Abstract
Epidemiological and pathological studies have suggested that infection with the oral pathogen Porphyromonas gingivalis can potentiate atherosclerosis and human coronary heart disease. Furthermore, infection with invasive, but not noninvasive P. gingivalis has been demonstrated to accelerate atherosclerosis in apolipoprotein E-deficient (ApoE−/−) mice and to accelerate local inflammatory responses in aortic tissue. In the present study, using high-density oligonucleotide microarrays, we have defined the gene expression profile of human aortic endothelial cells (HAEC) after infection with invasive and noninvasive P. gingivalis. After infection of HAEC with invasive P. gingivalis strain 381, we observed the upregulation of 68 genes. Genes coding for the cytokines Gro2 and Gro3; the adhesion molecules intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule (VCAM)-1, and ELAM-1 (E-selectin); the chemokine interleukin-8 (IL-8); and the proinflammatory molecules IL-6 and cyclooxygenase-2 were among the most highly upregulated genes in P. gingivalis 381-infected HAEC compared to uninfected HAEC control. Increased mRNA levels for signaling molecules, transcriptional regulators, and cell surface receptors were also observed. Of note, only 4 of these 68 genes were also upregulated in HAEC infected with the noninvasive P. gingivalis fimA mutant. Reverse transcription-PCR, enzyme-linked immunosorbent assay, and fluorescence-activated cell sorting analysis confirmed the expression of ICAM-1, VCAM-1, E-/P-selectins, IL-6, and IL-8 in HAEC infected with invasive P. gingivalis. We also demonstrated that increased expression of ICAM-1 and VCAM-1 in aortic tissue of ApoE−/− mice orally challenged with invasive P. gingivalis but not with the noninvasive P. gingivalis fimA mutant by immunohistochemical analysis. Taken together, these results demonstrate that P. gingivalis fimbria-mediated invasion upregulates inflammatory gene expression in HAEC and in aortic tissue and indicates that invasive P. gingivalis infection accelerates inflammatory responses directly in the aorta.
Atherosclerosis, formerly considered a lipid storage disease, actually involves an ongoing inflammatory response. Modified lipoproteins and local or distant infections have been proposed to contribute to the inflammatory process in atherosclerosis (36). Cross-sectional and epidemiologic studies have demonstrated that patients with chronic inflammatory periodontitis, compared to nondiseased patients, are at increased risk for developing atherosclerosis (1, 9). Porphyromonas gingivalis, the major etiological agent of adult periodontal disease (15, 26), has been identified in diseased human atherosclerotic tissues (13) and has been shown to increase the mean area and extent of atherosclerotic lesions in apolipoprotein-E-knockout (ApoE−/−) mice (11, 23, 24). However, a P. gingivalis fimA-deficient mutant, which does not adhere to or invade endothelial cells (5), failed to accelerate atherosclerosis in ApoE−/− mice despite a measurable bacteremia and localization of the mutant to the aorta (11). Furthermore, only invasive bacteria were found to accelerate a local inflammatory response in aortic lesions of vascular tissues (11). These results support that P. gingivalis invasion is critical for accelerated atheroma development.
We have previously demonstrated that invasive strains of P. gingivalis, but not a noninvasive fimA mutant, stimulate the expression of cell adhesion molecules, including intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and P-/E-selectin on the cell surface of human umbilical vein endothelial cells (HUVEC) (21). In addition, we reported that P. gingivalis can modulate the expression of chemokines such as interleukin-8 (IL-8), in HUVEC, through a fimbria-mediated mechanism (29). These findings suggested that live and invasive bacteria are required for the induction of inflammatory molecules in endothelial cells. Our initial studies focused on the expression of a subset of endothelial cell genes in response to invasive bacterial infection. However, a high-throughput analysis of the complete host response to P. gingivalis infection of endothelial cells is still lacking.
Since it has been reported that endothelial cells obtained from different anatomical sites do not react similarly (28), the aims of the present study were (i) to utilize DNA microarray analysis to characterize the primary responses of human aortic endothelial cells (HAEC), a more relevant cell type to atherosclerosis progression, to P. gingivalis, (ii) to identify host genes differentially regulated by invasive and noninvasive P. gingivalis challenge, and (iii) to confirm specific molecules identified by microarray analysis in aortic tissue using an mouse model of P. gingivalis infection-accelerated atherosclerosis. We demonstrate that P. gingivalis infection of HAEC upregulates expression of several classes of molecules known to play a role in atheroma development and that this response is mediated via fimbria-induced invasion. Furthermore, elevated levels of cellular adhesion molecules which were identified by microarray were also detected in aortic tissue obtained from ApoE−/− mice orally challenged with invasive, but not noninvasive, P. gingivalis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The P. gingivalis wild-type strain 381 and the isogenic fimA mutant (DPG3) (26) were used throughout these studies to determine the role of P. gingivalis invasion in the regulation of mRNA profiles in a cell culture system. The P. gingivalis strains were routinely maintained on brain heart infusion (BHI) blood agar plates (Difco, Sparks, Md.) and BHI broth containing 0.5% yeast extract (Difco), hemin (10 μg ml−1), and vitamin K (1 μg ml−1). DPG3 strain was maintained on similar medium containing erythromycin (10 μg ml−1). For all experiments, bacterial cells were incubated under anaerobic conditions. Heat-killed P. gingivalis was prepared by heating a bacterial suspension for 10 min at 60°C.
Cell culture and infection with P. gingivalis.
HAEC were purchased from Cascade Biologics, Inc. (Portland, Oreg.) and were maintained in Media-200 supplemented with low-serum growth supplement (20 μl ml−1; Cascade Biologics, Inc.) at 37°C in 5% CO2. Confluent second- to fourth-passage HAEC were used in all experiments. Endothelial cells were plated 24 h before infection in a six-well plate at a cell density of 3 × 105 cells per well, as determined by cell counting with a hemocytometer. The multiplicity of infection (MOI) for P. gingivalis was calculated based on the number of HAEC per well when seeded. Wild-type P. gingivalis and DPG3 were grown to an optical density of 1.0, were washed and resuspended in HAEC growth medium to a final concentration of 3 × 107 cells ml−1. The bacterial inoculum (1 ml) was added to confluent HAEC monolayers (MOI = 100) and incubated at 37°C in 5% CO2 for 1 h. For microarray analysis, after 1 h infection, nonadherent bacteria were removed by washing, and HAEC infected with P. gingivalis were cultured in fresh medium for an additional 5 h. When the total incubation period reached 6 h post-P. gingivalis infection, HAEC were harvested, and total RNA was isolated and processed as described for the microarray analysis. For reverse transcription-PCR (RT-PCR) and protein assays, after 1 h infection, supernatants were collected for enzyme-linked immunosorbent assay (ELISA) as described in the cytokine assay listed below, and cells were harvested either for RT-PCR or for fluorescence-activated cell sorting (FACS) analysis as described below. For the 6- and 24-h experiments, nonadherent bacteria were removed by washing, and HAEC infected with P. gingivalis were cultured in fresh medium for an additional 5 or 23 h. When the total incubation period reached 6 or 24 h post-P. gingivalis infection, supernatants and HAEC were harvested for RT-PCR or protein analysis. Unchallenged HAEC were used as a control. Total RNA was isolated from infected or control cells by using the RNeasy minikit (QIAGEN, Valencia, Calif.) and treated with RNase-free DNase (QIAGEN) to remove contaminating genomic DNA according to the manufacturer's instructions. Bacterial adherence and invasion was determined as previously described (5).
DNA microarray and data analysis.
The HG-U95Av2 arrays, representing approximately 10,000 full-length human genes, were used in the present study (Affymetrix, Santa Clara, Calif.). The poly(A)+ RNA was purified from total RNA collected from HAEC 6 h after P. gingivalis infection by using Oligotex polystyrene-latex resin (QIAGEN). Subsequently, synthesis of cDNA and cRNA, target hybridization, washing and scanning was carried out according to the Affymetrix protocol. Affymetrix GeneChips were scanned, and the resulting image files were used to calculate and normalize the hybridization intensity data utilizing the Microarray Suite 5.0 software (Affymetrix). Briefly, the fluorescence of each array was normalized by global scaling with a target intensity of 500. The statistical algorithm within this software was used for the absolute analysis of each individual microarray. The single-array analysis measures a relative level of expression of a transcript (signal) and determines whether a transcript is present (P) or absent (A). Absolute analysis of each microarray was followed by comparison analysis using GeneSpring software (Silicon Genetics, Redwood City, Calif.). The comparison estimates the magnitude of change (i.e., the fold change of the normalized data) and the direction of the change (increase, decrease, or no change) of a transcript across the two arrays. Each experiment was performed twice, and only transcripts showing the same detection call in these duplicates (P/P or A/A) were included here. For the comparison analysis, mean data for two sets of replicate samples were used in the comparison. For most data sets, the results were reported as the average fold change from the comparisons. A given transcript was designated as “upregulated” when the average fold change increased at least twofold in expression level between two sets of replicate samples. A given transcript was designated as “downregulated” when the average fold change decreased at least twofold in expression level (ratio of ≤0.5) between two sets of replicate samples. The range of the transcript ratio for downregulated genes was observed to be between 0.2 and 1.0 as described in Table 3.
TABLE 3.
Analysis of HAEC mRNA downregulated expression after P. gingivalis infectiona
| Category and common name | GenBank accession no. | Description | Relative mRNA expression ratio (infected/control)
|
|
|---|---|---|---|---|
| Wt/ctl | DPG3/ctlb | |||
| Cell cycle control | ||||
| RBQ-1 | X85133 | RBQ-1 mRNA | 0.3 | 0.6 |
| CDC2 | Y00272 | Cell cycle control gene CDC2 | 0.3 | 0.2 |
| DUSP4 | U48807 | MAP kinase phosphatase (MKP-2) | 0.4 | 0.7 |
| RANBP6 | AF039023 | Ran-GTP binding protein | 0.5 | 0.7 |
| Stress-related gene | ||||
| Unknownc | D14874 | Adrenomedullin precursor | 0.4 | 0.7 |
| NR3C2 | M16801 | Mineralcorticoid receptor (hMR) | 0.5 | 1.0 |
| Others | ||||
| Unknown | AF007155 | Clone 23763 unknown mRNA | 0.2 | 0.6 |
| POLR2K | AI744294 | cDNA, 3′ end | 0.3 | 0.3 |
| Unknown | AL040446 | cDNA, 5′ end | 0.3 | 0.7 |
| Apg4B | AB023160 | KIAA0943 | 0.4 | 0.5 |
| Unknown | AF070641 | clone 24421 | 0.4 | 0.9 |
| PPP1R3C | N36638 | cDNA, 5′ end/clone = IMAGE-268833 | 0.4 | 0.6 |
| DKFZP | AL050390 | Mrna | 0.4 | 0.8 |
| D43538 | MTG8a protein | 0.5 | 0.6 | |
| KIAA0241 | D87682 | KIAA0241 gene | 0.5 | 0.8 |
| LYL1 | M22637 | LYL-1 protein | 0.5 | 0.7 |
P. gingivalis 381 (wild type) or DPG3 (fimA mutant) were added to HAEC cultures at an MOI of 100, followed by incubation at 37°C for 1 h; nonadherent bacteria were removed by washing, and HAEC infected with P. gingivalis were cultured in fresh culture medium for an additional 5 h. When the total incubation period reached 6 h post-P. gingivalis infection, HAEC were harvested, and total RNA was extracted and analyzed by using microarray as described in Materials and Methods. Control cultures were incubated with culture medium only. Genes whose expression in P. gingivalis 381-infected HAEC were at least twofold lower (ratio of ≤0.5) than those in uninfected HAEC are listed. Gene names in italics designate genes whose expression in P. gingivalis 381-infected and DPG3-infected HAEC cultures was at least twofold lower (ratio of ≤0.5) than those in uninfected HAEC.
See Table 2, footnote b.
Unknown, no common name available.
RT-PCR.
Total RNA was treated with DNase as described above and the absence of genomic contamination was confirmed by gel electrophoresis and PCR amplification with glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primers (Table 1). RT-PCR was used to study the kinetics of selected genes and to verify the patterns of gene expression revealed by microarray analysis. Total RNA was isolated as described above. RT-PCRs were carried out by using SuperScript One-Step RT-PCR with Platinum Taq polymerase (Invitrogen Life Technologies, Carlsbad, Calif.). A 100- or 200-ng sample of DNase-treated total RNA was used as a template in each reaction. The primers used for these reactions are listed in Table 1.
TABLE 1.
Primers and amplification conditions for RT-PCR analysisa
| Gene | Primer type | Sequence | ATb(°C) | Size (bp) | RNA amt (ng) | Cycle no. |
|---|---|---|---|---|---|---|
| ICAM-1 | Sense | 5′-TATGGCAACGACTCCTTCT-3′ | 55 | 238 | 200 | 30 |
| Antisense | 5′-CATTCAGCGTCACCTTGG-3′ | |||||
| VCAM-1 | Sense | 5′-ATGACATGCTTGAGCCAGG-3′ | 55 | 260 | 200 | 30 |
| Antisense | 5′-GTGTCTCCTTCTTTGACACT-3′ | |||||
| E-selectin | Sense | 5′-CTCTGACAGAAGAAGCCAAG-3′ | 55 | 255 | 200 | 30 |
| Antisense | 5′-ACTTGAGTCCACTGAAGCCA-3′ | |||||
| GRO2 | Sense | 5′-CCGAAGTCATAGCCACACTC-3′ | 60 | 528 | 100 | 36 |
| Antisense | 5′-GGCCATTTTCTTGGATTCCT-3′ | |||||
| GRO3 | Sense | 5′-GAACTGCGCTGCCAGTG-3′ | 60 | 543 | 100 | 30 |
| Antisense | 5′-AGGTGGCTGACACATTATGG-3′ | |||||
| COX-2 | Sense | 5′-TTCAAATGAGATTGTGGAAAAATTGCT-3′ | 55 | 305 | 100 | 36 |
| Antisense | 5′-AGATCATCTCTGCCTGAGTATCTT-3′ | |||||
| IL-6 | Sense | 5′-ATGAACTCCTTCTCCACAAGCGC-3′ | 60 | 628 | 100 | 25 |
| Antisense | 5′-GAAGAGCCCTCAGGCTGGACTG-3′ | |||||
| IL-8 | Sense | 5′-ATGACTTCCAAGCTGGCCGTGGCT-3′ | 60 | 294 | 100 | 30 |
| Antisense | 5′-TCTCAGCCCTCTTCAAAAACTTCTC-3′ | |||||
| GAPDH | Sense | 5′-GGTGAAGGTCGGAGTCAACGG-3′ | 55 | 520 | 100 | 36 |
| Antisense | 5′-GGTCATGAGTCCTTCCACGAT-3′ |
The full names of the genes and their accession numbers are given in Table 2. Sense and antisense primers, annealing temperatures, the expected sizes of the specific PCR product, the RNA amounts per reaction, and the number of PCR cycles used for PCR analysis of the indicated genes are shown.
AT, annealing temperature.
Cytokine assay and FACS analysis.
For the cytokine assay, concentrations of IL-8 and IL-6 in cell culture supernatants were determined by using commercially available ELISA kits (BD Biosciences, San Diego, Calif.) as described according to manufacturer's instructions. For FACS analysis, cells were grown in triplicate in six-well dishes and were incubated with P. gingivalis as described above. Cells were dissociated with trypsin and EDTA, washed with phosphate-buffered saline and fixed with ice-cold 4% paraformaldehyde at 4°C. Fixed cells were then reacted with fluorescein isothiocyanate-conjugated mouse immunoglobulin G1 (IgG1) of anti-human ICAM-1, VCAM-1, E-/P-selectin, P-selectin monoclonal antibodies, and isotype-specific fluorescein isothiocyanate-conjugated mouse IgG1 (Serotec, Oxford, United Kingdom) was used as a negative control. HAEC were analyzed by flow cytometry using FACScan flow cytometer (Becton Dickinson, Cockeysville, Md.). A total of 10,000 events were counted for each condition.
Oral infection to ApoE−/− mice.
Five-week-old male ApoE−/− mice (Jackson Laboratories, Bar Harbor, Maine) were cared for in accordance with NIH and Boston University Institutional Animal Care and Use Committee approved procedures and received standard chow diet and water ad libitum. ApoE−/− mice were challenged orally with wild-type (invasive) or fimA mutant (noninvasive) P. gingivalis five times per week over 3 weeks as described previously (11). This infection regimen was originally described by Lalla et al. (23). In this model, the intent is to mimic chronic P. gingivalis exposure, as is typically seen during human infections (11). Unchallenged mice served as age-matched controls. All animals (n = 6 for each group) were monitored daily until sacrifice (6 weeks after the final oral challenge) and appeared healthy throughout the course of the present study.
Immunohistochemistry.
Cryosections (10 μM) of dissected aorta sinus from ApoE−/− mice were incubated with the following primary antibodies and isotype-matched antibodies which were used to probe 10-μm cryosections: (i) rat anti-mouse ICAM-1 antibody and isotype-matched control rat IgG2a (Serotec, Kidlington, Oxford, United Kingdom) and (ii) rat anti-mouse VCAM-1 antibody and isotype-matched control rat IgG1 (Serotec). Immuno-enzyme staining was performed by biotin-streptavidin-peroxidase method (Dako, Carpinteria, Calif.).
Statistical analysis.
All statistical analyses were performed by using One-way analysis of variance with Tukey-Kramer multiple-comparisons test. Differences in the data were considered significant when the probability value was <5.0% (P < 0.05).
RESULTS
Innate immune response genes are upregulated after invasive P. gingivalis infection of HAEC.
We previously determined that at an MOI of 100 optimal invasion of P. gingivalis 381 into endothelial cells occurred after a 1- to 2-h exposure period and did not increase with an extended incubation period up to 4 h (5). Likewise, we determined that intracellular P. gingivalis 381 remained viable within cells (5). Since we were interested in defining the response of HAEC to invasive P. gingivalis, HAEC were exposed to P. gingivalis 381 for 1 h, extracellular bacteria were then washed, and cultures incubated for an additional 5 h. The invasion efficiency of P. gingivalis 381 into HAEC, after 1- and 6-h infections, was similar to that observed previously in bovine aortic endothelial cells (5) (data not shown).
Using microarray technology, we observed the expression of 5,209 genes in uninfected HAEC as assessed at 6 h. In P. gingivalis 381-infected HAEC we observed the upregulation of 68 genes compared to uninfected HAEC control cultures (Table 2). The majority of these genes were involved in proinflammatory responses and included: (i) genes encoding cytokines, cellular receptors, adhesion molecules, and enzymes; (ii) genes involved in angiogenesis; (iii) apoptotic and antiapoptotic genes; and (iv) genes involved in nuclear factor-κB (NF-κB) signal transduction. Among the most highly expressed genes in P. gingivalis 381-infected HAEC were genes belonging to the chemokine family and cell adhesion molecules that are involved in the recruitment and trafficking of lymphocytes to sites of vascular inflammation. In the group of genes encoding cellular receptors we found that the proteinase-activated receptor 2 (PAR-2) gene, which has been previously reported to be involved in the development of atherosclerotic plaque (30), was upregulated among other upregulated genes encoding inflammatory cytokine receptors such as IL-18R1 and gamma interferon receptor alpha chain. The second group of genes that was identified were genes involved in angiogenesis, such as fibroblast growth factor 5 and epidermal growth factor. Several genes involved in cell apoptosis were also upregulated after infection with invasive P. gingivalis and included tumor necrosis factor alpha (TNF-α)-inducible primary response gene 3 (TNFAIP3), TNFAIP2 (B94 protein), TNF receptor-associated factor 1 (TRAF1), TNFSF10 (TNF-related apoptosis inducing ligand; TRAIL), CFLAR (caspase-like apoptosis regulatory protein 2), and TANK (TRAF-interacting protein I-TRAF). In addition, we also observed upregulation of antiapoptotic genes, including the BIRC3 (human inhibitor of apoptosis protein 1) and BCL2A1 (human Bcl-1 related; Bfl-1) genes, after infection with invasive P. gingivalis. In the third group of upregulated genes were those encoding molecules of nucleic acid binding, with the majority of these involved in NF-κB signal transduction. These genes included MAD-3, I-REL, CEBPD (NF-IL-6-beta), Jun-B, p50-NF-κB (NF of κ light polypeptide gene enhancer), JUNB (transactivator jun-B), NFKBIE (IκB epsilon), NFkB1 (NF-κB DNA-binding subunit), and NFkB2 (NF-κB subunit).
TABLE 2.
Analysis of HAEC mRNA upregulated expression after P. gingivalis infectiona
| Category and common name | GenBank accession no. | Description | Relative mRNA expression ratio (infected/control)b
|
|
|---|---|---|---|---|
| Wt/ctl | DPG3/ctl | |||
| Cytokines | ||||
| GRO3 | M36821 | Cytokine GRO-gamma | 61.7 | - |
| SCYA20 | U64197 | Chemokine exodus-1 | 60.6 | 1.4 |
| GRO2 | M36820 | Cytokine GRO-beta | 30.6 | 1.3 |
| MGSA | X54489 | Gene for melanoma growth stimulatory activity | 16.7 | 1.5 |
| SCYA2 | M26683 | Gamma interferon treatment inducible mRNA | 13.5 | - |
| JE | M28225 | JE gene encoding a monocyte secretory protein | 13.3 | 1.8 |
| SCYA2 | M26683 | Gamma interferon treatment-inducible mRNA | 12.7 | 2.0 |
| C-X3-C Chemokine | U84487 | Small inducible cytokine subfamily D | 10.5 | 1.0 |
| IL-8 | M17017 | β-Thromboglobulin-like protein | 5.5 | 1.4 |
| IL-8 | M28130 | IL-8 | 4.5 | 1.2 |
| IL-6 | X04430 | Beta 2 interferon | 4.4 | 1.0 |
| Cytokine receptors | ||||
| RDC1 | U67784 | Orphan G protein-coupled receptor | 6.0 | 1.0 |
| F2RL1 | U67058 | Proteinase activated receptor-2 protein | 3.4 | 1.8 |
| U19247 | Gamma interferon receptor alpha chain | 2.7 | 1.0 | |
| IL18R1 | U43672 | Putative transmembrane receptor IL-1Rrp | 2.3 | - |
| F2RL1 | U34038 | Proteinase-activated receptor 2 | 2.2 | 1.4 |
| Adhesion molecules | ||||
| SELE | M24736 | Endothelial leukocyte adhesion molecule 1 | 27.8 | 2.4 |
| VCAM1 | M73255 | Vascular cell adhesion molecule 1 | 20.3 | 1.7 |
| VCAM1 | M30257 | Vascular cell adhesion molecule 1 | 19.6 | 0.9 |
| ICAM1 | M24283 | Human major group rhinovirus receptor | 12.6 | 1.4 |
| Vitronectin | M14648 | Vitronectin receptor alpha | 2.2 | 1.7 |
| Enzymes | ||||
| CYP2J2 | U37143 | Cytochrome P450 monooxygenase | 5.2 | 0.9 |
| F3 | J02931 | Placental tissue factor | 5.2 | 0.6 |
| hCox-2 | U04636 | Cyclooxygenase-2 | 5.0 | 1.9 |
| SERPINB2 | Y00630 | Plasminogen activator-inhibitor 2, PAI-2 | 3.7 | 1.7 |
| PPAP2B | AF017786 | Phosphatidic acid phosphohydrolase homolog (Dri42) | 2.0 | 0.9 |
| Angiogenesis growth factors | ||||
| FGF5 | M37825 | Fibroblast growth factor 5 | 3.3 | 1.4 |
| EGF | M60278 | Epidermal growth factor-like growth factor | 3.2 | 1.1 |
| Apoptosis and cell death | ||||
| TNFAIP3 | M59465 | TNF-α inducible protein A20 | 22.2 | 1.4 |
| BIRC3 | U45878 | Human inhibitor of apoptosis protein 1 | 12.2 | 1.4 |
| BCL2A1 | U27467 | Human Bcl-2 related (Bfl-1) | 7.5 | 2.1 |
| TNFAIP2 | M92357 | B94 protein | 6.9 | 1.0 |
| RICK | AF117829 | 8q21.3-RICK gene | 5.7 | - |
| TRAF1 | U19261 | Epstein-Barr virus-induced protein | 5.6 | - |
| GG2-1 | AF099935 | MDC-3.13 isoform 2 | 2.5 | 1.0 |
| TNFSF10 | U37518 | TNF-related apoptosis inducing ligand TRAIL | 2.3 | 1.3 |
| CFLAR | AF005775 | Caspase-like apoptosis regulatory protein 2 | 2.1 | 1.5 |
| TANK | U59863 | TRAF-interacting protein 1-TRAF | 2.2 | 1.4 |
| Nucleic acid binding | ||||
| IRF1 | L05072 | interferon regulatory factor 1 | 6.7 | - |
| ATF3 | L19871 | activating transcription factor 3 | 6.0 | - |
| MAD-3 | M69043 | MAD-3 mRNA encoding IκB-like activity | 5.6 | 1.0 |
| I-REL | M83221 | I-Rel | 5.6 | 1.6 |
| CEBPD | M83667 | NF-IL6-beta protein | 5.3 | 0.8 |
| Jun-B | X51345 | jun-B | 4.7 | 1.6 |
| p50-NF-κB | S76638 | Nuclear factor of κ light polypeptide gene enhancer | 4.3 | 1.1 |
| JUNB | M29039 | Transactivator jun-B | 4.2 | 0.8 |
| ELL2 | U88629 | RNA polymerase II elongation factor ELL2 | 3.0 | 1.7 |
| ARHB | M12174 | Ras-related rho | 2.8 | 1.3 |
| ELL2 | U88629 | RNA polymerase II elongation factor ELL2 | 2.8 | 1.8 |
| AREB6 | D15050 | Transcription factor AREB6 | 2.8 | 1.3 |
| NFKBIE | U91616 | I kappa B epsilon (IκBe) | 2.7 | 1.4 |
| PSMA1 | M64992 | P30-33K | 2.3 | 2.1 |
| NFKB1 | M58603 | NF-κB DNA-binding subunit | 2.3 | 1.1 |
| NFKB2 | X61498 | NF-κB subunit | 2.2 | - |
| NFKB1 | M58603 | NF-κB | 2.1 | 0.9 |
| Others | ||||
| CD69 | Z22576 | CD69 | 14.1 | - |
| RHO6 | U69563 | Homo sapienscDNA | 10.0 | 1.8 |
| Diubiquitin | AL031983 | Diubiquitin | 8.8 | - |
| B61 | M57730 | Human B61 | 6.9 | 0.6 |
| CIG49 | AF026939 | CIG49 | 3.9 | - |
| HDAC9 | AB018287 | KIAA0744 | 3.5 | - |
| Pending | AL023584 | HIV type 1 enhancer-binding protein 2 | 3.4 | 1.2 |
| KIAA0277 | D87467 | KIAA0277 | 3.1 | 1.0 |
| ARFGAP1 | AA402332 | cDNA, 3′ end | 3.0 | 2.2 |
| ARHGEF11 | AB002378 | KIAA0380 | 2.4 | 2.2 |
| N143 | AJ002572 | transcriptional unit N143 | 2.2 | 1.9 |
| MSX1 | M97676 | homeobox protein region 7 (HOX7) | 2.1 | 1.3 |
| MT2A | AI547258 | Homo sapiens cDNA, 5′ end | 2.0 | 1.0 |
P. gingivalis 381 (wild-type) or DPG3 (fimA mutant) were added to HAEC cultures at an MOI of 100, followed by incubation at 37°C for 1 h. The nonadherent bacteria were removed by washing, and HAEC infected with P. gingivalis were cultured in fresh culture medium for an additional 5 h. When the total incubation period reached 6 h after P. gingivalis infection, HAEC were harvested, and total RNA was extracted and analyzed by using microarray as described in Materials and Methods. Control cultures were incubated with culture medium only. Genes whose expression in P. gingivalis 381-infected HAEC were at least twofold higher than those in uninfected HAEC are listed. Gene names in boldface designate genes whose expression in P. gingivalis 381-infected HAEC were at least twofold higher than in DPG3-infected HAEC cultures. Gene names in italics designate genes whose expression in P. gingivalis 381-infected and DPG3-infected HAEC cultures was at least twofold higher than those in uninfected HAEC. -, The call for the gene in DPG3-infected HAEC was absent or undetectable.
Wt/Ctl and DPG3/Ctl, ratios of wild-type to control and DPG3 to control, respectively.
It should be noted that the majority of genes expressed by HAEC under these experimental conditions were not affected by P. gingivalis infection, indicating that the endothelial cell mRNA response to P. gingivalis 381 infection was specific and relatively well defined. We also observed in P. gingivalis 381-infected HAEC the downregulation of 16 genes compared to uninfected HAEC control (Table 3). These included genes encoding cellular receptors expressed in stress conditions and genes involved in cell cycle control. The range of the transcript ratio for downregulated genes was observed to be between 0.2 and 1.0.
Fimbria-mediated invasion is required to stimulate inflammatory gene expression in HAEC.
To determine whether fimbria-mediated invasion was required for the observed response that was seen after infection of HAEC with invasive P. gingivalis, we next examined the transcriptional profile of HAEC in response to a noninvasive P. gingivalis strain. Invasion efficiencies of 0.0144 and 0.0001% were observed after infection of HAEC with P. gingivalis 381 and the DPG3, respectively (data not shown). The invasion efficiencies for strains 381 and DPG3 were in agreement with our previous studies (5). Among the 68 genes induced after infection with the wild-type P. gingivalis, only four genes (SCYA2, gamma interferon treatment-inducible mRNA; SELE, endothelial leukocyte adhesion molecule 1; PSMA1, prosomal protein P30-33K; and KIAA0380) were also upregulated in P. gingivalis DPG3-infected HAEC (Table 2). Among the downregulated genes in P. gingivalis 381-infected HAEC, only three genes (cell cycle control gene CDC2, KIAA0943, and POLR2K) were also found to be downregulated in DPG3-infected HAEC (Table 3), although these differences were not as apparent as the differences observed in the upregulated genes. These results indicated that P. gingivalis fimbria-mediated invasion is crucial for the stimulation of inflammatory gene expression in HAEC.
Expression of adhesion molecules and cytokines in P. gingivalis-infected HAEC.
To confirm the data obtained with human arrays and to further investigate the role of invasive P. gingivalis infection of the endothelium and its putative atherosclerotic changes, we selected a panel of inflammatory genes previously shown to play essential roles in the pathobiology of atherosclerosis (14, 33, 37). These genes include the chemokines GRO2, GRO3, and IL-8; the cell adhesion molecules ICAM-1, VCAM-1, and E-selectin; and the inflammatory molecules IL-6 and COX-2. RT-PCR analysis confirmed the upregulation of these genes representing inflammatory molecules previously reported to be involved in the initial process of an atherosclerotic lesion (Fig. 1). As observed by microarray analysis, these genes were not upregulated in HAEC infected with the P. gingivalis DPG3 but were expressed in HAEC infected with P. gingivalis 381. These genes were upregulated at 6 h and also at 24 h in HAEC infected with P. gingivalis 381 but not upregulated in HAEC infected with the DPG3, with the exception of the SELE (E-selectin) gene. Microarray analysis indicated that SELE was the only gene upregulated in both P. gingivalis 381-infected HAEC and P. gingivalis DPG3-infected HAEC at 6 h.
FIG. 1.
RT-PCR analysis of inflammatory genes subsets expressed by HAEC in response to P. gingivalis infection. P. gingivalis 381 (wild type) or DPG3 (fimA mutant) were added to HAEC cultures at an MOI of 100 and then incubated at 37°C for 1 h. Nonadherent bacteria were removed by washing, and HAEC infected with P. gingivalis were either used at this point or cultured in fresh culture medium for an additional 5 or 23 h. When the total incubation period reached 6 h, or 24 h post-P. gingivalis infection, HAEC were harvested, and total RNA was extracted for mRNA expression analysis by using RT-PCR. Control cultures were incubated with culture medium only. The results are representative of three independent experiments: 1 h, HAEC were harvested 1 h post-P. gingivalis infection; 6 h, HAEC were infected with P. gingivalis for 1 h, and nonadherent bacteria were then removed by washing and were subsequently cultured in fresh culture medium for an additional 5 h; 24 h, HAEC were infected with P. gingivalis for 1 h, and nonadherent bacteria were then removed by washing and subsequently cultured in fresh culture medium for additional 23 h. Lanes: −, uninfected HAEC; 381, wild-type P. gingivalis; DPG3, fimA mutant. GAPDH was used as a control gene.
We next determined cell adhesion molecule expression by flow cytometry (for gene products expressed on the cell membrane) and IL-6 and IL-8 expression by ELISA (for gene products secreted into the culture supernatants). IL-6 and IL-8 levels in P. gingivalis 381-infected HAEC were not increased at 1 h, but the production of IL-6 and IL-8 increased significantly at 6 h after infection and was continually expressed 24 h after initial stimulation (Fig. 2). These increased levels were significantly higher than those for P. gingivalis DPG3-infected HAEC at 6 and 24 h postinfection (Fig. 2).
FIG. 2.
IL-6 and IL-8 production by HAEC in response to P. gingivalis infection. P. gingivalis 381 (wild-type) or DPG3 (fimA mutant) were added to HAEC cultures at an MOI of 100 and incubated at 37°C for 1 h. Supernatants were harvested for IL-6 and IL-8 ELISA analysis. For the 6- and 24-h experiments, nonadherent bacteria were removed by washing at 1 h, and HAEC were cultured in fresh culture medium for an additional 5 or 23 h. When the total incubation period reached 6 h, or 24 h post-P. gingivalis infection, supernatants were harvested for IL-6 and IL-8 ELISA analysis. Control cultures were incubated with culture media only. The right panel depicts the IL-6 or IL-8 ELISA results at 1 h post-P. gingivalis infection. Representative findings are shown as mean and SDs from three independent experiments. 1 h, HAEC were harvested 1 h post-P. gingivalis infection; 6 h, HAEC were infected with P. gingivalis for 1 h, and nonadherent bacteria were then removed by washing and subsequently cultured in fresh culture medium for an additional 5 h; 24 h, HAEC were infected with P. gingivalis for 1 h, and nonadherent bacteria were then removed by washing and were subsequently cultured in fresh culture medium for an additional 23 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001 [compared to control], #, P < 0.01; ##, P < 0.001 [compared to DPG3].
We next examined the protein expression of cell adhesion molecules that have been demonstrated to be involved in adhesion, rolling, and transmigration of lymphocytes into the vascular endothelial lining. The expression of ICAM-1, VCAM-1, E-selectin, and P-selectin were all upregulated at 1 and 6 h postinfection with invasive P. gingivalis (Fig. 3); however, at 24 h, only ICAM-1 and VCAM-1 expression remained elevated on the HAEC cell surface, whereas E-selectin and P-selectin expression were similar to that observed in uninfected HAEC. The addition of heat-killed P. gingivalis 381 did not result in stimulation of adhesion molecule expression on HAEC, nor did the addition of DPG3 (Fig. 3). These results indicate that stimulation of adhesion molecules on HAEC by P. gingivalis requires live, fimbriated bacteria.
FIG. 3.
ICAM-1, VCAM-1, E-/P-selectin, and P-selectin expression on HAEC in response to P. gingivalis infection. P. gingivalis 381 (wild-type), DPG3 (fimA- mutant), or heat-killed P. gingivalis 381 were added to HAEC cultures at an MOI of 100, followed by incubation at 37°C for 1 h, and nonadherent bacteria were removed by washing. HAEC infected with P. gingivalis were either fixed at this point or were cultured in fresh culture medium for an additional 5 h or 23 h. When the total incubation period reached 6 or 24 h post-P. gingivalis infection, HAEC were harvested and analyzed for ICAM-1, VCAM-1, E-/P-selectin, or P-selectin by FACS. Control cultures were incubated with culture medium only. These results are representative of four independent experiments. 1 h, HAEC were harvested 1 h post-P. gingivalis infection; 6 h, HAEC were infected with P. gingivalis for 1 h, and nonadherent bacteria were then removed by washing and cultured in fresh culture medium for an additional 5 h; 24 h, HAEC were infected with P. gingivalis for 1 h, and nonadherent bacteria were then removed by washing and cultured in fresh culture medium for an additional 23 h.
Characterization of ICAM-1 and VCAM-1 expression in aortic arch tissue after infection with P. gingivalis.
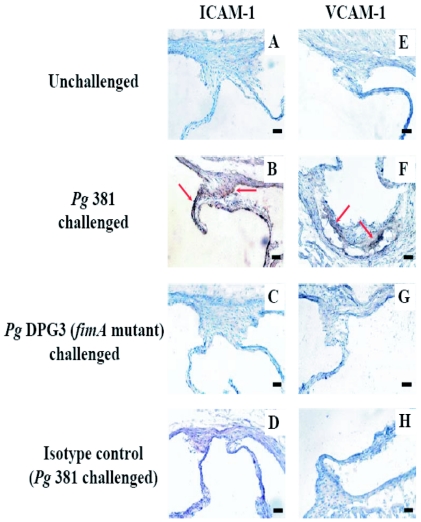
To address the biologic significance of the microarray data, we next determined whether oral challenge of ApoE−/− mice with P. gingivalis stimulated cellular adhesion molecule expression directly in aortic tissue. We have previously determined that after oral challenge with P. gingivalis the organism can be detected in the bloodstream and in aortic tissue by PCR (11). Likewise, we have previously determined that at 6 weeks after oral challenge ApoE−/− mice challenged with P. gingivalis 381-accelerated atherosclerosis. In the present study, P. gingivalis was also detected in aortic tissue by PCR at 24 h after final oral challenge (data not shown). At 6 weeks after oral challenge, immunohistochemical analysis revealed elevated levels of ICAM-1 and VCAM-1 in representative plaque lesions in aortic tissue obtained from ApoE−/− mice orally challenged with invasive P. gingivalis (Fig. 4). Sections of aortic arch tissue obtained from unchallenged ApoE−/− mice and ApoE−/− mice that were challenged with the noninvasive P. gingivalis expressed low levels of ICAM-1 and VCAM-1 (Fig. 4). The expression of ICAM-1 and VCAM-1 in plaque samples obtained at 6 weeks after the final oral challenge correlated with plaque accumulation in the respective groups of mice as we reported previously (11; data not shown). These results demonstrate that ApoE−/− mice orally challenged with invasive P. gingivalis present with an inflammatory response characterized by increased expression of cell adhesion molecules and that this response correlates with the later stages of atheroma development.
FIG. 4.
Expression of ICAM-1 and VCAM-1 in the aortic sinus from ApoE−/− mice orally challenged with P. gingivalis was confirmed at 6 weeks after challenge. The aortic sinus cross-sections (n = 6 for each group) of mice without challenge (unchallenged; A and E), mice challenged with invasive P. gingivalis 381 (B, D, F, and H), or mice challenged with noninvasive P. gingivalis fimA mutant DPG3 (C and G) were isolated and single immunoenzyme stained as described in Materials and Methods. Representative images stained with anti-ICAM-1 (A to C), anti-VCAM-1 (E, F, and G), and isotype-matched control (D and H) antibodies were shown. The sections were counterstained with hematoxylin. Red arrows point out indicated marker-positive stained areas. Scale bar, 50 μm.
DISCUSSION
The initial step of bacterial adherence and invasion to host cells typically requires the bacterial surface component fimbriae (31). P. gingivalis had been shown to invade bovine heart and aortic endothelial cells, and the expression of fimbriae is essential for this process (5, 31). Moreover, we have previously demonstrated that invasive strains of P. gingivalis, but not a noninvasive fimA mutant, stimulate the expression of cell adhesion molecules on the cell surface of HUVEC (21) and that P. gingivalis can modulate the expression of chemokines in HUVEC, through a fimbria-mediated mechanism (29). Our initial studies focused on the expression of a subset of endothelial cell genes in response to invasive bacterial infection. Since it has been reported that endothelial cells obtained from different anatomic sites do not react similarly (28), we utilized DNA microarray analysis to characterize the primary responses of HAEC, a more relevant cell type to atherosclerosis progression, to P. gingivalis. In the present study, using human DNA microarrays, we have further defined the overall pattern of gene expression in HAEC after 1 h infection with invasive, fimbriated P. gingivalis at an MOI of 100. The transcription profiles of HAEC infected with either invasive or noninvasive P. gingivalis revealed that the primary response of HAEC to invasive P. gingivalis was specifically linked to genes involved in inflammatory and atherogenic responses. The genes that were upregulated in HAEC, in response to internalized P. gingivalis 381, were primarily genes encoding chemokines, adhesion molecules and enzymes involved in inflammation. These results are consistent with our previous studies with HUVEC (21, 29), which showed that infection of HUVEC with invasive P. gingivalis 381 stimulates transcription of chemokines and cytokines. Although differences in HUVEC versus HAEC were observed with regard to the magnitude of the IL-8 response, this is most likely due to the different anatomic sites from which these cells were obtained, as well as to the fact that the HUVEC represent an immortalized cell line, whereas the HAEC are primary cells. Growth factors involved in angiogenesis were also found among the group of chemokine genes upregulated in response to wild-type P. gingivalis challenge. Notably, most of these upregulated genes have been reported to be associated with atherosclerotic changes of the endothelium (14, 33, 37). Since many pathophysiological studies have demonstrated that inflammation of the endothelium may be a key factor involved in the initiation and progression of atherosclerosis (36), the results of the present study support the hypothesis that P. gingivalis invasion in HAEC can lead to inflammatory and potentially atherosclerotic changes. In further support of these in vitro results, we have recently reported that only invasive P. gingivalis, but not noninvasive P. gingivalis, accelerates atherosclerotic lesion formation in an ApoE−/− mouse model of atherosclerosis (11).
Only 68 genes, among a total of 10,000 genes examined in HAEC infected with invasive P. gingivalis 381, were upregulated after infection. Most of these genes encode chemokine family members and adhesion molecules that are known to be involved in lymphocyte recruitment and trafficking to sites of vascular inflammation. These findings support our studies, as well as other reports involving the interactions of other infectious pathogens, such as cytomegalovirus and Chlamydia pneumoniae, with endothelial cells (17, 34). Both human epidemiological studies and animal studies have implicated C. pneumoniae, a common respiratory pathogen, as an additional risk factor for the development and acceleration of atherosclerotic lesions. In vitro studies have also demonstrated that C. pneumoniae infection of vascular endothelial cells can induce the expression of many molecules that are important mediators of atherosclerosis (17), including cytokines, adhesion molecules, and chemokines associated with activation of NF-κB, as well as molecules with procoagulant activity, and those which promote the oxidation of low-density lipoprotein (3, 7, 8, 19, 22, 27, 39). In the study of Coombes and Mahony (3), cDNA arrays were used to characterize the mRNA expression profile for 268 human genes after infection of the human microvascular endothelial cell line HMEC-1 with C. pneumoniae. In agreement with our results, these authors also observed that genes encoding IL-8, epidermal growth factor, fibroblast growth factor, and the alpha interferon receptor were upregulated after infection with C. pneumoniae (3).
Studies of the molecular signals that regulate the trafficking of leukocytes to sites of atherosclerotic lesions have recently focused on chemokines. IL-8 has been observed to be elevated in the serum of patients with acute coronary syndromes and increased in human atheroma-associated cells such as monocytes or monocyte-derived macrophages compared to normal vessels (35, 41). Although IL-8 is thought to act predominantly on neutrophils, recent reports have demonstrated that IL-8 can rapidly cause rolling monocytes to adhere firmly to endothelial monolayers expressing E-selectin under flow conditions mimicking a vascular model (25). Chemokines have also been shown to mediate endothelial cell chemotactic and proliferative activities and stimulate angiogenesis (2).
In addition to the elevated levels of IL-8 in HAEC infected with invasive P. gingivalis, we also observed increased expression of the adhesion molecules ICAM-1, VCAM-1, and E-selectin. The accumulation of blood-borne leukocytes within inflamed atherosclerotic tissues in response to antigenic stimulation is a major step in the progression of atherosclerosis (32). Our results demonstrate that the molecules involved in the initial process of leukocyte binding to the activated endothelium, such as IL-8, in addition to several Glu-Lys-Arg (ELR)+ chemokine and cell adhesion molecules ICAM-1, VCAM-1, P- and E-selectins (4, 20, 32, 33) were all upregulated in response to invasive bacterial infection. In the present study, we demonstrated that only invasive P. gingivalis induced the expression of ICAM-1 and VCAM-1 on the surface of HAEC. These results were further confirmed in vivo by using an ApoE−/− mouse model of atherosclerosis. In these studies we demonstrated that oral infection with invasive P. gingivalis resulted in increased expression of ICAM-1 and VCAM-1 in aortic tissue. Furthermore, mice orally infected with noninvasive P. gingivalis did not present with expression of ICAM-1 or VCAM-1 in aortic tissue.
Apoptosis or programmed cell death is considered to be an important event in the development of atherosclerosis (40), since many studies have identified increased apoptosis of vascular cells in atherosclerotic plaques compared to normal vessels (10, 12, 16, 38). However, the exact mechanisms and consequences of apoptosis in the development and progression of atherosclerosis are still controversial (38). In the present study, both apoptosis-inducing genes and antiapoptosis factor genes were upregulated by wild-type P. gingivalis infection. Previous reports demonstrated that stimulation of human endothelial cells with TNF-α directly results in both pro- and antiapoptotic signals; TNF-α-induced apoptosis of endothelial cells is mediated, in part, by the degradation of Bcl-2 and the activation of caspase-3 (6), and TNF-α is also capable of protecting against apoptosis acting through the transcription factor NF-κB via the induction of A1, a Bcl-2 homologue (18). This gene (BclA1; Bcl-related Bfl1) was upregulated in P. gingivalis-infected HAEC, and this indicates that P. gingivalis infection may initiate divergent survival and death pathways in HAEC.
Interestingly, only 16 and 68 genes, among a total of 10,000 genes examined in HAEC infected with invasive P. gingivalis 381, were also downregulated and upregulated after infection, respectively. These numbers of regulated genes indicate that the endothelial cell transcription profile of mRNA expression response to P. gingivalis 381 infection was specific and relatively well defined. Little is known regarding gene downregulation during endothelial cell infection by bacteria or the impact of this on the development of atherosclerosis. We did observe that several HAEC genes were downregulated during wild-type P. gingivalis challenge; however, the importance of these observations to the overall inflammatory response of HAEC to infection requires further investigation.
Collectively, our findings indicate that fimbria-dependent adherence and invasion of P. gingivalis into HAEC induces the expression of genes involved in inflammatory and atherogenic responses by HAEC. Furthermore, these results were confirmed by using an ApoE−/− mouse model of atherosclerosis after challenge with invasive P. gingivalis. Since the vascular endothelium is essential for the recruitment of leukocytes during atherogenesis, studies aimed at the inflammatory activation of endothelial cells by P. gingivalis may begin to elucidate the mechanisms of infection-accelerated atherosclerosis and provide potential targets for therapeutic intervention.
Acknowledgments
This study was supported by Public Health Service grant PO1DE13191 from the National Institute of Dental and Craniofacial Research to C.A.G. and in part, by National Science Council grant NSC 91-2314-B-038-011 to H.-H.C. (Taiwan).
Editor: V. J. DiRita
REFERENCES
- 1.Beck, J., R. Garcia, G. Heiss, P. S. Vokonas, and S. Offenbacher. 1996. Periodontal disease and cardiovascular disease. J. Periodontol. 67:1123-1137. [DOI] [PubMed] [Google Scholar]
- 2.Belperio, J. A., M. P. Keane, D. A. Arenberg, C. L. Addison, J. E. Ehlert, M. D. Burdick, and R. M. Strieter. 2000. CXC chemokines in angiogenesis. J. Leukoc. Biol. 68:1-8. [PubMed] [Google Scholar]
- 3.Coombes, B. K., and J. B. Mahony. 2001. cDNA array analysis of altered gene expression in human endothelial cells in response to Chlamydia pneumoniae infection. Infect. Immun. 69:1420-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies, M., J. Gordon, A. J., A. J. H. Gearing, R. Pigott, N. Woolf, D. Katz, and A. Kyriakopoulos. 1993. The expression of adhesion molecules ICAM-1, PECAM, and E-selectin in human atherosclerosis. J. Pathol. 171:223-229. [DOI] [PubMed] [Google Scholar]
- 5.Deshpande, R. G., M. B. Khan, and G. C. A. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimmeler, S., K. Breitschopf, J. Haendeler, and A. M. Zeiher. 1999. Dephosphorylation targets Bcl-2 for ubiquitin-dependent degradation: a link between the apoptosome and the proteasome pathway. J. Exp. Med. 189:1815-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dittrich, R., C. Dragonas, A. Mueller, T. Maltaris, J. Rupp, M. W. Beckmann, and M. Maass. 2004. Endothelial Chlamydia pneumoniae infection promotes oxidation of LDL. Biochem. Biophys. Res. Commun. 319:501-505. [DOI] [PubMed] [Google Scholar]
- 8.Fryer, R. H., E. P. Schwobe, M. L. Woods, and G. M. Rodgers. 1997. Chlamydia species infect human vascular endothelial cells and induce procoagulant activity. J. Investig. Med. 45:168-174. [PubMed] [Google Scholar]
- 9.Genco, R. J. 1998. Periodontal disease and risk for myocardial infarction and cardiovascular disease. Cardiovasc. Rev. Rep. 19:34-40. [Google Scholar]
- 10.Geng, Y. J., and P. Libby. 1995. Evidence for apoptosis in advanced human atheroma: colocalization with interleukin-1β-converting enzyme. Am. J. Pathol. 147:251-266. [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson III, F. C., C. Hong, H.-H. Chou, H. Yumoto, J. Chen, E. Lien, J. Wong, and C. A. Genco. 2004. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation 109:2801-2806. [DOI] [PubMed] [Google Scholar]
- 12.Han, D. K., C. C. Haudenschild, M. K. Hong, B. T. Tinkle, M. B. Leon, and G. Liau. 1995. Evidence for apoptosis in human atherogenesis and in a rat vascular injury model. Am. J. Pathol. 147:267-277. [PMC free article] [PubMed] [Google Scholar]
- 13.Haraszthy, V. I., J. J. Zambon, M. Trevisan, M. Zeid, and R. J. Genco. 2000. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 71:1554-1560. [DOI] [PubMed] [Google Scholar]
- 14.Holm, T., J. K. Damas, K. Holven, I. Nordoy, F. R. U. Brosstad, T., T. Wahre, J. Kjekshus, S. S. Froland, H. G. Eiken, N. O. Solum, L. Gullestad, M. Nenseter, and P. Aukrust. 2003. CXC-chemokines in coronary artery disease: possible pathogenic role of interactions between oxidized low-density lipoprotein, platelets, and peripheral blood mononuclear cells. J. Thromb. Haemost. 1:257-262. [DOI] [PubMed] [Google Scholar]
- 15.Holt, S. C., J. Ebersole, J. Felton, M. Brunsvold, and K. S. Kornman. 1988. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science 239:55-57. [DOI] [PubMed] [Google Scholar]
- 16.Isner, J. M., M. Kearney, S. Bortman, and J. Passeri. 1995. Apoptosis in human atherosclerosis and restenosis. Circulation 91:2703-2711. [DOI] [PubMed] [Google Scholar]
- 17.Kalayoglu, M. V., P. Libby, and G. I. Byrne. 2002. Chlamydia pneumoniae as an emerging risk factor in cardiovascular disease. JAMA 288:2724-2731. [DOI] [PubMed] [Google Scholar]
- 18.Karsan, A., E. Yee, and J. M. Harlan. 1996. Endothelial cell death induced by tumor necrosis factor-alpha is inhibited by the Bcl-2 family member, A1. J. Biol. Chem. 271:27201-27204. [DOI] [PubMed] [Google Scholar]
- 19.Kaukoranta-Tolvanen, S. S., T. Ronni, M. Leinonen, P. Saikku, and K. Laitinen. 1996. Expression of adhesion molecules on endothelial cells stimulated by Chlamydia pneumoniae. Microb. Pathog. 21:407-411. [DOI] [PubMed] [Google Scholar]
- 20.Kerr, J. R. 1999. Cell adhesion molecules in the pathogenesis of and host defense against microbial infection. J. Clin. Pathol. Mol. Pathol. 52:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khlgatian, M., H. Nassar, H.-H. Chou, F. C. Gibson., and C. A. Genco. 2002. Fimbria-dependent activation of cell adhesion molecule expression in Porphyromonas gingivalis-infected endothelial cells. Infect. Immun. 70:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krull, M., A. C. Klucken, F. N. Wuppermann, O. Fuhrmann, C. Magerl, J. Seybold, S. Hippenstiel, J. H. Hegemann, C. A. Jantos, and N. Suttorp. 1999. Signal transduction pathways activated in endothelial cells following infection with Chlamydia pneumoniae. J. Immunol. 162:4834-4841. [PubMed] [Google Scholar]
- 23.Lalla, E., I. B. Lamster, M. A. Hofmann, L. Bucciarelli, A. P. Jerud, S. Tucker, Y. Lu, P. N. Papapanou, and A. M. Schmidt. 2003. Oral infection with a periodontal pathogen accelerates early atherosclerosis in apolipoprotein E-null mice. Arterioscler. Thromb. Vasc. Biol. 23:1405-1411. [DOI] [PubMed] [Google Scholar]
- 24.Li, L., E. Messas, L. E. Batista, R. A. Levine, and S. Amar. 2002. Porphyromonas gingivalis infection accelerates the progression of atherosclerosis in a heterozygous apolipoprotein E-deficient murine model. Circulation 105:861-867. [DOI] [PubMed] [Google Scholar]
- 25.Luscinskas, F. W., R. E. Gerszten, E. A. Garcia-Zepeda, Y. C. Lim, M. Yoshida, H. A. Ding, M. A. J. Gimbrone, A. D. Luster, and A. Rosenzweig. 2000. C-C and C-X-C chemokines trigger firm adhesion of monocytes to vascular endothelium under flow conditions. Ann. N. Y. Acad. Sci. 902:288-293. [DOI] [PubMed] [Google Scholar]
- 26.Maleck, R., A. Fisher, M. Caleca, C. Stinson, J. Van Oss, J.-Y. Lee, M.-I. Cho, R. J. Genco, E. R. T., and D. W. Dyer. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molestina, R. E., D. Dean, R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1998. Characterization of a strain of Chlamydia pneumoniae isolated from a coronary atheroma by analysis of the omp1 gene and biological activity in human endothelial cells. Infect. Immun. 66:1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris, T. E., P. A. Mattox, G. D. Shipley, C. R. Wagner, and J. D. Hosenpud. 1993. The pattern of cytokine messenger RNA expression in human aortic endothelial cells is different from that of human umbilical vein endothelial cells. Transpl. Immunol. 1:137-142. [DOI] [PubMed] [Google Scholar]
- 29.Nassar, H., H.-H. Chou, K. M., F. C. Gibson., T. E. Van Dyke, and C. A. Genco. 2002. Role for fimbriae and lysine-specific cysteine proteinase gingipain K in expression of interleukin-8 and monocyte chemoattractant protein in Porphyromonas gingivalis-infected endothelial cells. Infect. Immun. 70:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelken, N. A., S. J. Soifer, J. O'keefe, T. K. Vu, I. F. Charo, and S. R. Coughlin. 1992. Thrombin receptor expression in normal and atherosclerotic human arteries. J. Clin. Investig. 90:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Njoroge, T. G., R. J. Sojar, H. T. Hamada, N., and C. A. Genco. 1997. A role of fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noll, G. 1998. Pathogenesis of atherosclerosis: a possible relation to infection. Atherosclerosis 140(Suppl.):S3-S9. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien, K. D., M. D. Allen, and T. O. McDonald. 1993. Vascular cell adhesion molecule-1 is expressed in human coronary atherosclerotic plaques: implications for the mode of progression of advanced coronary atherosclerosis. J. Clin. Investig. 92:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricotta, D., G. Alessandri, C. Pollara, S. Fiorentini, F. Favilli, M. Tosetti, A. Mantovani, M. Grassi, E. Garrafa, L. Dei Cas, C. Muneretto, and A. Caruso. 2001. Adult human heart microvascular endothelial cells are permissive for non-lytic infection by human cytomegalovirus. Cardiovasc. Res. 49:440-448. [DOI] [PubMed] [Google Scholar]
- 35.Romuk, E., B. Skrzep-Poloczek, C. Wojciechowska, A. Tomasik, E. Birkner, J. Wodniecki, B. Gabrylewicz, A. Ochala, and M. Tendera. 2002. Selectin-P and interleukin-8 plasma levels in coronary heart disease patients. Eur. J. Clin. Investig. 32:657-661. [DOI] [PubMed] [Google Scholar]
- 36.Ross, R. 1999. Atherosclerosis-an inflammatory disease. N. Engl. J. Med. 340:115-126. [DOI] [PubMed] [Google Scholar]
- 37.Scalia, R., J. Z. Appel, and A. M. Lefer. 1998. Leukocyte-endothelium interaction during the early stages of hypercholesterolemia in the rabbit: role of P-selectin, ICAM-1, and VCAM-1. Thromb. Vasc. Biol. 18:1093-1100. [DOI] [PubMed] [Google Scholar]
- 38.Stoneman, V. E., and M. R. Bennett. 2004. Role of apoptosis in atherosclerosis and its therapeutic implications. Clin. Sci. 107:343-354. [DOI] [PubMed] [Google Scholar]
- 39.Summersgill, J. T., R. E. Molestina, R. D. Miller, and J. A. Ramirez. 2000. Interactions of Chlamydia pneumoniae with human endothelial cells. J. Infect. Dis. 181(Suppl. 3):S479-S482. [DOI] [PubMed] [Google Scholar]
- 40.Vink, A., D. P. de Kleijn, and G. Pasterkamp. 2004. Functional role for Toll-like receptors in atherosclerosis and arterial remodeling. Curr. Opin. Lipidol. 15:515-521. [DOI] [PubMed] [Google Scholar]
- 41.Wang, N., I. Tabas, R. Winchester, S. Ravalli, L. E. Rabbani, and A. Tall. 1996. Interleukin 8 is induced by cholesterol loading of macrophages and expressed by macrophage foam cells in human atheroma. J. Biol. Chem. 271:8837-8842. [DOI] [PubMed] [Google Scholar]