Abstract
The cyclic dinucleotide second messenger cyclic diguanylate (c-diGMP) has been implicated in regulation of cell surface properties in several bacterial species, including Vibrio cholerae. Expression of genes required for V. cholerae biofilm formation is activated by an increased intracellular c-diGMP concentration. The response regulator VieA, which contains a domain responsible for degradation of c-diGMP, is required to maintain a low concentration of c-diGMP and repress biofilm formation. The VieSAB three-component signal transduction system was, however, originally identified as a regulator of ctxAB, the genes encoding cholera toxin (CT). Here we show that the c-diGMP phosphodiesterase activity of VieA is required to enhance CT production. This regulation occurred at the transcriptional level, and ectopically altering the c-diGMP concentration by expression of diguanylate cyclase or phosphodiesterase enzymes also affected ctxAB transcription. The c-diGMP phosphodiesterase activity of VieA was also required for maximal transcription toxT but did not influence the activity of ToxR or expression of TcpP. Finally, a single amino acid substitution in VieA that increases the intracellular c-diGMP concentration led to attenuation in the infant mouse model of cholera. Since virulence genes including toxT and ctxA are repressed by a high concentration of c-diGMP, while biofilm genes are activated, we suggest that c-diGMP signaling is important for the transition of V. cholerae from the environment to the host.
Cyclic diguanylate (c-diGMP) [bis(3′,5′)-cyclic diguanylic acid] is a prokaryotic cyclic dinucleotide second messenger that has been implicated in controlling properties of the cell surface in diverse bacterial species (8, 23). c-diGMP was originally characterized in Gluconacetobacter xylinus (formerly Acetobacter xylinum), an organism that synthesizes a cellulose extracellular matrix (44, 45). In G. xylinus, the c-diGMP concentration is controlled by six proteins that have competing diguanylate cyclase or phosphodiesterase activities to synthesize and degrade c-diGMP, respectively (49). Each of these cyclase and phosphodiesterase proteins may be activated by environmental signals that serve as the first messenger; for example, the G. xylinus PDEA1 phosphodiesterase is responsive to oxygen (6). The c-diGMP second messenger then controls production of the cellulose matrix by allosterically activating the cellulose synthase enzyme (54). The protein domains responsible for synthesis and degradation of c-diGMP, named GGDEF and EAL, respectively, for their conserved amino acid residues, are widespread in bacterial genomes (13). There is evidence that proteins with the GGDEF and EAL domains control production of extracellular polysaccharide matrices in several bacterial species, including Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Yersinia pestis, and Vibrio cholerae (12, 14, 25, 48, 51).
In V. cholerae, the causative agent of the severe diarrheal disease cholera, c-diGMP concentration affects transcription of Vibrio exopolysaccharide synthesis (vps) genes that are required for biofilm formation (51). The ability of V. cholerae to form biofilms may contribute to persistence of the organism in environmental reservoirs between epidemics, since bacteria within a biofilm are more resistant to chemical stresses (53, 55). Although the mechanism has not yet been elucidated, increased concentration of the c-diGMP second messenger activates vps gene expression. The response regulator VieA contains an EAL domain that is a putative c-diGMP phosphodiesterase and that is required to maintain a low level of c-diGMP under growth conditions that do not favor biofilm formation (51). Mutations at conserved residues in the VieA EAL domain result in an elevated c-diGMP concentration and activation of vps transcription. In contrast, the VieA phosphoreceiver and helix-turn-helix domains are not necessary for regulation of vps transcription (51).
Although VieA negatively regulates expression of genes required for biofilm formation, it was originally identified as a putative positive regulator of virulence genes. In humans, V. cholerae causes disease by colonizing the small intestine and secreting cholera toxin (CT), an ADP-ribosylating toxin that elicits profuse watery diarrhea (24). Expression of CT and the toxin-coregulated pilus (TCP), the primary colonization factor, are controlled by a complex network of transcriptional activators (reviewed in references 30 and 43). Both ctxAB, the genes encoding CT, and the tcp operon are activated by an AraC family transcription factor, ToxT (2, 5, 18, 21). Expression of toxT is, in turn, controlled by two pairs of inner membrane proteins, ToxR/S and TcpP/H (10, 17, 31). TcpP and TcpH are encoded adjacent to the tcp operon, and their expression is activated by two proteins, AphA and AphB, that are responsive to environmental signals (28, 29). In addition to indirectly controlling ctxAB expression by activating transcription of toxT, ToxR has been shown to directly activate transcription from the ctxAB promoter in Escherichia coli (39). The transmembrane domain of ToxR was recently implicated in the ToxT-independent induction of CT expression by bile salts (22). The VieSAB three-component system was identified as an additional regulator of virulence genes in a screen for in vivo activators of ctxAB (34). The VieSAB system is required for maximal expression of ctxAB both in vivo and during growth under in vitro conditions that induce virulence factor expression, but the exact regulatory mechanism was not determined (52).
Here we show that the VieA response regulator is required in the V. cholerae classical biotype for maximal activation of CT production during growth in vitro under conditions that induce virulence gene expression. Specifically, the EAL domain of VieA must be functional to maintain the low intracellular c-diGMP concentration that enhances CT production. We show that transcription of both ctxA and toxT is repressed by an increased intracellular concentration of c-diGMP and that control of the c-diGMP concentration by VieA is necessary for wild-type levels of colonization in an animal model of infection. Since we have previously implicated the c-diGMP second messenger as an activator of vps gene expression and biofilm formation, we propose that c-diGMP modulates gene expression during the transition from environmental reservoirs to the human host.
MATERIALS AND METHODS
Growth conditions.
Bacteria were grown in Luria-Bertani (LB) broth at 37°C with aeration unless otherwise noted. M9 salts plus 0.5% glycerol, trace metals (1 ml/liter of 5% MgSO4, 0.5% MnCl2 · 4H2O, 0.5% FeCl3, 0.4% trinitriloacetic acid) (3), and l-asparagine, l-arginine, l-glutamate, and l-serine, each at 25 mM (M9 + NRES), was prepared as described previously (38). A similar medium with morpholinepropanesulfonic acid (MOPS) salts as the base (46) was used for 32P labeling of nucleotides. Expression from the ParaBAD promoter was induced by addition of arabinose to a 0.1% final concentration. Antibiotics were used at the following concentrations: streptomycin (Sm), 100 μg/ml; ampicillin (Ap), 50 μg/ml; chloramphenicol (Cm), 5 μg/ml.
Strain and plasmid construction.
All strains and plasmids used in this study are listed in Table 1. Plasmids with oriR6K were propagated in E. coli DH5αλpir; all other plasmids were propagated in E. coli DH5α. Oligonucleotide primers are listed in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 | 16, 27 |
| DH5αλpir | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 λ::pir | 16, 27 |
| SM10λpir | thi recA thr leu tonA lacY supE RP4-2-Tc::Mu λ::pir | Laboratory strain |
| AC-E1277 | SM10λpir (pAT1276), Apr | This work |
| V. cholerae strains | ||
| O395 | Classical biotype, Smr | 50 |
| AC-V61 | O395 lacZ::res-tet-res, spontaneous partial deletion of lacZ, Smr | 4 |
| AC-V1386 | AC-V61 ΔvieA, Smr | 51 |
| AC-V1596 | AC-V61 vieA(E170A), Smr | 51 |
| AC-V744 | O395 ΔtoxR, Smr | 26 |
| AC-V449 | O395 ΔtcpP, Smr | 57 |
| AC-V1296 | AC-V61 toxT-lacZ-tcpJ, Smr | This work |
| AC-V1382 | AC-V1386 toxT-lacZ-tcpJ, Smr | This work |
| AC-V1848 | AC-V1596 toxT-lacZ-tcpJ, Smr | This work |
| AC-V1302 | AC-V1296 toxR::pGP704 Smr Apr | This work |
| AC-V1460 | AC-V61 pBAD33, Smr Cmr | 51 |
| AC-V1726 | AC-V61 pAT1662, Smr Cmr | 51 |
| AC-V1463 | AC-V1386 pBAD33, Smr Cmr | 51 |
| AC-V1464 | AC-V1386 pAT1337, Smr Cmr | 51 |
| AC-V1660 | AC-V1386 pAT1568, Smr Cmr | This work |
| AC-V1661 | AC-V1386 pAT1615, Smr Cmr | This work |
| AC-V1843 | AC-V1386 pAT1647, Smr Cmr | This work |
| AC-V1109 | O395 pMMB67EH, Smr Apr | This work |
| AC-V1815 | AC-V1596 pMMB67EH, Smr Apr | This work |
| AC-V1824 | AC-V1596 pAT1822, Smr Apr | This work |
| AC-V1869 | AC-V61 vpsR::pGP704 Smr Apr | This work |
| AC-V1870 | AC-V1596 vpsR::pGP704 Smr Apr | This work |
| Plasmids | ||
| pCVD442 | oriR6K mobRP4 sacB, Apr | 11 |
| pAT1276 | pCVD442::toxT-lacZ-tcpJ, Apr | This work |
| pGP704 | oriR6K mobRP4, Apr | 38 |
| pAC396 | pGP704::‘toxR', Apr | 38 |
| pAT1868 | pGP704::‘vpsR', Apr | This work |
| pCR-Script | fl (+) ori, Apr | Stratagene |
| pGEM-T | fl ori, Apr | Promega |
| pAT856 | pGEM-T::‘rpoB', Apr | 52 |
| pAT853 | pGEM-T::‘ctxA', Apr | 52 |
| pDSM701 | pGEM-T::‘toxT', Apr | 52 |
| pDSM702 | pGEM-T::‘ompT', Apr | 36 |
| pDSM703 | pGEM-T::‘ompU', Apr | 36 |
| pAT1809 | pGEM-T::‘tcpP', Apr | This work |
| pMMB67EH | IncQ broad-host-range cloning vector, Apr | 40 |
| pAT1822 | pMMB67EH::vieA, Apr | This work |
| pBAD33 | pACYC184 ori, araC ParaBAD, Cmr | 15 |
| pAT1337 | pBAD33::vieA, Cmr | 48 |
| pAT1568 | pBAD33::VieA-EAL-His6, Cmr | 51 |
| pAT1615 | pBAD33::VieAEAL(E170A)-His6, Cmr | 51 |
| pAT1647 | pBAD33::‵RocS-His6, Cmr | 51 |
| pAT1662 | pBAD33::VCA0956-His6, Cmr | 51 |
TABLE 2.
Oligonucleotide primers used in this studya
| Oligonucleotide name | Sequence 5′ - 3′ |
|---|---|
| toxTF1 | GCTCTAGATGTCAAACCATATCAGCCTAG |
| TZJR1 | AACTGCAGAGGATCAAGTAAACGTATTCC |
| 5JF1 | GGGGTACCCTACAGTATTTAACTCAACCA |
| 3JR1 | GCGAGCTCCTTCATTTTAGCTCACATTAA |
| TZJF1 | AACTGCAGTTTANGATACATTTTTATGACCATGATTACGGATTCA |
| CZBR1 | GGGGTACCTTATTTTTGACACCAGACCAA |
| tcpPF1 | TCAATTTCCCGATAACCTTT |
| tcpPR1 | AGTCAGCTTCATCAACAACG |
| F38 | GATGGAGATTTTGGCTTCA |
| vieAR | AACTGCAGTAGGTACAGCCATAACTCTCG |
| tcpEF2 | GGAGTTATCTATGACCCTGTT |
| lacZR | GAGGGGACGACGACAGTATC |
| oriR | CAGCAGTTCAACCTGTTG |
| toxRR2 | ATGGCATCGTTAGGGTTAG |
| vpsRF2 | TGGCATGCTATGCCTATGAAGCGTTTG |
| vpsRR2 | GCGAGCTCGTAGCACGATATCCGATGC |
| vpsRF3 | CATGCTGAAGTTGGAAAA |
Restriction enzyme sites are indicated by underlining.
Plasmid pAT1276 was constructed by ligating toxT, lacZ, and tcpJ fragments into pCVD442. The toxT fragment (1.2 kbp) was amplified by PCR from O395 genomic DNA using primers toxTF1 and TZJR1; the tcpJ fragment (800 bp) was similarly amplified using primers 5JF1 and 3JR1. The E. coli lacZ gene was amplified with primers TZJF1 and CZBR1. All three products were cloned into pCR-Script and subsequently removed by digestion with the following enzymes: for toxT, XbaI/PstI; for lacZ, PstI/KpnI; for tcpJ, KpnI/SacI. The products were ligated to XbaI/SacI-digested pCVD442, and the construct was confirmed by enzyme digestions, PCR, and sequencing.
Plasmid pAT1809 for generation of tcpP RNA probe was constructed by PCR amplifying an internal fragment of tcpP (347 bp) from O395 genomic DNA, using primers TcpPF1 and TcpPR1 and cloning into pGEM-T. The template for in vitro transcription of the antisense probe was generated from this plasmid by PCR using primers TcpPF1 and T7R. Plasmid pAT1822 for complementation of the vieA(E170A) mutation was constructed by PCR amplification of the vieA gene and 1,517 bp 5′ to the vieA start codon using primers F38 and VieAR. The product was subcloned into pCR-Script, removed by digestion with SacI/SalI, and ligated into similarly digested pMMB67EH. Plasmid pAT1868 for generating plasmid integrations at the vpsR locus was constructed by PCR amplification of a 507-bp internal fragment of the vpsR gene using primers vpsRF2 and vpsRR2. The PCR product was digested with SphI/SacI and ligated into similarly digested pGP704.
Strains AC-V1296, AC-V1382, and AC-V1848 were generated by allelic exchange with plasmid pAT1276 in the appropriate strain background as previously described (33). Integration of lacZ downstream of toxT was confirmed by PCR with primers tcpEF2 and lacZR. Strain AC-V1302 was constructed by mating strain AC-V1296 with E. coli SM10λpir containing plasmid pAC396 and selecting Smr Apr exconjugates as described previously (33). Integration of the plasmid at the toxR locus was confirmed by colony PCR using primers oriR/toxRR2. Strains AC-V1869 and AC-V1870 were similarly constructed by mating strains AC-V61 and AC-V1596, respectively, with E. coli SM10λpir containing plasmid pAT1868 and selecting Smr Apr exconjugates. Integration of the plasmid at the vpsR locus was confirmed by colony PCR using primers oriR and vpsRF3. Strains AC-V1660, AC-V1661, AC-V1843, AC-V1109, AC-V1815, and AC-V1824 were constructed by electroporation of the appropriate plasmid into the indicated strain background. Strains O395, AC-V1386, and AC-V1596 were made electrocompetent as described previously (52).
Cholera toxin ELISA.
Strains were grown overnight in M9 + NRES at 30°C with aeration. The final optical density at 600 nm (OD600) was determined, cells were pelleted by centrifugation, and the supernatant was filter sterilized (0.45-μm polyvinylidene difluoride filter). The concentration of CT in supernatants was determined by GM1 enzyme-linked immunosorbent assay (ELISA) using polyclonal goat antiserum to the CT B subunit (List Biolabs) and antigoat immunoglobulin G conjugated to alkaline phosphatase (Sigma) as described previously (20). A purified CT B subunit of known concentration (List Biolabs) was used to determine the amount of CT in the samples.
RNase protection assay (RPA) and c-diGMP quantitation.
RPAs were performed essentially as described previously (52) using RNA isolated from strains grown in MOPS + NRES at 30°C with aeration to mid-exponential phase (OD600 = 0.6). The rpoB riboprobe was included in all hybridizations to serve as a loading control as described previously (52). Data were collected with a PhosphorImager and analyzed using ImageQuant (Molecular Dynamics). Band intensities were quantitated and normalized to the intensity of the rpoB band. The reported severalfold differences in transcript level are the average from three independent experiments. Determination of c-diGMP concentration was done using aliquots of the same cultures essentially as described previously (51) except that bacteria were labeled with [32P]orthophosphate for 30 min. Quantitation of c-diGMP spots observed by two-dimensional thin-layer chromatography (2D-TLC) was done using ImageQuant. Values were corrected for background and were normalized to the intensity of the GDP spot. The intensity of the GDP spot did not vary appreciably (less than 1.5-fold differences) in the strain backgrounds tested, with the exception of the GGDEF-overexpressing strain, in which it was substantially reduced.
Immunoblots for TcpA and TcpP.
Strains were grown in M9 + NRES at 30°C with aeration to an OD600 of 0.6. Cells were pelleted by centrifugation, resuspended in sodium dodecyl sulfate sample buffer, and boiled for 5 min. Equivalent volumes of each sample were separated by electrophoresis on sodium dodecyl sulfate-polyacrylamide gels. Proteins were transferred to Hybond nitrocellulose (Amersham) or Immobilon-P polyvinylidene difluoride (Millipore) membranes and blotted with rabbit polyclonal antisera against the TcpA or TcpP proteins. Donkey antirabbit immunoglobulin conjugated to horseradish peroxidase was used as a secondary antibody. Immunoreactive bands were detected with the ECL kit (Amersham) and exposure to film (Kodak).
β-Galactosidase assays.
Strains containing the toxT-lacZ-tcpJ transcriptional fusion were struck for single colonies on LB plates and incubated overnight at 37°C. Colonies were resuspended in M9 + NRES, the OD600 was determined, and 6 ml M9 + NRES in 16- by 1.5-cm tubes was inoculated to a starting OD600 of 0.1. Cultures were incubated at 30°C with vigorous shaking. At 2-h intervals, the OD600 was determined and 0.5 ml was harvested for β-galactosidase assays, which were done according to the Miller method (37).
In vivo competition assays.
Competition assays with the infant mouse model of cholera were performed essentially as described previously (33) except that plate-grown bacteria were used as the inocula. The lacZ vie(AE170A) mutant and wild-type O395 strains were struck for single colonies on LB plates and incubated overnight at 37°C. For each strain, approximately 30 colonies were scraped off plates into 1 ml of LB and the OD600 was determined. Equivalent amounts of each strain were mixed and adjusted to a final OD600 of 0.01 (approximately 107 CFU/ml). Five-day-old CD-1 infant mice were intragastrically inoculated with 50 μl of this mix. Competitions to assess complementation of the vieA(E170A) mutant were performed against strain AC-V1109. Similar competitions were performed in vitro by inoculating bacteria into LB and incubating overnight at 37°C with aeration. Care of and procedures done on mice complied with relevant federal guidelines as well as Tufts University animal use policies.
RESULTS
VieA regulates cholera toxin production.
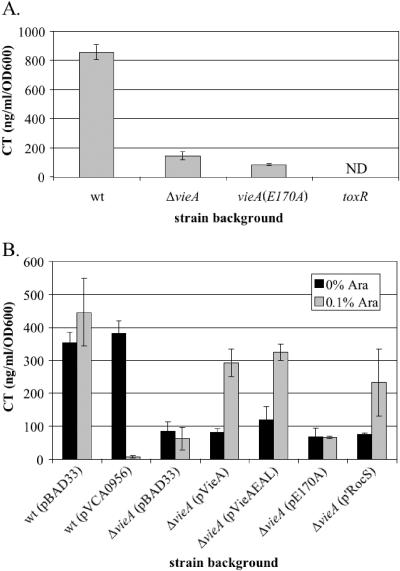
We have previously shown that the VieSAB three-component signal transduction system regulates production of CT in the V. cholerae El Tor biotype (52). Unlike the El Tor biotype, which requires a complex growth condition for in vitro induction of virulence genes, the V. cholerae classical biotype expresses virulence factors during growth at 30°C in either LB (pH 6.5) or a defined medium (M9) plus the L amino acids asparagine, arginine, glutamate, and serine (NRES) (38). Although the classical biotype does not require the VieSAB system for expression of CT under the LB (pH 6.5) inducing condition (data not shown), we decided to test whether the VieSAB system is necessary for CT production in M9 + NRES because the VieS sensor kinase contains two motifs in its predicted periplasmic domain with homology to amino acid binding proteins. While the VieB response regulator was dispensable (data not shown), VieA was necessary for maximal CT production in M9 + NRES as assessed by GM1 ELISA (Fig. 1A). Specifically, the putative c-diGMP phosphodiesterase EAL domain of VieA was required to enhance CT production. The E170A point mutation at a highly conserved residue of the EAL domain does not affect expression of VieA as assessed by immunoblotting with polyclonal antiserum against VieA (data not shown) but causes an eightfold reduction in the amount of CT produced, similar to the sixfold reduction observed for the in-frame deletion of vieA (Fig. 1A). These results suggest that VieA activates CT production indirectly by lowering the intracellular concentration of c-diGMP.
FIG. 1.
CT production is regulated by c-diGMP. GM1 ELISAs to quantify production of the CT B subunit were done with culture supernatants from the indicated strains grown overnight in M9 + NRES. Purified CT B subunit of known concentration was used to determine the concentration in the samples. ND, not detected. A. Strains AC-V61 (wild type), AC-V1386 (ΔvieA), AC-V1596 [vieA(E170A)], and AC-V744 (ΔtoxR). B. Strains were grown in the absence (black bars) or presence (gray bars) of 0.1% arabinose. The wild-type and ΔvieA mutant strain backgrounds carrying the indicated plasmids (strains AC-V1460, AC-V1726, AC-V1463, AC-V1464, AC-V1660, AC-V1661, and AC-V1843).
VieA regulates cholera toxin through control of c-diGMP.
To confirm that VieA regulates CT production by controlling intracellular c-diGMP concentration, the c-diGMP concentration was ectopically altered by expression of proteins with the GGDEF or EAL domains from an arabinose-inducible promoter. To increase the c-diGMP concentration, the GGDEF protein VCA0956, which synthesizes c-diGMP both in vivo and in vitro (48; A.D. Tischler and A. Camilli, unpublished data), was expressed. Since c-diGMP is barely detectable in the wild-type strain, we used the ΔvieA mutant, which exhibits an elevated c-diGMP concentration (51), and attempted to complement the mutation by expressing either the VieA EAL domain or an N-terminally truncated form of the RocS protein (‵RocS; VC0653) that includes an EAL domain. Both of these proteins have putative c-diGMP phosphodiesterase activity, since they were previously shown to complement the ΔvieA strain with respect to vps gene expression (51). As a negative control, the VieA EAL domain containing the E170A point mutation was expressed. All of these proteins were tagged with a His6 epitope to facilitate determination of expression levels by immunoblotting. Each protein was undetectable in the absence of induction, and all were expressed at similar levels following induction with arabinose (data not shown).
Ectopic expression of the GGDEF and EAL proteins by arabinose induction from a PBAD promoter had the expected effects on CT production. Expression of the GGDEF protein VCA0956 in wild-type V. cholerae, which, like mutations in vieA, causes an increase in the c-diGMP concentration, reduced CT production to a virtually undetectable level (Fig. 1B). Conversely, expression of either full-length VieA or the VieA EAL domain complemented the ΔvieA mutation; CT production was fully restored to the level for the wild-type vector control (Fig. 1B). In contrast, complementation was not observed when the VieA EAL domain containing the E170A point mutation was expressed (Fig. 1B). Finally, expression of ′RocS restored CT production to near the wild-type level (Fig. 1B). These results confirm that VieA regulates CT expression indirectly through its influence on the level of c-diGMP.
Cholera toxin is regulated at the transcriptional level.
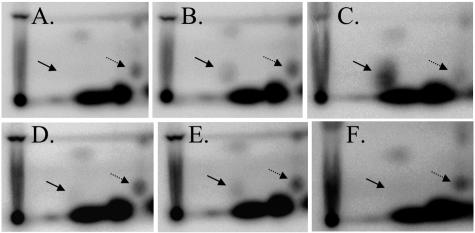
Since production of CT was decreased in strains with increased intracellular c-diGMP concentration, we tested whether transcription of the ctxAB genes was similarly affected. In order to monitor the c-diGMP concentration simultaneously with levels of ctxA transcription, these experiments were performed in a phosphate-limiting MOPS minimal medium that allows radioactive labeling of nucleotides with 32P (51). Strains were grown in MOPS plus NRES to mid-exponential phase (OD600 = 0.6), and samples of each culture were taken for nucleotide labeling and RNA extraction. 2D-TLC of nucleotide extracts confirmed the predicted changes in the c-diGMP concentration. Specifically, the vieA(E170A) and ΔvieA mutations caused sixfold increases in the c-diGMP concentration, relative to that of the wild type (Fig. 2A and B and data not shown). Similarly, expression of the VCA0956 GGDEF protein resulted in a 28-fold increase in the level of c-diGMP relative to that of the uninduced control (Fig. 2C and data not shown). In contrast, expression of the VieA EAL domain or ‵RocS in the ΔvieA mutant background lowered the c-diGMP concentration (Fig. 2D and F). While ‵RocS expression resulted in a fivefold decrease in the c-diGMP level relative to that of the uninduced control, the VieA EAL domain reduced the c-diGMP concentration only two-fold. Finally, expression of the E170A mutant VieA EAL domain in the ΔvieA mutant background had no effect on the level of c-diGMP (Fig. 2E).
FIG. 2.
2D-TLC of 32P-labeled nucleotide extracts to detect c-diGMP. Strains were grown in MOPS plus NRES to an OD600 of 0.6 and labeled for 30 min with [32P]orthophosphate. Nucleotides were extracted with formic acid, spotted on TLC plates (origin at the bottom left corner), and developed in 0.2 M NH4HCO3, pH 7.8, in the first dimension (bottom to top) and 1.5 M KH2PO4, pH 3.65, in the second dimension (left to right). c-diGMP and GDP spots are indicated by the solid and dashed arrows, respectively. Only the relevant portion of the TLC plate is shown. A. AC-V61, wild type. B. AC-V1596, vieA(E170A). C-F. Strains were grown with 0.1% arabinose to induce expression from the pBAD promoter. C. AC-V1726, wild type (pVCA0956). D. AC-V1660, ΔvieA (pVieA-EAL). E. AC-V1661, ΔvieA (pE170A). F. AC-V1843, ΔvieA (p‵RocS).
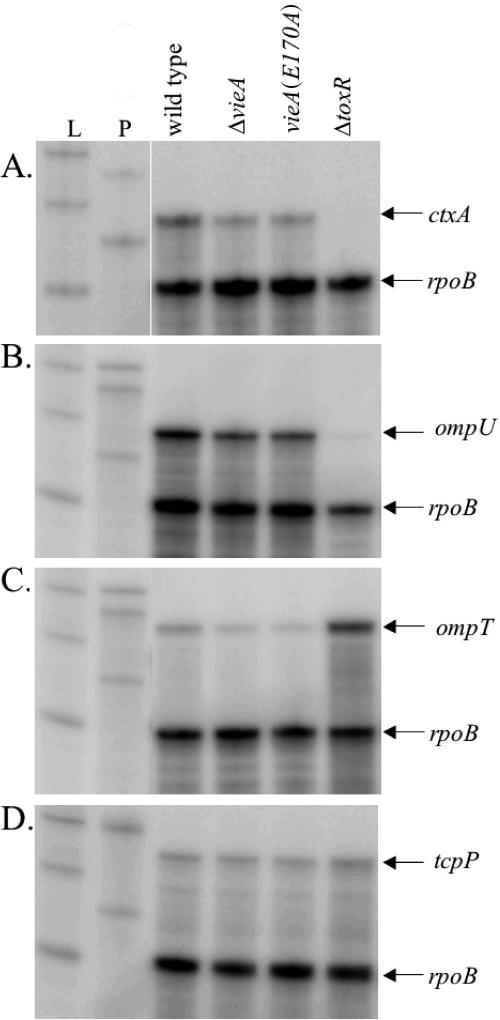
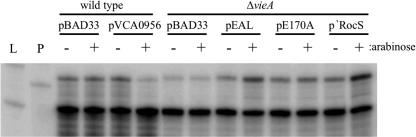
The amount of ctxA transcript was measured for each strain by RPA. A fourfold decrease in ctxA transcript was consistently observed for both the ΔvieA and vieA(E170A) mutants compared to the wild type, suggesting that ctxA transcription is repressed by an increased c-diGMP concentration (Fig. 3A). To confirm that the c-diGMP concentration affects transcription of ctxA, similar RPA experiments were performed using strains overexpressing either the VCA0956 GGDEF protein or the various EAL domains. In quantifying the level of ctxA transcript for these experiments, we compared the arabinose-induced sample to the relevant uninduced control for each strain rather than comparing all samples to the vector control due to differences in growth rates of the strains that may have influenced gene expression. We believe this is an appropriate comparison, since all proteins expressed from the pBAD33 vector were undetectable in the absence of arabinose induction (data not shown). Consistent with the results of the CT ELISAs, a sevenfold decrease in ctxA transcript was observed when the VCA0956 GGDEF protein was expressed (Fig. 4). Conversely, expression of the VieA EAL domain or ‵RocS restored ctxA transcription to near wild-type levels (3-fold and 4.5-fold increases, respectively) (Fig. 4). Overexpression of the E170A mutant VieA EAL domain, however, gave no increase in ctxA transcription relative to that for the uninduced control (Fig. 4).
FIG. 3.
VieA regulates transcription of ctxA. The wild-type (AC-V61), ΔvieA (AC-V1386), vieA(E170A) (AC-V1596), and ΔtoxR (AC-V744) strains were grown to an OD600 of 0.6 in MOPS plus NRES, RNA was extracted, and indicated transcripts were detected by RNase protection assay. L indicates RNA ladder; P indicates undigested probes (only 0.1% of probe used in RPAs was loaded). All samples were normalized to the rpoB loading control and are the average for three independent experiments.
FIG. 4.
Ectopic expression of the GGDEF or EAL protein affects ctxA transcription. Strains with the indicated plasmids were grown in MOPS plus NRES with or without 0.1% arabinose to an OD600 of 0.6, RNA was isolated, and the ctxA and rpoB transcripts were detected by RPA. L indicates RNA ladder; P indicates undigested probes (only 0.1% of probe used in RPAs was loaded). All samples were normalized to the rpoB loading control and are the average for three independent experiments.
c-diGMP concentration affects transcription of toxT.
Our previous work in the El Tor biotype suggested that the VieSAB system might indirectly affect ctxAB transcription by controlling transcription of the positive regulator ToxT (52). To test whether toxT transcription is regulated by VieA in the classical biotype, RPAs were done on the same RNA samples used to assess ctxA regulation. In these mid-exponential-phase RNA samples, the level of toxT transcript was quite low, preventing accurate quantitation of the effect of c-diGMP on toxT regulation (data not shown).
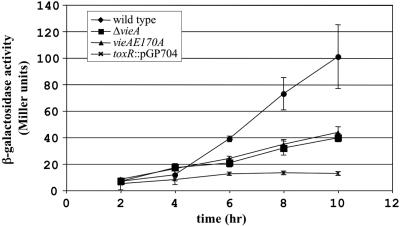
As an alternative method to assess toxT expression, a nonpolar toxT-lacZ transcriptional fusion was constructed and introduced into the wild-type, ΔvieA, and vieA(E170A) strain backgrounds. Transcriptional activity was assessed for each of these strains over time during growth in M9 + NRES by performing β-galactosidase assays. A toxR plasmid insertion mutant was included in these experiments as a negative control. Although all strains grew at indistinguishable rates (data not shown), reporter fusion activity increased 10-fold during exponential-phase growth for the wild-type strain but only fourfold for the ΔvieA or vieA(E170A) mutant (Fig. 5). The activity of the fusion was significantly different from that of the wild type at 6, 8, and 10 h in both the ΔvieA and vieA(E170A) mutants as assessed by unpaired, two-tailed t tests (P < 0.05). These results suggest that the ability of VieA to lower the c-diGMP concentration is required for maximal expression of ToxT, which in turn activates ctxA transcription.
FIG. 5.
toxT-lacZ fusion activity is reduced in vieA mutants. Strains containing the toxT-lacZ fusion were grown in M9 + NRES at 30°C with shaking. β-Galactosidase activity was determined at 2-h intervals by the Miller method. Each result presented is the average ± standard error from three independent experiments.
c-diGMP does not affect regulators above ToxT.
ToxT is positively regulated by two DNA binding proteins, ToxR and TcpP, both of which localize to the inner membrane (10, 17, 31). In addition to regulating expression of virulence factors, ToxR is responsible for the inverse regulation of the outer membrane porins, OmpT and OmpU. ToxR is a repressor of ompT transcription (35), while it activates expression of ompU (7). Since ToxR is constitutively expressed under standard growth conditions (9), environmental signals that activate expression of virulence genes presumably affect the ability of ToxR to bind DNA. To examine whether the c-diGMP concentration alters toxT transcription by changing the activation state of ToxR, RPAs to examine ompU and ompT transcription were done. Although a slight (approximately 1.5-fold) decrease in ompU transcript was observed in the ΔvieA and vieA(E170A) mutants relative to the wild-type level, a similar 2.5-fold decrease was observed for the ompT transcript (Fig. 3B and C). If ToxR activity was affected by the c-diGMP concentration, an increase in the level of ompT transcript would be expected, concomitant with the decrease in ompU transcription. These results suggest that ToxR activation is not affected by the c-diGMP concentration.
TcpP is regulated at the transcriptional level by two activators, AphA and AphB (28, 29). To test whether decreased toxT expression might be caused by reduced tcpP transcription, RPAs were performed. Although the amount of tcpP transcript detected was low, it was reproducibly detected at levels similar to that for the wild type in both the ΔvieA and vieA(E170A) mutants (less that 1.5-fold reduction) (Fig. 3D). In addition, overexpression of EAL or GGDEF domains did not affect the level of tcpP transcript (data not shown).
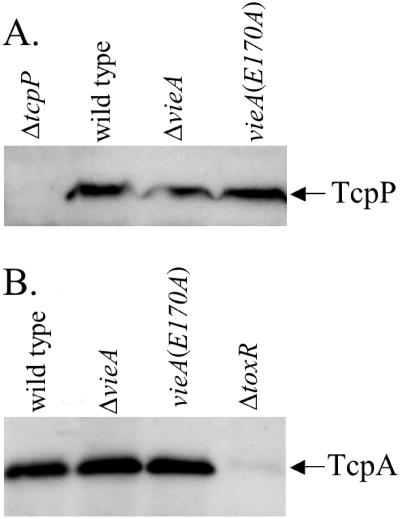
It was recently reported that TcpP is a target for proteolysis in the absence of its partner protein, TcpH (1). The protease responsible for degradation of TcpP has not yet been identified, but we have observed that several secreted proteases are up-regulated in a vieA mutant (A.D. Tischler, J. T. Pratt, and A. Camilli, unpublished data). To test whether TcpP protein expression is affected by an increased c-diGMP concentration at either the level of translation or protein stability, immunoblots were performed on vieA mutant strains grown in M9 + NRES. We consistently observed similar levels of TcpP protein in the wild-type and vieA mutant strains (Fig. 6A), suggesting that translation and stability of TcpP are unaffected by c-diGMP.
FIG. 6.
TcpP and TcpA protein levels are not affected by vieA mutations. A. TcpP was detected by immunoblotting for strains AC-V61 (wild type), AC-V1386 (ΔvieA), AC-V1596 [vieA(E170A)], and AC-V449 (ΔtcpP) grown to an OD600 of 0.6 in M9 + NRES at 30°C. B. TcpA was detected by immunoblotting for strains AC-V61 (wild type), AC-V1386 (ΔvieA), AC-V1596 [vieA(E170A)], and AC-V744 (ΔtoxR) grown to an OD600 of 0.6 in M9 + NRES at 30°C.
c-diGMP does not affect TcpA production.
ToxT is a master regulator of virulence gene expression, activating not only transcription of the ctxAB genes but also that of many other genes, including those required for synthesizing the TCP (2). To test whether other genes regulated by ToxT are also affected by the c-diGMP concentration, expression of TcpA, the major pilin subunit, was assessed by immunoblotting. Contrary to our expectations, TcpA production was not affected by either mutation in vieA (Fig. 6B).
Control of c-diGMP concentration by VieA is required for intestinal colonization.
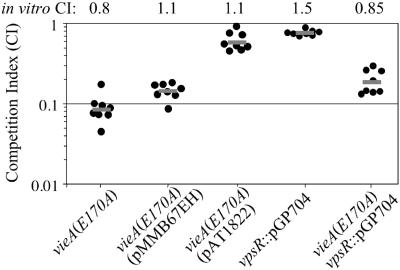
Because control of the c-diGMP concentration by VieA is required in the V. cholerae classical biotype for in vitro expression of ctxA and toxT but not for tcpA expression, it was unclear whether this regulation would affect virulence. To address this question, competition assays were performed in the infant mouse model of cholera. In contrast to the El Tor biotype, in which the VieSAB system is not required for colonization (33), the classical biotype requires the function of the VieA response regulator for full colonization. Specifically, the c-diGMP phosphodiesterase activity of VieA is necessary, since the vieA(E170A) mutant exhibited a 10-fold decrease in colonization fitness (Fig. 7).
FIG. 7.
The vieA(E170A) mutant is attenuated for colonization of the infant mouse. Competition assays were performed with the infant mouse model of cholera. Strains AC-V1596 [vieA(E170A)], AC-V1869 (vpsR::pGP704), and AC-V1870 [vieA(E170A), vpsR::pGP704] were competed against strain O395. Strains AC-V1815 and AC-V1824 [vieA(E170A) containing the pMMB67EH or pAT1822 complementing plasmid, respectively] were competed against strain AC-V1109 (O395 carrying pMMB67EH). The competition index is the ratio of mutant to wild-type bacteria recovered from the small intestine, corrected for the ratio present in the inoculum. Each dot represents an individual mouse; the gray line represents the geometric mean of the data. The competition indices from in vitro competitions in LB are indicated above the graph.
To confirm that the colonization defect of the vieA(E170A) point mutant is due to the vieA(E170A) mutation and not a secondary mutation, the mutant was complemented with plasmid pAT1822, which encodes vieA on the low-copy-number vector pMMB67EH. While the pMMB67EH vector did not affect the competition index, the pAT1822 complementing plasmid restored the vieA(E170A) mutant to near wild-type levels of colonization (Fig. 7). We suspect that the competition index of the complemented strain is not restored to a value of 1 because the complementing plasmid is unstable. Although pAT1822 was present in 100% of colony isolates from the inoculum, it had been lost by an average of 32% of the isolates after growth within the animal, as determined by replica plating to medium containing Ap. In contrast, the pMMB67EH vector control was lost by less that 1% of the wild-type control strain after passage through the animal. This analysis implies that vieA expression from pAT1822 is slightly deleterious to strain fitness during infection, perhaps as a result of overexpression or incorrect temporal regulation.
Since we have previously implicated the c-diGMP phosphodiesterase activity of VieA in regulation of the vps genes that are required for exopolysaccharide synthesis and biofilm formation (51), we hypothesized that overexpression of the vps genes in the vieA(E170A) mutant might be responsible for the colonization defect. To test this possibility, a plasmid insertion mutation was constructed in vpsR, which encodes a positive regulator that is necessary for vps gene expression (56). This mutation abrogates vps gene expression as assessed by measuring β-galactosidase activity of a vps-lacZ fusion (data not shown). In addition, the vpsR::pGP704 mutation in an otherwise wild-type background does not affect colonization of the infant mouse small intestine in a competition assay (Fig. 7). The vieA(E170A) vpsR::pGP704 double mutant still exhibits a fivefold defect in colonization fitness (Fig. 7), suggesting that misregulation of vps gene expression cannot account for the colonization defect of the vieA(E170A) mutant. Taken together, the above results suggest that c-diGMP plays a role in modulating in vivo expression of virulence factors that mediate colonization.
DISCUSSION
Virulence gene expression in V. cholerae is regulated by a complex cascade of transcriptional activators. Here we show that in addition to the known regulatory factors, the cyclic nucleotide second messenger c-diGMP negatively regulates transcription of the toxT and ctxA genes under conditions that stimulate virulence factor expression. Specifically, the putative phosphodiesterase activity of the VieA EAL domain is required for maximal expression of ctxA and toxT during growth under a minimal medium plus amino acids condition. The ability of VieA to control the c-diGMP concentration is also critical during infection, since the vieA(E170A) EAL domain point mutant was attenuated 10-fold in the infant mouse model of cholera. This represents the first demonstration with any organism that the c-diGMP second messenger regulates expression of virulence factors.
Our results implicate c-diGMP as a signal that represses expression of virulence factors and suggest possible mechanisms of transcriptional regulation by this second messenger. Since an increased concentration of c-diGMP repressed transcription of toxT, regulators of toxT may be directly affected by c-diGMP. We ruled out the possibility that ToxR is responsive to the c-diGMP concentration by examining expression of genes encoding the outer membrane proteins OmpU and OmpT, which are regulated directly by ToxR (7, 35). Transcription of ompU and ompT was only modestly affected by mutations in VieA that perturb the c-diGMP concentration. Additionally, levels of both the ompU and ompT transcripts were reduced in the vieA mutants, inconsistent with the inverse regulation of these genes by ToxR. ToxR cooperates with TcpP to activate transcription at the toxT promoter (31). We demonstrate that both tcpP transcript and TcpP protein levels are unchanged in response to the c-diGMP concentration. It is possible, however, that TcpP protein activity is affected by c-diGMP, resulting in reduced toxT transcription.
Alternatively, the activity of ToxT may be affected by the c-diGMP concentration. ToxT is a member of the AraC family of transcriptional regulators (18); like other members of this family, ToxT has an N-terminal domain that may function as a binding site for a small molecule effector such as c-diGMP. Indeed, it has been suggested that ToxT responds directly to environmental stimuli including temperature and bile (47). ToxT positively regulates its own expression by activating transcription from the tcpA promoter, which lies approximately 10.6 kb 5′ of toxT; the resultant read-through transcription is thought to contribute to sustained ToxT expression (57). Since we cannot distinguish between transcripts initiating at the toxT and tcpA promoters using the toxT-lacZ fusion, it is possible that the differences in fusion activity that we observe are due to changes in activation of the tcpA promoter by ToxT. However, it is unlikely that transcription from the tcpA promoter is negatively regulated in a specific manner by c-diGMP, since we observed no change in the production of TcpA in vieA mutant strains compared to that for the wild type.
A final hypothesis to explain virulence gene regulation by c-diGMP is that an as yet unidentified regulatory factor is responsible for sensing the intracellular concentration of c-diGMP and controlling virulence gene expression. This hypothesis is attractive given the unexpected differences we observed in the expression of ctxA and tcpA. Perhaps a repressor that acts specifically at the toxT and ctxA promoters controls transcription in response to c-diGMP.
Since we have shown that transcription of toxT is altered in vieA mutant strains that exhibit increased levels of c-diGMP, it is surprising that TcpA production was unaffected in these mutants. Although production of CT and TCP is generally coordinately regulated, since both the ctxA and tcpA promoters are activated by ToxT, differences in the regulation of these two promoters have previously been reported. For example, both ctxA and tcpA are repressed by H-NS, a histone-like nucleoid structuring protein, but repression is less stringent at the tcpA promoter (41). ToxT is thought to act, in part, by alleviating H-NS-mediated repression (58). Perhaps the concentration of the ToxT protein in strains with an increased concentration of c-diGMP is sufficient to derepress the tcpA promoter but not the ctxA promoter. In addition, differences in the timing of ctxA and tcpA transcription have been observed during infection (32). Specifically, while tcpA expression is induced within 1 h postinoculation, ctxA transcription is not induced until 4 h postinoculation. Perhaps changes in the concentration of ToxT within the cell account for these differences in the timing of transcriptional induction. Alternatively, it is possible that additional factors, including those that control and/or respond to the c-diGMP concentration, contribute to repression of ctxA early in the infection process.
In addition to regulating virulence gene expression under in vitro inducing conditions, control of the c-diGMP concentration by VieA is needed for wild-type levels of colonization of the host small intestine. In competition assays with the infant mouse model of cholera, we demonstrate that a VieA mutant that lacks c-diGMP phosphodiesterase activity is reduced 10-fold in colonization fitness. We were able to complement this defect in trans, confirming that it is the mutation in vieA that is responsible for the defect, rather than a secondary mutation. The reduced colonization fitness of the vieA(E170A) mutant was somewhat surprising, however, given that we observed no change in expression of the major colonization factor TcpA under in vitro growth conditions that induce virulence gene expression. In addition to affecting expression of the virulence factors toxT and ctxAB, mutations in vieA that increase the concentration of c-diGMP also result in increased expression of the Vibrio exopolysaccharide (vps) genes that are required for biofilm formation (51). To test whether misregulation of the vps genes is responsible for the colonization defect of the vieA(E170A) mutant, we constructed a mutation in vpsR, a positive regulator of vps gene expression. This mutation did not affect the ability of the vieA(E170A) mutant to colonize the small intestine of the infant mouse, suggesting that misregulation of the vps genes is not responsible for the colonization defect. It is possible that although mutations in vieA do not affect expression of TcpA in vitro, the timing of transcriptional activation or the assembly of TcpA subunits into a functional pilus may be altered in vivo. Finally, it is possible that VieA and the c-diGMP second messenger regulate expression of other factor(s) required for colonization.
In contrast to the Vibrio exopolysaccharide (vps) genes, which we have previously demonstrated are activated by an increased intracellular concentration of c-diGMP (51), virulence genes are negatively regulated by c-diGMP. This inverse regulation of environmental and virulence genes by c-diGMP is intriguing. It suggests that the c-diGMP second messenger may play an important role in the transition between growth in the environment and survival within the host small intestine. Our data show that the V. cholerae classical biotype requires regulation of the c-diGMP concentration by VieA to achieve wild-type levels of colonization in the infant mouse model. Although the VieSAB three-component system is not necessary for colonization by El Tor V. cholerae (33), it is possible that proteins with redundant function can fulfill this role of VieA in the El Tor biotype. The V. cholerae genome encodes 22 proteins that are homologous to putative c-diGMP phosphodiesterases, and each of these may contribute to reducing the intracellular c-diGMP concentration upon entry into the host. In fact, the VC0130 gene, which encodes a putative c-diGMP phosphodiesterase, was recently identified in a screen for in vivo-induced genes in the El Tor biotype (42). Perhaps the VC0130 protein is required for regulation of the c-diGMP concentration to activate virulence gene expression in El Tor V. cholerae.
Although this is the first report that virulence genes are transcriptionally regulated in response to the c-diGMP concentration, it is likely that c-diGMP regulates virulence phenotypes in other bacterial species. Coding sequences for the conserved GGDEF and EAL protein domains that are responsible for synthesis and degradation of c-diGMP are present in the genomes of the majority of pathogens with completed sequences. There is preliminary evidence that proteins with these domains control virulence-related processes in P. aeruginosa (12) and Y. pestis (19, 25), but whether these proteins control the c-diGMP concentration remains to be determined. It is possible that regulation of virulence by c-diGMP will become a common theme in pathogenic bacterial species.
ADDENDUM IN PROOF
After acceptance of our paper, a paper (K. B. Hirst, M. MacCross, M. U. Shiloh, K. H Darwin, S. Singh, R. A. Jones, S. Ehrt, Z. Zhang, B. L. Gaffney, S. Gandotra, D. W. Holden, D. Murray, and C. Nathan, Mol. Microbiol 56:1234-1245, 2005) demonstrating that the Salmonella EAL domain protein CdgR is required for resistance to oxidative stress both in vitro and during growth in host pathogens was published. Mutation of cdgR also enhanced killing of macrophages by Salmonella. CdgR was required to lower the cellular level of cyclic diguanylate, suggesting that cyclic diguanylate also regulates virulence properties in Salmonella.
Acknowledgments
We are grateful to V. DiRita and R. Taylor for providing antisera against TcpP and TcpA, respectively.
This material is based on work supported under an NSF Graduate Research Fellowship to A.D.T. This research was supported by NIH grant AI45746 to A.C. and the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928). A.C. is an investigator of the Howard Hughes Medical Institute.
Editor: V. J. DiRita
REFERENCES
- 1.Beck, N. A., E. S. Krukonis, and V. J. DiRita. 2004. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J. Bacteriol. 186:8309-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, R. C., and R. K. Taylor. 1995. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol. Microbiol. 16:425-439. [DOI] [PubMed] [Google Scholar]
- 3.Callahan, L. T., R. C. Ryder, and S. H. Richardson. 1971. Biochemistry of Vibrio cholerae virulence. II. Skin permeability factor/cholera enterotoxin production in a chemically defined medium. Infect. Immun. 4:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camilli, A., and J. J. Mekalanos. 1995. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol. Microbiol. 18:671-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champion, G. A., M. N. Neely, M. A. Brennan, and V. J. DiRita. 1997. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol. Microbiol. 23:323-331. [DOI] [PubMed] [Google Scholar]
- 6.Chang, A. L., J. R. Tuckerman, G. Gonzalez, R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Gilles-Gonzalez. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40:3420-3426. [DOI] [PubMed] [Google Scholar]
- 7.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 8.D'Argenio, D. A., and S. I. Miller. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497-2502. [DOI] [PubMed] [Google Scholar]
- 9.DiRita, V. J., M. N. Neely, R. K. Taylor, and P. M. Bruss. 1996. Differential expression of the ToxR regulon in classical and El Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc. Natl. Acad. Sci. USA 93:7991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 13.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, B., C. Latasa, C. Solano, F. G. Portillo, C. Gamazo, and I. Lasa. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264-277. [DOI] [PubMed] [Google Scholar]
- 15.Guzman, L., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Häse, C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins, D. E., E. Nazareno, and V. J. DiRita. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J. Bacteriol. 174:6974-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch, B. J., R. D. Perry, and T. J. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 20.Holmgren, J., and A.-M. Svennerholm. 1973. Enzyme-linked immunosorbent assays for cholera serology. Infect. Immun. 7:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hulbert, R. R., and R. K. Taylor. 2002. Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter. J. Bacteriol. 184:5533-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung, D. T., and J. J. Mekalanos. 2005. Bile acids induce cholera toxin expression in a ToxT-independent manner. Proc. Natl. Acad. Sci. USA 102:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 24.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirillina, O., J. D. Fetherston, A. G. Bobrov, J. Abney, and R. D. Perry. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:75-88. [DOI] [PubMed] [Google Scholar]
- 26.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolter, R., M. Imizuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 28.Kovacikova, G., W. Lin, and K. Skorupski. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol. Microbiol. 53:129-142. [DOI] [PubMed] [Google Scholar]
- 29.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 30.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6:186-190. [DOI] [PubMed] [Google Scholar]
- 31.Krukonis, E. S., R. R. Yu, and V. J. DiRita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 32.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. H., M. J. Angelichio, J. J. Mekalanos, and A. Camilli. 1998. Nucleotide sequence and spatiotemporal expression of the Vibrio cholerae vieSAB genes during infection. J. Bacteriol. 180:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, C. C., J. A. Crawford, V. J. DiRita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 36.Merrell, D. S., C. Bailey, J. B. Kaper, and A. Camilli. 2001. The ToxR-mediated organic acid tolerance response of Vibrio cholerae requires OmpU. J. Bacteriol. 183:2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator toxR is a transmembrane DNA-binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 40.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-48. [DOI] [PubMed] [Google Scholar]
- 41.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osorio, C. G., J. A. Crawford, J. Michalsky, H. Martinez-Wilson, J. B. Kaper, and A. Camilli. 2005. Second-generation recombination-based in vivo expression technology for large-scale screening for Vibrio cholerae genes induced during infection of the mouse small intestine. Infect. Immun. 73:972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reidl, J., and K. E. Klose. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol. Rev. 26:125-139. [DOI] [PubMed] [Google Scholar]
- 44.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 45.Ross, P., Y. Aloni, H. Weinhouse, D. Michaeli, P. Weinberger-Ohana, R. Mayer, and M. Benziman. 1986. Control of cellulose synthesis in Acetobacter xylinum, a unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149:101-117. [Google Scholar]
- 46.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schuhmacher, D. A., and K. E. Klose. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 181:1508-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition of sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 49.Tal, R., H. C. Wong, R. Calhoon, D. Gelfand, A. L. Fear, G. Volman, R. Mayer, P. Ross, D. Amikam, H. Weinhouse, A. Cohen, S. Sapir, P. Ohana, and M. Benziman. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA fusions to identify a pilus colonization factor co-ordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tischler, A. D., S. H. Lee, and A. Camilli. 2002. The Vibrio cholerae vieSAB locus encodes a pathway contributing to cholera toxin production. J. Bacteriol. 184:4104-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinhouse, H., S. Sapir, D. Amikam, Y. Shilo, G. Volman, P. Ohana, and M. Benziman. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416:207-211. [DOI] [PubMed] [Google Scholar]
- 55.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, R. R., and V. J. DiRita. 1999. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J. Bacteriol. 181:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]