamide in hybridization solution), and hybridized DNA was detected with an alkaline phosphatase-labeled antidigoxigenin monoclonal antibody (Boehringer Mannheim).
Motility assay.
Tryptone swarm plates containing 1% Bacto tryptone, 3% NaCl, and 0.3% Bacto agar were used in motility assays. To document swarming motility, plates were inoculated with 2 μl of an overnight culture of cells normalized to an optical density of 1.0 at 600 nm. Plates were incubated at 28°C for 14 h, and motility was assessed qualitatively by examining circular swarms formed by the growing motile bacterial cells.
Antibodies and immunoblot analysis.
Preparation of polyclonal antibodies against lateral and polar flagellins was carried out as previously described (28). Serum was mainly collected 1 week after the final injection. After leaving for 2 h at room temperature, blood was centrifuged at 8,000 × g for 10 min. The serum was collected and stored at −30°C. Whole-cell preparations of wild-type and mutant strains (106 CFU) were loaded onto a sodium dodecyl sulfate (SDS)-10% polyacrylamide electrophoresis gel (PAGE). After electrophoresis, proteins were electrophoretically transferred to an Immobilon-P transfer membrane (Millipore, Bedford, MA). The membrane was blocked with 5% skimmed milk in Tris-buffered saline (20 mM Tris, 137 mM NaCl, pH 7.6; TBS) containing 0.1% Tween 20 (TBST) and probed with anti-flagellin antibody (polar or lateral) diluted to 1:3,000 for 30 min at room temperature. The secondary antibody was anti-rabbit immunoglobulin conjugated to horseradish peroxidase (Amersham Life Sciences). The blots were developed with the enhanced chemiluminescence (ECL) Western blotting kit (Amersham) according to the manufacturer's instructions.
Construction of deletion mutants. The vpaH and lafTU deletion mutants of V. parahaemolyticus TH3996 were constructed using the following primers (sequences in parentheses): Vp-M1 (5′-ACCTGAGGATCCCATGAGCG-3′), Vp-M2 (5′-GGTCTTGGCTTACGCATTTGCTCTGCTCTA-3′), Vp-M3 (5′-TAGAGCAGAGCAAATGCGTAAGCCAAGACC-3′), and Vp-M4 (5′-GACTTTTAACCTGCAGGTAAA-3′) for the vpaH mutant and TU-M1 (5′-TCTTGGATCCGACGCCACCTC-3′), TU-M2 (5′-AAAATGGCGTAAGCTTACGCGCTGTTTCTA-3′), TU-M3 (5′-TAGAAACAGCGCGTAAGCTTACGCCATTTT-3′), and TU-M4 (5′-TCGCTTTCTGCAGCTCGCTGT-3′) for the lafTU mutant.
For construction of the vpaH mutant, two DNA fragments were amplified by PCR with V. parahaemolyticus TH3996 chromosomal DNA as a template using the primer pairs Vp-M1 and Vp-M2 and Vp-M3 and Vp-M4, respectively. Primers Vp-M2 and Vp-M3 included a complementary 15-bp sequence at their 3′ and 5′ ends, respectively. These two fragments were used as templates in a second PCR using primers Vp-M1 and Vp-M4, resulting in the construction of a fragment with a 111-bp deletion in the vpaH gene. The fragment containing the deletion was purified and cloned into the pT7Bule T-vector (Novagen, Inc.).
This fragment was then removed from the pT7Bule T-vector by digestion with BamHI and PstI and cloned into the R6K-ori suicide vector pYAK1 (22), which contains the sacB gene conferring sensitivity to sucrose. This plasmid was introduced into E. coli SM10λpir (29), which was then conjugated with V. parahaemolyticus TH3996. vpaH deletion mutants were obtained by screening with Thiosulfate-citrate-bile-sucrose agar (Nissui Seiyaku, Tokyo, Japan) plates containing 5 μg/ml chloramphenicol and then selected on LB plates supplemented with 3% NaCl and 10% sucrose. The lafTU deletion mutant was constructed in a similar manner to that described for the construction of the vpaH deletion mutant.
Adherence assay.
HeLa cells (2 × 105) were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Sigma) in 12-well dishes, which were maintained in a humidified atmosphere of 5% CO2 in air at 37°C overnight. Overnight cultures of V. parahaemolyticus strains were diluted 1:100 in fresh LB medium, grown at 37°C to the late-log phase, and then centrifuged and suspended in phosphate-buffered saline (PBS, pH 7.2). Bacterial cell suspensions were used to infect HeLa cells at a multiplicity of infection of 10. After 1.5 h, nonadherent bacteria were removed by washing five times with PBS (pH 7.2). Monolayers and cell-associated bacteria were then recovered by treatment with 0.2% Triton X-100 for 5 min at room temperature. The recovered bacteria were plated on LB agar plates after dilution. Results are presented as the ratio of the adherence of the tested strains to that of the wild-type controls.
Biofilm formation.
Measurement of biofilm formation was conducted with a slight modification of a previously described method (46). Briefly, borosilicate glass tubes were utilized as surfaces for bacterial attachment. Six hundred microliters of LB medium supplemented with 3% NaCl broth, inoculated with a 1:100 dilution of overnight cultures grown in LB medium supplemented with 3% NaCl broth, was placed in each tube. These were incubated at 28°C for 20 h without shaking. After the bacterial cultures were poured out, tubes were rinsed vigorously with distilled water to remove nonadherent cells, filled with 700 μl of a 0.1% crystal violet solution (Wako Pure Chemical Industries, Ltd., Osaka, Japan), allowed to incubate for 30 min, and again rinsed vigorously with distilled water. Biofilm formation was quantified by measuring the optical density at 570 nm of a solution produced by extracting cell-associated dye with 1 ml of dimethyl sulfoxide. All assays were performed in triplicate.
Complementation of the vpaH gene.
The vpaH gene containing a putative promoter region from the TH3996 strain was amplified by PCR using the following oligonucleotide primers: 5′-ACTAGTGGATCCAAATTGTTC-3′ and 5′-GTGGATTGTTGTCGACTTACT-3′. The amplified fragment was cloned into a pT7Blue T-vector and digested with BamHI and SalI. The digested fragment was then cloned into a pSA19CP vector (30), which was introduced into 3996-M1 or RIMD2212189 by electroporation.
Transmission electron microscopy.
Bacterial suspensions were placed on Formvar-coated copper grids and negatively stained with a 2% solution of potassium phosphotungstate. Bacteria were then viewed on a Hitachi H7100 transmission electron microscope.
Nucleotide sequence accession number.
The nucleotide sequence data reported here will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB190437.
RESULTS
Cloning and analysis of the hns homologue gene.
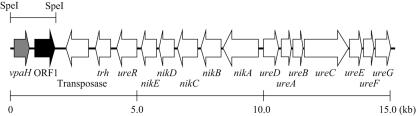
We previously cloned and sequenced a ca. 16-kb DNA region containing the trh gene and the gene cluster for urease production from a clinical isolate of the V. parahaemolyticus TH3996 strain (34). This DNA region is uniquely present in trh-positive V. parahaemolyticus strains and is characteristic of foreign DNA with a lower G+C content (41%) than the genome average (45.4%).
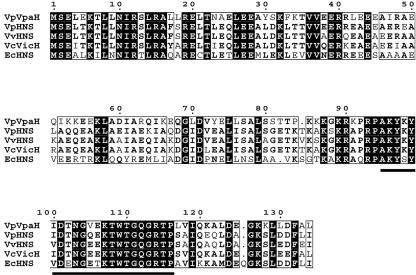
In this study, we cloned and sequenced upstream of this region for further analysis. Nucleotide sequencing of a ca. 1.8-kb SpeI fragment revealed that the G+C content of the region was 33.4%, a G+C content lower than the average of the whole genome in this bacterium (24). Therefore, we suggest that this DNA region is also of foreign origin. Two open reading frames (ORF) encoding proteins of 134 and 232 amino acids were found on the fragment (Fig. 1). The former ORF encoded a putative protein that had approximately 55.2% identity with V. cholerae VicH, 54.4% identity with V. parahaemolyticus RIMD 2210633 H-NS, and 41% identity with E. coli H-NS (Fig. 2).
FIG. 1.
Genetic organization of the DNA region containing the vpaH and trh genes of V. parahaemolyticus TH3996.
FIG. 2.
Multiple sequence alignment of the VpaH and H-NS proteins. The amino acid sequence alignment was compared using the computer program CLUSTAL W available at http://pbil.ibcp.fr. Identical residues are indicated by black boxes, and similar residues are indicated by white boxes. Residues conserved in the DNA-binding domain are indicated by the bold line below the boxes.
The H-NS gene was present in all the V. parahaemolyticus strains tested (n = 46) by dot blot hybridization using a digoxigenin-labeled hns probe (data not shown). We amplified the hns gene in TH3996 by PCR and determined the sequence. The sequence was completely identical with the hns gene of RIMD2210633 and obviously distinct from the ORF in the 1.8-kb SpeI fragment of TH3996. Therefore, we designated the putative protein encoded by the ORF VpaH, for V. parahaemolyticus H-NS-like protein. The vpaH gene was present about 2.6 kb downstream of the trh gene in the opposite direction of the trh transcription (Fig. 1). The N-terminal domain of VpaH was predicted to be mainly α-helical, while the C-terminal domain is probably a mixed α-helical and β-sheet structure (data not shown). A consensus motif of the DNA-binding domain in the C-terminal [AP]-K-Y-x (5,6)-[GS]-[ED]-x (0, 2)-T-W-[TS]-G-[QR]-G-[RK]-[TAK]-[PL] was conserved in the VpaH protein (Fig. 2). These observations suggest that VpaH is structurally conserved along with other members of a class of regulatory proteins, such as H-NS.
To determine the distribution of vpaH among V. parahaemolyticus strains, chromosomal DNA was prepared from a variety of clinical and environmental V. parahaemolyticus strains. Chromosomal DNA from 46 strains was examined by dot blot hybridization using a specific probe for vpaH. The vpaH gene was present only in the trh-positive strains (n = 18) but not in KP-positive strains (n = 13) or in trh-negative environmental strains (n = 15) (Table 2).
TABLE 2.
Distribution of the vpaH gene among V. parahaemolyticus strains
| Genotype | No.
|
|
|---|---|---|
| Testeda | vpaH positive (%) | |
| Clinical strains | ||
| tdh+ | 13 | 0 |
| trh+ | 8 | 8 (100) |
| tdh+trh+ | 10 | 10 (100) |
| Environmental strains, tdh deficient and trh deficient | 15 | 0 |
Dot hybridization was carried out at 42°C under high stringency conditions.
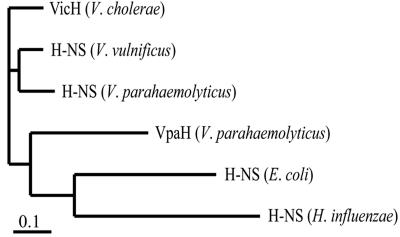
To look at the phylogenetic relationship of vpaH, we compared the amino acid sequences of VpaH and other homologues of the H-NS proteins in V. parahaemolyticus, V. cholerae, V. vulnificus, E. coli, and Haemophilus influenzae (Fig. 3). The phylogenetic branch of vpaH was separate from the hns genes of other vibrio species. Thus, these data suggest that vpaH is not very closely related to the hns genes of other vibrios. Taken together, these results suggest that not only trh and ure (34), but also vpaH, were transmitted into V. parahaemolyticus strains in the past by lateral transfer.
FIG. 3.
Amino acid sequence comparison representing the phylogenetic relationship of VpaH and other H-NS proteins. The scale bar represents 10% divergence at the amino acid level.
Construction and characterization of a vpaH deletion mutant.
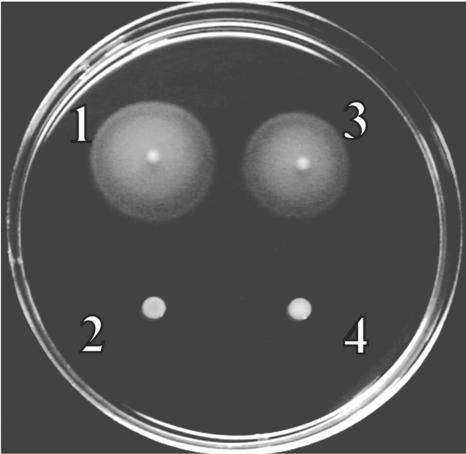
Mutations in the hns gene abolish motility in S. enterica serovar Typhimurium (17) and E. coli (4). In order to investigate whether vpaH is involved in bacterial motility, a vpaH deletion mutant was constructed by homologous recombination as described in the Materials and Methods section. Deletion of vpaH was confirmed by PCR and Southern hybridization (data not shown). The growth curve of the vpaH deletion mutant was indistinguishable from that of the wild-type strain in in vitro culture conditions, such as growth in LB medium at 37°C (data not shown). We tested the motility of the vpaH mutant strain using the semisolid motility plate assay. The vpaH mutant strain was nonmotile on the medium (Fig. 4). However, motility was fully restored by in trans complementation of the vpaH gene (Fig. 4). In V. parahaemolyticus, the lateral flagella are involved in movement over solid surfaces or in viscous environments and such movement is called swarming (39). Therefore, these results strongly suggest that the nonmotile phenotype of the vpaH mutant strain is due to a defect of lateral flagella.
FIG. 4.
Motility assay of wild-type and mutant strains. Bacteria were inoculated on a semisolid tryptone motility agar (1% tryptone, 3% NaCl, 0.3% agar) and grown at 28°C for 14 h. 1, wild-type strain; 2, vpaH deletion mutant strain; 3, vpaH complementation strain; 4, lafTU deletion mutant strain.
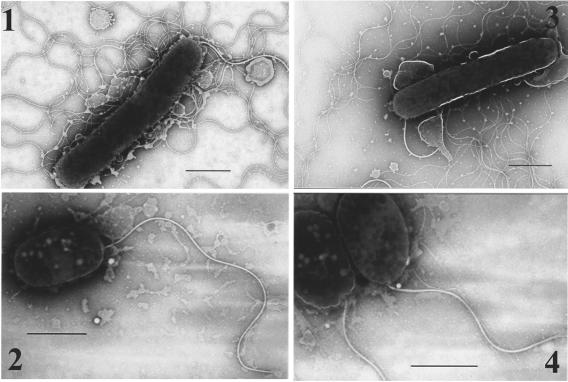
In order to elucidate whether this motility defect was caused by the loss of motor function or by changes in the overall structure of the lateral flagella, bacterial cells were observed by electron microscopy. Lateral flagella were observed on the wild-type and vpaH-complemented strains (Fig. 5). In contrast, we could not observe any lateral flagella on the vpaH mutant strain. Polar flagella were observed in all the strains analyzed (Fig. 5).
FIG. 5.
Transmission electron microscopy images of wild-type and mutant strains. Bacteria were grown on the LB plate supplemented with 3% NaCl at 37°C overnight and gently placed onto Formvar-coated copper grids and negatively stained using 2% potassium phosphotungstate. 1, wild-type strain; 2, vpaH deletion mutant strain; 3, vpaH complementation strain; 4, lafTU deletion mutant strain. Bars, 1 μm.
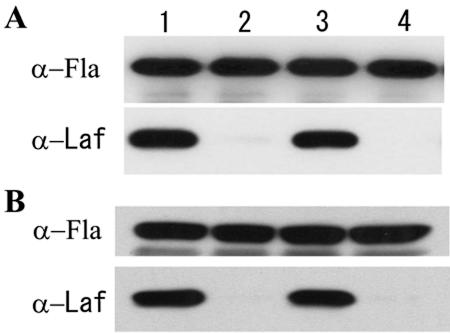
The expression of lateral and polar flagella in the vpaH mutant strain was assessed by Western blot analysis using rabbit polyclonal antisera specific for lateral or polar flagella. Expression of lateral flagella by the vpaH-deficient mutant was not detected by antilateral flagellin antibody. However, it was detected in the vpaH-complemented strain and the wild-type strain. Expression of polar flagella was unaffected in the vpaH-deficient mutant (Fig. 6). For comparison, we constructed a deletion mutant of lafTU, the lateral flagella motor genes, by homologous recombination (data not shown). The lafTU deletion mutant showed a phenotype similar to the vpaH mutant, with no motility on semisolid motility medium and failure to express the lateral flagella as previously reported (27) (Fig. 4, 5, and 6). These results suggest that the vpaH mutant is defective in lateral flagella biogenesis.
FIG. 6.
Western immunoblot analysis of whole-cell preparations of wild-type and mutant strains. Whole-cell proteins were obtained from bacteria after incubation at 37°C (A) and 28°C (B) for 14 h on LB plates. Whole-cell proteins (106 CFU) were loaded onto an SDS-10% PAGE gel, transferred to a nitrocellulose membrane, and probed with an α-Laf or α-Fla antibody, followed by anti-rabbit secondary antibodies, and then developed with the ECL blotting kit (Amersham). Lane 1, wild-type strain; lane 2, vpaH deletion mutant strain; lane 3, vpaH complementation strain; lane 4, lafTU deletion mutant strain.
The cell morphology of swimmer (28) and swarmer cells of V. parahaemolyticus is distinct. Beside lateral flagella formation, swarmer cells are elongated compared with swimmer cells when grown on a surface or in a viscous environment (26). Cells of the vpaH and lafTU mutant strains were round compared with cells of the wild-type strain (Fig. 4). These electron microscope observations were consistent with previous reports (26).
Phenotype of the vpaH deletion mutant strain.
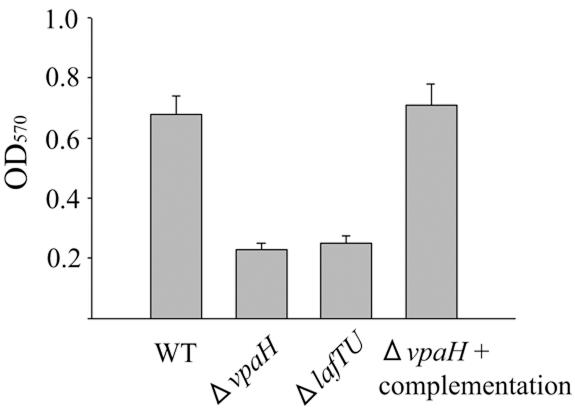
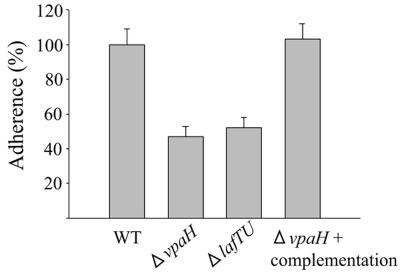
Swarming motility was thought to contribute to the bacterial colonization of host cell surfaces and play a significant role in biofilm formation (15). Therefore, we examined whether vpaH was involved in bacterial adherence to human epithelial cells and biofilm formation. The mutants vpaH and lafTU were impaired in their capacity to form biofilms, and both had about a 60% lower ability than the wild-type strain (Fig. 7). In adherence assays, vpaH and lafTU mutant strains showed 50% reduction in their adherence to HeLa cells compared with the wild-type strain (Fig. 8). These data demonstrate that the lateral flagella of TH3996 are involved in biofilm formation and adherence to HeLa cells, again suggesting that vpaH is involved in the regulation of lateral flagella biogenesis.
FIG. 7.
Ability of wild-type and mutant strains to form biofilms. Wild-type and mutant strain biofilms were formed in LB medium supplemented with 3% NaCl at 28°C for 20 h. Bacteria attached to borosilicate glass tubes were removed by washing five times with distilled water and then stained with crystal violet. The optical density at 570 nm was measured to quantify the amount of dimethyl sulfoxide-solubilized dye. Data shown are the averages of three independent experiments. Error bars represent standard deviations.
FIG. 8.
Adherence of wild-type and mutant strains to HeLa cells. HeLa cells (2 × 105) were infected with wild-type and mutant strains at a multiplicity of infection of 10. After 1.5 h, nonadherent bacteria were removed by washing five times with PBS. The monolayers and cell-associated bacteria were then recovered by treatment with 0.2% Triton X-100 for 5 min at room temperature. The recovered bacteria were plated on LB agar plates after dilution. Results are presented as the ratio of the adherence of the tested strains to that of the wild-type controls. Error bars represent standard deviations.
Introduction of vpaH into V. parahaemolyticus, which does not possess the vpaH gene.
To elucidate the function of vpaH in lateral flagella biogenesis, we introduced the vpaH gene from strain TH3996 into the V. parahaemolyticus strain RIMD2212189, which does not possess vpaH. The introduction resulted in the increased production of the flagellin protein of lateral flagella compared with that of the wild-type strain (Fig. 9). These data suggest that VpaH acts as a positive regulator of lateral flagella biogenesis in V. parahaemolyticus.
FIG. 9.
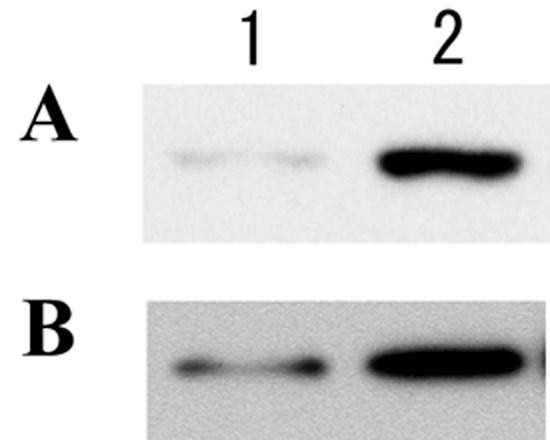
Increased lateral flagella expression by introduction of the vpaH gene into the RIMD2212189 strain, which does not possess the vpaH gene. Bacteria were grown overnight at 37°C (A) or 28°C (B) on LB plates supplemented with 3% NaCl. Whole-cell proteins (2 × 105 CFU) were loaded onto an SDS-10% PAGE gel, transferred to a nitrocellulose membrane, and probed with an antilateral flagella antibody, followed by an anti-rabbit secondary antibody, and then developed by ECL (Amersham). Lane 1, wild-type strain; lane 2, vpaH-introduced strain.
DISCUSSION
Histone-like DNA binding proteins, such as H-NS, the heat-unstable protein, and the integration host factor, in association with topoisomerases, play important roles in the maintenance of bacterial nucleoid organization (9, 10, 11). H-NS influences the expression of a myriad of seemingly unrelated genes by organizing promoter and regulatory regions into nucleoprotein complexes in response to environmental signals. The expression of genes influenced by H-NS is typically responsive to environmental parameters known to influence DNA topology, such as temperature, pH, and osmolarity (1). In V. parahaemolyticus, little is known about the H-NS and H-NS-like proteins.
In this study, we cloned and characterized a gene encoding an H-NS-like protein, VpaH, in V. parahaemolyticus. The vpaH gene was present only in trh-positive V. parahaemolyticus strains and in close proximity downstream of the trh gene. The G+C content of the vpaH gene was 38.6%, which is lower than the average of the whole genome in this bacterium. Phylogenetically, vpaH was not very closely related to the hns genes of other vibrios such as V. cholerae, V. parahaemolyticus, and V. vulnificus. Taken together, these data suggest that the vpaH gene was laterally transmitted into V. parahaemolyticus strains as the trh gene was (34).
Numerous phenotypes have been associated with mutations in hns genes. For example, motility is positively controlled by H-NS in E. coli (4) and S. enterica serovar Typhimurium (17). H-NS is a positive transcriptional regulator of the flhDC operon, the master operon that controls the expression of all other flagella components (23). Therefore, mutations in the hns gene of E. coli lower expression of the flagella genes, resulting in the loss of intact flagella and motility (4). V. parahaemolyticus possesses two flagella systems, polar and lateral. The polar flagella system propels the bacterium in liquid environments (swimming motility), while the lateral flagella system moves the bacterium on a solid surface or in viscous environments (swarming motility) (28). In this study, deletion of the vpaH gene in the TH3996 strain caused a dramatic reduction in motility of the organism on semisolid medium and a failure of lateral flagella production (Fig. 4, 5, and 6). This strongly suggests that VpaH is a positive regulator of the lateral flagella system in the TH3996 strain.
According to the completed genome sequence of TDH-positive V. parahaemolyticus strain RIMD2210633, the lateral flagella system of V. parahaemolyticus does not contain the flhDC operon (24). Therefore, the loss of lateral flagella and motility in the vpaH mutant strain does not occur through the flhDC operon. This feature is different from the phenotype of the hns mutant of E. coli.
Regarding bacterial motility, two different nonmotile mutant classes can be distinguished. Mot-deficient cells are immobile because of a defect in the motor proteins, although they possess flagella filaments. In contrast, Fla-deficient cells are unable to generate flagella because of a deficiency in biogenesis, transport, or assembly, resulting in a lack of motility (23). Most nonmotile bacterial mutants result from the latter case. The vpaH deletion mutant of V. parahaemolyticus belongs to the Fla-deficient class of mutants because of its inability to generate lateral flagella.
In V. parahaemolyticus, production of lateral flagella is thought to be an adaptation for living on surfaces, and their expression has been associated with better adherence and colonization (3). Gavin et al. recently demonstrated that lateral flagella are required for the adherence of Aeromonas species to human epithelial cells as well as the formation of biofilm and therefore could be involved in colonization of the human host (12). Biofilm formation, an important process for microbial survival in the environment and within host organisms, starts from initial attachment to a surface (7). In Pseudomonas aeruginosa and E. coli, initial attachment to a surface is accelerated by flagella (33, 35). In the present study, we demonstrated that mutant strains of vpaH or lafTU had a decreased ability to adhere to HeLa cells. Additionally, mutations in vpaH and lafTU caused a reduction in biofilm formation compared with the wild-type strain. These cell adherence and biofilm formation phenotypes both demonstrate the importance of lateral flagella.
To gain further insight into VpaH function in lateral flagella biogenesis, we introduced the vpaH gene into a vpaH noncoding strain. This resulted in an increase in the production of the flagellin protein of lateral flagella compared with the parental strain (Fig. 9). These data suggest that VpaH can act as a positive regulator of lateral flagella biogenesis not only in trh-positive strains but also in other V. parahaemolyticus strains, such as KP-positive and environmental strains. In KP-positive and environmental strains, however, lateral flagella are expressed even without the possession of vpaH, so the mode of regulation should be, at least in part, different from that of strain TH3996. Stewart and McCarter (41) reported that LafK controls the expression of early genes and FliAL controls late flagella gene expression in the lateral flagella gene system of V. parahaemolyticus. The V. parahaemolyticus strains used in the study by Stewart and McCarter (41) seem to be trh negative, so we do not think VpaH is involved in the regulatory system proposed by them.
In conclusion, we have demonstrated that vpaH, a gene of foreign origin, acts as a positive regulator in the biogenesis of lateral flagella in V. parahaemolyticus TH3996. This is an example of a laterally transmitted gene regulating an important bacterial function that existed before the acquisition of the regulator gene.
Acknowledgments
We thank the staff of Kansai International Airport Quarantine Station for the V. parahaemolyticus strains.
This work was supported by the Research for the Future Program (00L01411) of the Japan Society for the Promotion of Science and Grants-in-Aid for Scientific Research on Priority Areas and Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Editor: V. J. DiRita
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Atlung, T., S. Sund, K. Olesen, and L. Brondsted. 1996. The histone-like protein H-NS acts as a transcriptional repressor for expression of the anaerobic and growth phase activator AppY of Escherichia coli. J. Bacteriol. 178:3418-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belas, M. R., and R. R. Colwell. 1982. Adsorption kinetics of laterally and polarly flagellated Vibrio spp. J. Bacteriol. 151:1568-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella Escherichia coli. J. Bacteriol. 176:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake, P. A., R. E. Weaver, and D. G. Hollis. 1980. Diseases of humans (other than cholera) caused by vibrios. Annu. Rev. Microbiol. 34:341-367. [DOI] [PubMed] [Google Scholar]
- 6.Daniels, N. A., L. Mackinnon, R. Bishop, S. Altekruse, B. Ray, R. M. Hammond, S. Thompson, S. Wilson, N. H. Bean, P. M. Griffin, and L. Slutsker. 2000. Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181:1661-1666. [DOI] [PubMed] [Google Scholar]
- 7.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donato, G. M., M. J. Lilivelt, and T. H. Kawula. 1997. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J. Bacteriol. 179:6618-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman, C. J. 1995. DNA topology and the global control of bacterial gene expression: implications for the regulation of virulence gene expression. Microbiology 141:1271-1280. [DOI] [PubMed] [Google Scholar]
- 10.Drlica, K. 1984. The biology of bacterial DNA topoisomerases. Microbiol. Rev. 48:273-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drlica. K., and J. Rouviere-Yaniv. 1987. Histone-like proteins of bacteria. Microbiol. Rev. 51:301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin, R., A. A. Rabaan, S. Merino, J. M. Tomas, I. Gryllos, and J. G. Shaw. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43:383-397. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 173:1176-1183. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, J. A., D. Pickard, C. F. Higgins, A. Khan, S. N. Chatfield, T. Ali, C. J. Dorman, C. E. Hormaeche, and G. Dougan. 1994. Role of hns in the virulence phenotype of pathogenic salmonellae. Mol. Microbiol. 13:133-140. [DOI] [PubMed] [Google Scholar]
- 15.Harshey, R. M. 1994. Bees aren't the only ones: swarming in Gram-negative bacteria. Mol. Microbiol. 13:389-394. [DOI] [PubMed] [Google Scholar]
- 16.Higgins, C. F., C. J. Dorman, D. A. Stirling, L. Waddell, I. R. Booth, G. May, and E. Bremer. 1988. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569-584. [DOI] [PubMed] [Google Scholar]
- 17.Hinton, J. C. D., D. S. Santos, A. Seirafi, C. J. Hulton, G. D. Pavitt, and C. F. Higgins. 1992. Expression and mutational analysis of the nucleoid-associated protein H-NS of Salmonella typhimurium. Mol. Microbiol. 6:2327-2337. [DOI] [PubMed] [Google Scholar]
- 18.Honda, T., and T. Iida. 1993. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysin. Rev. Med. Microbiol. 4:106-113. [Google Scholar]
- 19.Honda, T., Y. Ni, and T. Miwatani. 1988. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56:961-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda, T., Y. Takeda, T. Miwatani, K. Kato, and Y. Nimura. 1976. Clinical features of patients suffering from food poisoning due to Vibrio parahaemolyticus, especially on changes in electrocardiograms. J. Jpn. Assoc. Infect. Dis. 50:216-223. [DOI] [PubMed] [Google Scholar]
- 21.Hulton, C. S. J., A. Seirafi, J. C. D. Hinton, J. M. Sidebotham, L. Waddell, G. D. Pavitt, T. Owen-Hughes, A. Spassky, H. Buc, and C. F. Higgins. 1990. Histon-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell 63:631-642. [DOI] [PubMed] [Google Scholar]
- 22.Kodama, T., Y. Akeda, G. Kono, A. Takahashi, K. Imura, T. Iida, and T. Honda. 2002. The EspB protein of enterohaemorrhagic Escherichia coli interacts directly with α-catenin. Cell. Microbiol. 4:213-222. [DOI] [PubMed] [Google Scholar]
- 23.Macnab, R. M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131-158. [DOI] [PubMed] [Google Scholar]
- 24.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, et al. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 25.Maurelli, A. T., and P. J. Sansonetti. 1988. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc. Natl. Acad. Sci. USA 85:2820-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarter, L., and M. Silverman. 1990. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 7:1057-1062. [DOI] [PubMed] [Google Scholar]
- 27.McCarter, L. L., and M. E. Wright. 1993. Identification of genes encoding components of the swarmer cell flagella motor and propeller and a sigma factor controlling differentiation of Vibrio parahaemolyticus. J. Bacteriol. 175:3361-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarter, L., M. Hilmen, and M. Silverman. 1988. Flagella dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345-351. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura, T., H. Hamashima, and K. Okamoto. 2000. Carboxy terminal region of haemolysin of Aeromonas sobria triggers dimerization. Microb. Pathog. 28:25-36. [DOI] [PubMed] [Google Scholar]
- 31.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Byrne, C. P., and C. J. Dorman. 1994. Transcription of the Salmonella typhimurium spv virulence locus is regulated negatively by the nucleoid-associated protein H-NS. FEMS Microbiol. Lett. 121:99-105. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole, G. A., and R. Kolter. 1998. Flagella and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 34.Park, K. S., T. Iida, Y. Yamaichi, T. Oyagi, K. Yamamoto, and T. Honda. 2000. Genetic characterization of DNA region containing the trh and ure genes of Vibrio parahaemolyticus. Infect. Immun. 68:5742-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 36.Sakazaki, R., K. Tamura, T. Kato, Y. Obara, S. Yamai, and K. Hobo. 1968. Studies on the enteropathogenic, facultatively halophilic bacteria. Vibrio parahaemolyticus. III. Enteropathogenicity. Jpn. J. Med. Sci. Biol. 21:325-331. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Shindo, H., T. Iwaki, R. Ieda, H. Kurumizaka, C. Ueguchi, T. Mizuno, S. Morikawa, H. Nakamura, and H. Kuboniwa. 1995. Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli. FEBS Lett. 360:125-131. [DOI] [PubMed] [Google Scholar]
- 39.Shinoda, S., and K. Okamoto. 1977. Formation and function of Vibrio parahaemolyticus lateral flagella. J. Bacteriol. 129:1266-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirai, H., H. Ito, T. Hirayama, Y. Nakabayashi, K. Kumagai, Y. Takeda, and M. Nishibuchi. 1990. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect. Immun. 58:3568-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, B. J., and L. L. McCarter. 2003. Lateral flagella gene system of Vibrio parahaemolyticus. J. Bacteriol. 185:4508-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, F. L., T. Iida, and J. Swings. 2004. Biodiversity of vibrios. Microbiol. Mol. Bio. Rev. 68:403-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J. D., T. J. Gibson, F. Plewniak. F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueguchi, C., T. Suzuki, T. Yoshida, K. Tanaka, and T. Mizuno. 1996. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J. Mol. Biol. 263:149-162. [DOI] [PubMed] [Google Scholar]
- 45.Varshavsky, A. J., S. A. Nedospasov, V. V. Bakayev, T. G. Bakayeva, and G. P. Georgiev. 1977. Histone-like proteins in the purified Escherichia coli deoxyribonucleoprotein. Nucleic Acids Res. 4:2725-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams, R. M., and S. Rimsky. 1997. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol. Lett. 156:175-185. [DOI] [PubMed] [Google Scholar]
- 48.Williams, R. M., S. Rimsky, and H. Buc. 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 178:4335-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]