Abstract
Nerve activity controls fiber size and fiber type in skeletal muscle, but the underlying molecular mechanisms remain largely unknown. We have previously shown that Ras–mitogen-activated protein kinase and calcineurin control fiber type but not fiber size in regenerating rat skeletal muscle. Here we report that constitutively active protein kinase B (PKB), also known as Akt, increases fiber size and prevents denervation atrophy in regenerating and adult rat muscles but does not affect fiber type profile. The coexistence of hypertrophic muscle fibers overexpressing activated PKB with normal-size untransfected fibers within the same muscle points to a cell-autonomous control of muscle growth by PKB. The physiological role of this pathway is confirmed by the finding that PKB kinase activity and phosphorylation status are significantly increased in innervated compared with denervated regenerating muscles in parallel with muscle growth. Muscle fiber hypertrophy induced by activated PKB and by a Ras double mutant (RasV12C40) that activates selectively the phosphoinositide 3-kinase–PKB pathway is completely blocked by rapamycin, showing that the mammalian target of rapamycin kinase is the major downstream effector of this pathway in the control of muscle fiber size. On the other hand, nerve activity-dependent growth of regenerating muscle is only partially inhibited by dominant negative PKB and rapamycin, suggesting that other nerve-dependent signaling pathways are involved in muscle growth. The present results support the notion that fiber size and fiber type are regulated by nerve activity through different mechanisms.
During embryonic development and regeneration skeletal muscle fibers can initially differentiate in the absence of neural influence; however, their subsequent survival, growth, and diversification require nerve activity. The maintenance of muscle fiber size and fiber type properties in the adult also depends on innervation, as clearly shown by the effects of denervation, cross-reinnervation, and electrostimulation. A major open issue in muscle biology is to define the transduction pathways that couple the electrical signals induced by motor neuron activity to the transcriptional and posttranscriptional changes underlying the remodeling of the muscle phenotype. The identification of these pathways has important clinical implications and may lead to the development of new drugs able to prevent or delay muscle wasting and loss of muscle force, a major cause of disability resulting from aging, disuse, and neuromuscular disorders.
Signaling through the calcineurin and Ras pathways has been implicated in nerve activity-dependent muscle gene regulation. Skeletal muscle-specific expression of constitutively active calcineurin in transgenic mice induces increased proportion of slow fibers but not muscle hypertrophy (1). The effect of intramuscular injection of activated calcineurin on slow muscle gene expression is controversial (2, 3). The physiological role of calcineurin has been investigated in vivo by using pharmacological inhibition with cyclosporin A (CsA) and FK506. These inhibitors were found to induce a slow-to-fast fiber type switch in adult muscle (4) and prevent the fast-to-slow conversion induced by functional overload (5), but this finding was not confirmed in another study (6). The effect of pharmacological inhibition of calcineurin on skeletal muscle growth is also controversial. Some researchers have reported that muscle hypertrophy induced by functional overload and muscle weight recovery after atrophy is inhibited by CsA and FK506 (5, 7), although this hypertrophy may vary according to muscle type and stage of muscle growth (8), whereas others found no inhibition (6, 9). A genetic inhibitory strategy was recently used to show that overexpression of the calcineurin peptide inhibitor cain/cabin-1 blocks, like CsA and FK506, the expression of the slow fiber phenotype but not fiber growth during muscle regeneration (10).
Ras-dependent pathways appear to affect both fiber size and fiber type in vivo (11). A Ras double mutant (RasV12S35), which activates the mitogen-activated protein kinase, specifically the extracellular signal-regulated kinase, is able to induce the slow phenotype, but not muscle growth, in regenerating denervated muscle. In contrast, a Ras double mutant (RasV12C40) that activates phosphoinositide 3-kinase (PI3K) and its downstream target, the serine-threonine protein kinase B (PKB), also known as Akt, is able to induce muscle growth, but not slow fiber type specification. In the present study, we show that constitutively active PKB induces muscle fiber hypertrophy in regenerating muscle. On the other hand, nerve activity-dependent muscle growth is inhibited by dominant negative PKB and rapamycin, showing that the mammalian target of rapamycin (mTOR) kinase is a major downstream effector of the PI3K–PKB pathway in muscle growth regulation.
Methods
Muscle Transfection, Electrostimulation, and Treatment with Rapamycin or FK506.
Adult male Wistar rats (200–250 g) were used in all experiments. Regenerating innervated or denervated muscles were injected with plasmid DNA (50 μg) at day 3 after muscle injury induced by bupivacaine treatment as described (12). The following constructs, all containing a hemagglutinin (HA) tag, were used: the Ras double mutant RasV12C40 (13), a constitutively active (c.a.) PKB, containing a myristoylation/palmitylation signal to the N-terminal end (14), and a catalytically inactive dominant negative PKB, containing a CAAX motif to the C-terminal end (15). Muscles were removed 7 days later, i.e., day 10 after muscle injury, and frozen in isopentane cooled in liquid nitrogen. In some experiments, rats were injected i.p. with 1.5 mg/kg rapamycin dissolved in 10% ethanol and 2% carboxymethylcellulose or with 1 mg/kg FK506 in saline once daily starting at day 3 after muscle injury and ending at day 10. Control animals were treated with vehicle. For electrostimulation experiments denervated regenerating soleus muscles were stimulated through electrodes implanted onto the muscles at 100 Hz (60 pulses every 60 sec), as described (16), from day 3 to day 10 after muscle injury. This impulse pattern was found to stimulate effectively muscle growth of regenerating muscles, whereas a pattern resembling that of slow motor neurons (10 Hz) was found to be much less efficient (unpublished observations). Electrostimulated animals were treated with rapamycin or vehicle from day 3 to day 10 after injury. Adult muscles were transfected by i.m. injection of plasmid DNA (20 μg) followed by electroporation to increase gene transfer efficiency (10). Denervated muscles were transfected immediately after section of the sciatic nerve and removed 7 days later.
Immunocytochemistry and Fiber Size Measurements.
Fibers expressing HA-tagged PKB and RasV12C40 were revealed in cryosections fixed with 4% paraformaldehyde and stained with anti-HA antibodies (Roche Molecular Biochemicals). Fibers expressing myosin heavy chain (MyHC)-slow were identified by immunofluorescence with the mAb BA-D5 in unfixed serial sections (17). Muscle fiber size was measured in all fibers transfected with PKB mutants and in an equal number of untransfected fibers from the same muscles. The percentage of slow fibers and fiber cross-sectional areas in muscles treated with rapamycin or FK506 were measured in at least five muscles per group and three distinct randomly chosen fields of each muscle cross section. Fiber cross-sectional areas were measured by using image software (Scion, Frederick, MD). All data are expressed as the mean ± SEM (error bars). Comparisons were made by using t test, with P < 0.05 being considered statistically significant.
Kinase Assay.
The kinase activity of PKB was measured by the immune complex kinase assay in muscle lysates essentially as described (18), using glycogen synthase 3 (GSK-3) fusion protein (GSK-3α peptide 7–25 fused to paramyosin) (Cell Signaling Technology, Beverly, MA) as substrate. The samples were analyzed by SDS/PAGE using a 15% gel. The extent of GSK-3 phosphorylation was quantitated on a PhosphorImager (Packard).
Electrophoresis and Western Blotting Analysis.
Muscle lysates containing 30 μg of protein were separated by SDS/PAGE using 10% gels, transferred to nitrocellulose membranes, and probed with antibodies against PKBα and PKBβ (Akt1 and 2) (Upstate Biotechnology), PKB-phospho-Ser-473, p70S6 kinase, S6, and phospho-S6 (Cell Signaling Technology). The blots were then incubated with secondary antibodies conjugated to horseradish peroxidase followed by enhanced chemiluminescence (Pierce). MyHCs were separated by SDS/PAGE as described (19) and revealed by Coomassie blue staining. Under the electrophoretic conditions used, the MyHC-slow isoform can be unambiguously identified as the high mobility band in the MyHC group of bands. The relative amount of MyHC-slow was quantitated by densitometric analysis and expressed as percentage of total MyHCs. MyHC-slow was also identified by Western blotting with the mAb BA-D5 (17). Troponin I-slow was identified in Western blots with a mAb (TI-3) that reacts with the slow, but not the fast, isoform of troponin I (20).
Results
Constitutively Active PKB Induces Fiber Hypertrophy in Regenerating and Adult Rat Muscles.
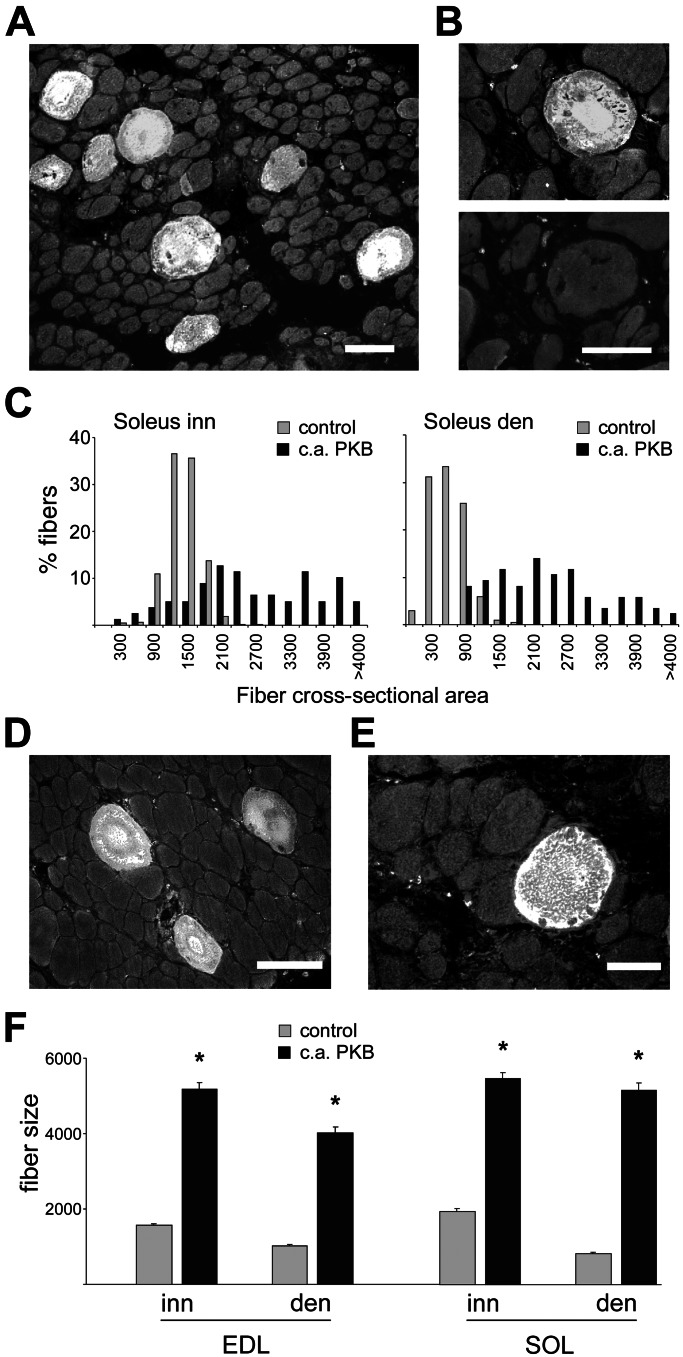
To explore the role of the PI3K–PKB pathway in fiber size and fiber type regulation, we first transfected regenerating muscles with membrane targeted, c.a. PKB mutant. Muscles were transfected by i.m. injection of plasmid DNA at day 3 after muscle injury, when they consist of thin myotubes (mean fiber size: 238 ± 7 μm2) and examined at day 10, when they consist of almost mature myofibers in the innervated soleus (mean fiber size: 1,224 ± 10 μm2, about 60% of adult soleus muscle fiber size) and relatively atrophic fibers in denervated soleus (mean fiber size: 459 ± 7 μm2). In the presence of the nerve, regenerating soleus fibers display by day 10 a slow phenotype, as shown by the expression of the MyHC slow gene. In contrast, denervated regenerating fibers display a default fast phenotype, characterized by the expression of fast MyHCs (11). Muscle fibers expressing c.a. PKB were identified by the presence of the HA epitope. As shown in Fig. 1 A–C, fibers overexpressing c.a. PKB are much larger in size compared with untransfected surrounding muscle fibers in both innervated (mean ± SEM: 2,634 ± 145 μm2) and denervated (2,227 ± 114 μm2) regenerating soleus. The same effect was seen in fibers transfected with c.a. PI3K (data not shown). Activated PKB stimulates muscle growth also in regenerating fast-twitch extensor digitorum longus (EDL) muscle (data not shown). The MyHC profile is unaffected by PKB; in particular, MyHC-slow is not induced in the transfected fibers of denervated soleus (Fig. 1B). We also examined the effect of c.a. PKB in nonregenerating adult muscles. Activated PKB was found to induce striking muscle fiber hypertrophy in both innervated and denervated soleus and EDL muscles (Fig. 1 D–F).
Figure 1.
Constitutively active PKB induces muscle fiber hypertrophy in regenerating (A–C) and adult (D–F) rat skeletal muscle. (A) Immunofluorescence analysis of a transverse section of regenerating denervated soleus muscle transfected with HA-tagged c.a. PKB and stained with anti-HA antibodies. Note that transfected fibers are much larger in size than surrounding untransfected fibers. (B) Serial sections of a regenerating denervated soleus muscle transfected with HA-tagged PKB and stained with antibodies specific for HA epitope (Upper) and MyHC-slow (Lower). Note that MyHC-slow is not expressed in the hypertrophic fiber expressing c.a. PKB. (C) Frequency histograms showing the distribution of cross-sectional areas (μm2) of fibers expressing c.a. PKB and surrounding untransfected fibers (control) in innervated (Left) and denervated (Right) regenerating soleus. A total of 1,920 muscle fibers were analyzed. (D and E) Denervated adult EDL (D) and soleus (E) muscles transfected with HA-tagged c.a. PKB. Hypertrophic fibers expressing c.a. PKB are seen in transverse sections stained with anti-HA. (F) Cross-sectional areas (μm2) of fibers expressing c.a. PKB and surrounding untransfected fibers (control) in innervated and denervated adult EDL and soleus muscles. Data are mean ± SEM. *, Significant difference from control group (P < 0.05). A total of 1,221 muscle fibers were analyzed. [Bars: 50 μm (A, B, and E); 100 μm (D).]
PKB Activity Is Increased in Regenerating Innervated Muscle.
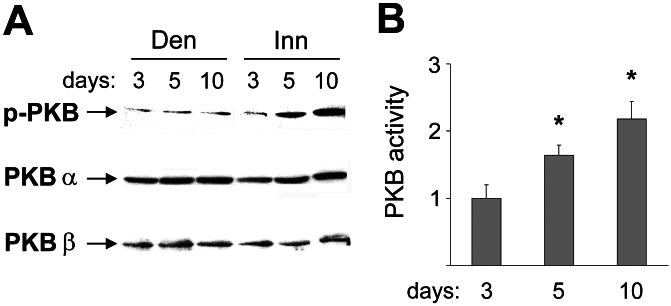
To determine whether the PKB pathway is activated in relation with muscle growth, we analyzed PKB protein expression and activity in innervated and denervated regenerating rat soleus muscle at days 3, 5, and 10 after muscle injury. Western blotting analysis shows no significant difference in the PKBα (Akt1) and PKBβ (Akt2) protein levels between innervated and denervated muscles (Fig. 2A). In contrast, the phosphorylation status of PKB, as determined by an antibody that reacts selectively with the phosphorylated (phospho-Ser-473) active form of PKB, is progressively increased at days 5 and 10 in innervated, but not in denervated, muscle (Fig. 2A). Accordingly, PKB kinase activity is significantly increased at days 5 and 10 in innervated compared with denervated muscles (Fig. 2B) in parallel with muscle growth.
Figure 2.
PKB activity is increased by nerve activity in regenerating soleus muscle. (A) Western blotting analysis with antibodies specific for PKBα and PKBβ shows that protein levels of PKBα and PKBβ are unchanged in innervated and denervated soleus. In contrast, the level of the phosphorylated active form of PKB (p-PKB) is progressively increased from day 3 to days 5 and 10 after injury in innervated compared with denervated muscle. The same result was observed in three different experiments. (B) PKB kinase activity, expressed as fold increase of innervated versus denervated muscle, is progressively increased from day 3 to days 5 and 10 after injury. Three innervated and three denervated muscles per day were examined. Data are mean ± SEM. *, Significant difference between innervated and denervated muscles (P < 0.05).
The Increase in Muscle Fiber Size Induced by Innervation and Electrostimulation in Regenerating Muscle Is Partially Inhibited by Dominant Negative PKB and Rapamycin.
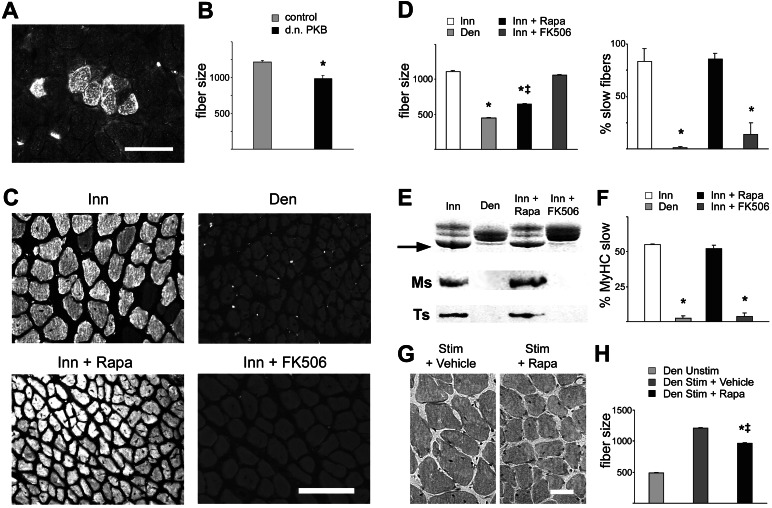
To determine whether PKB activity is required for muscle growth during regeneration, we transfected regenerating innervated muscles with a PKB mutant that codes for a catalytically inactive kinase and acts as dominant negative inhibitor of PKB activation (15). As shown in Fig. 3 A and B, muscle fibers expressing dominant negative PKB are significantly smaller in size compared with surrounding untransfected fibers. The role of PI3K–PKB signaling in muscle fiber growth in regenerating muscle was also examined by a pharmacological approach with rapamycin. Rapamycin binds FK506 binding protein 12 (FKBP12) and the rapamycin–FKBP12 complex blocks a major downstream effector of PKB, mTOR kinase. The drug was given from day 3 to day 10 after muscle injury, and this treatment was found to block completely the phosphorylation of targets known to be downstream of mTOR, such as p70S6K and the ribosomal protein S6 (data not shown). As a control for these experiments we used FK506, another immunosuppressive drug that also binds FKBP12; however, the FK506–FKBP12 complex blocks calcineurin, but not mTOR, activity. Muscle fiber growth in regenerating innervated soleus is markedly inhibited by rapamycin, whereas it is unaffected by FK506 (Fig. 3 C and D Left). Interestingly, rapamycin and FK506 have an opposite effect on fiber type differentiation in that the up-regulation of the MyHC-slow gene induced by slow motor neurons is blocked by FK506 (see also ref. 10) but not by rapamycin. Antibodies specific for MyHC-slow stain most soleus fibers in animals treated with rapamycin but only a minority of fibers in FK506-treated animals (Fig. 3 C and D Right). Accordingly, the up-regulation of MyHC-slow, as determined by SDS/PAGE and Western blotting analysis, is unaffected by rapamycin treatment but is blocked in FK506-treated animals (Fig. 3 E and F). The expression of another marker of slow fibers, troponin I-slow, is also blocked by FK506, but not rapamycin, treatment (Fig. 3E). Next, we examined the effect of rapamycin in muscles that were directly electrostimulated with a pattern of impulses that effectively induces muscle growth in denervated regenerating muscles. As shown in Fig. 3 G and H, the increase in fiber size induced by electrostimulation is partially inhibited by rapamycin treatment.
Figure 3.
Muscle fiber growth induced by innervation and electrostimulation in regenerating muscle is inhibited by dominant negative PKB and rapamycin. (A and B) Muscle fiber hypertrophy in regenerating skeletal muscle is inhibited by dominant negative PKB (d.n. PKB). Note smaller size of fibers expressing HA-tagged dominant negative PKB in regenerating innervated soleus muscle as visualized by immunofluorescence (A) and determined by measurements of cross-sectional areas (μm2) (B). A total of 555 muscle fibers were analyzed. *, Significant difference from control (P < 0.05). (C) Transverse sections of regenerating innervated soleus (Inn), denervated soleus (Den), innervated soleus from rats treated with rapamycin (Inn + Rapa), and innervated soleus from rats treated with FK506 (Inn + FK506). Immunofluorescence staining with anti-MyHC-slow. (D) Fiber size (cross-sectional areas in μm2) (Left) and percentage of slow fibers (Right) in the four experimental groups shown in C. Note that rapamycin affects muscle fiber size but not expression of MyHC-slow, whereas FK506 affects MyHC-slow expression but not fiber size. A total of 1,743 muscle fibers were analyzed for measurements of fiber size, and a total of 12,079 fibers were counted to determine the percentage of slow fibers. *, Significant difference from control innervated muscles (P < 0.05); ‡, significant difference from denervated muscles (P < 0.05). (E) SDS/PAGE profile of MyHCs (arrow: MyHC-slow band) (Top) and Western blotting analysis of the same samples with anti-MyHC-slow (Ms) (Middle) and anti-troponin I (Ts) (Bottom). Note that the expression of the two slow contractile proteins is prevented by FK506 but not by rapamycin treatment. (F) SDS/PAGE profile of MyHCs, densitometric values of MyHC-slow relative to all MyHCs in the four experimental groups. Data are mean ± SEM (n = 4). *, Significant difference from control innervated muscles (P < 0.05). (G and H) Rapamycin inhibits muscle growth induced by electrostimulation. Denervated regenerating soleus muscle were electrostimulated from day 3 to day 10 after injury. (G) Representative cross sections of electrostimulated soleus muscles treated with vehicle (Left) or rapamycin (Right) stained with hematoxylin-eosin. (H) Fiber size (cross-sectional areas in μm2) measurements show that electrostimulation induces a marked increase in fiber size and that this effect is partially, but significantly, inhibited by rapamycin treatment. Three muscles per group were examined. Data are mean ± SEM. A total of 2,117 muscle fibers were analyzed. *, Significant difference from denervated unstimulated muscles (P < 0.05); ‡, significant difference from vehicle-treated group (P < 0.05). [Bars: 50 μm (A and G); 100 μm (C).]
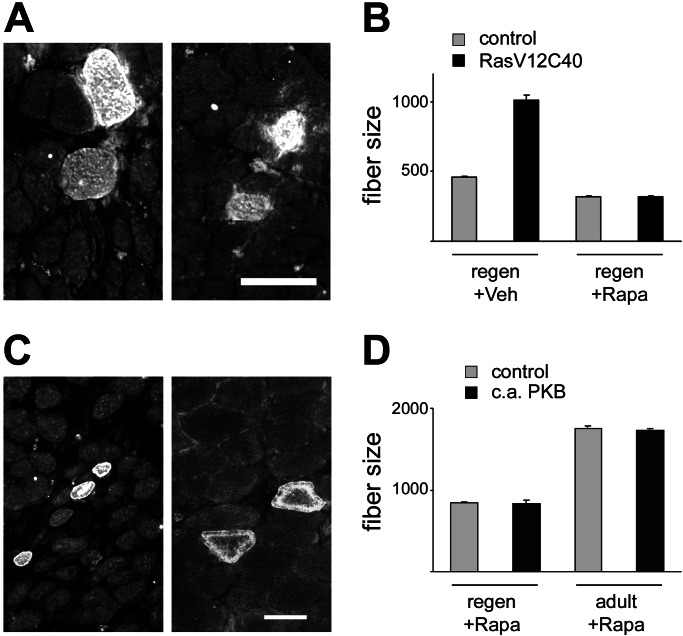
Muscle Fiber Hypertrophy Induced by Constitutively Active PKB and RasV12C40 Is Completely Blocked by Rapamycin.
The incomplete inhibition of the muscle growth-promoting effect of nerve activity by rapamycin could be caused by the fact that muscle growth is also controlled by PI3K–PKB-dependent but mTOR-independent pathways (see ref. 21). Alternatively, PI3K–PKB-independent pathways may also be involved. To address this issue, we examined the effect of rapamycin treatment on muscle fiber hypertrophy induced by RasV12C40 and c.a. PKB. As shown in Fig. 4, rapamycin blocks completely the increase in fiber size induced by RasV12C40 and c.a. PKB in regenerating muscles. Muscle fiber hypertrophy induced by c.a. PKB in adult muscles is also completely blocked by rapamycin (Fig. 4 C and D). These results indicate that (i) the increase in fiber size induced by RasV12C40 and c.a. PKB is mediated exclusively by mTOR and not by other PKB-dependent pathways, and (ii) other PI3K–PKB-independent pathways must also be involved in the control of muscle growth.
Figure 4.
Muscle fiber hypertrophy induced by RasV12C40 and constitutively active PKB is blocked by rapamycin. (A and B) Rapamycin blocks the increase in fiber size induced by RasV12C40. (A) Regenerating denervated soleus muscles were transfected with HA-tagged RasV12C40, and rats were treated with vehicle (Left) or rapamycin (Right). (B) Fiber size (cross-sectional areas in μm2) measurements show that muscle fibers transfected with RasV12C40 are much larger in size compared with surrounding untransfected fibers (control) and that this effect is completely inhibited by rapamycin treatment. Data are mean ± SEM. A total of 3,258 muscle fibers were analyzed. (C and D) Rapamycin blocks the increase in fiber size induced by c.a. PKB. (C) Regenerating innervated soleus (Left) and nonregenerating adult soleus muscle (Right) were transfected with HA-tagged c.a. PKB and rats were treated with rapamycin. Immunofluorescence analysis of sections stained with anti-HA antibody. (D) The mean cross-sectional area (μm2) of fibers expressing c.a. PKB is identical to that of control untransfected fibers in both regenerating and adult muscles. A total of 1,829 muscle fibers were analyzed. (Bars: A and C, 50 μm.)
Discussion
The PI3K–PKB pathway is a key mediator of cell survival and antiapoptotic signaling (22) and has recently emerged as an evolutionarily conserved controller of cell size (23). The PI3K–PKB pathway is required for skeletal myogenesis in culture (24–28) and has been implicated in myotube growth (29, 30). We have previously reported that a Ras double mutant, RasV12C40, which is known to activate selectively the PI3K–PKB pathway, is able to induce an increase in muscle fiber size and prevent denervation atrophy in vivo in regenerating rat skeletal muscle (11). Bodine et al. (6) recently showed that the PKB–mTOR pathway is required for muscle hypertrophy by using two models of overload hypertrophy, i.e., functional overload induced by elimination of synergistic muscles and restoration of muscle mass after atrophy induced by unloading. However, the role of PKB–mTOR in fiber type specification was not examined.
In the present study, we show that the PI3K–PKB pathway controls nerve activity-dependent muscle growth but not fiber type specification in regenerating skeletal muscle. Constitutively active PKB stimulates muscle fiber growth and this effect is completely prevented by rapamycin; therefore it must be mediated exclusively by mTOR and not by other downstream pathways nor by nonspecific effects of PKB overexpression. We find that activated PKB induces skeletal muscle hypertrophy and prevents denervation atrophy exclusively in transfected fibers in regenerating and adult skeletal muscle. The coexistence of hypertrophic muscle fibers overexpressing activated PKB with normal-size untransfected fibers within the same muscle points to a cell-autonomous control of muscle growth by PKB; that is, PKB appears to act directly in transfected fibers and not through local release of growth factors and autocrine/paracrine loops that could also affect surrounding untransfected fibers. The increase in fiber size during muscle development and compensatory hypertrophy is accompanied by proliferation of satellite cells and their subsequent fusion with muscle fibers (see ref. 31). It remains to be established whether a similar process is also induced by PKB overexpression. However, it should be stressed that activated PKB is exclusively expressed in muscle fibers and not in mononucleated cells since the earliest stages examined, i.e., 24 h posttransfection (data not shown), as previously found with other constructs (11). Therefore the primary effect of PKB must take place within the muscle fibers themselves and not in satellite cells.
The functional significance of PKB is supported by the finding that the endogenous PKB kinase activity is increased by innervation in parallel with muscle growth. In addition, dominant negative PKB and rapamycin partially inhibit the increase in fiber size induced by innervation, therefore the PKB–mTOR pathway is involved in nerve-dependent muscle growth. However, the finding that rapamycin blocks completely the increase in fiber size induced by RasV12C40 and c.a. PKB but only partially that induced by innervation and electrostimulation suggests that other unidentified rapamycin-insensitive signaling pathways also control fiber growth in regenerating muscle.
The mTOR kinase is known to stimulate translation initiation through the activation of the p70S6 kinase, which phosphorylates the ribosomal protein S6, and the inactivation of the 4E-BP-1 protein, an inhibitor of the translation initiation factor eIF4E (32). In regenerating muscle, like in other cell systems (see ref. 32), rapamycin does not act as a global repressor of protein synthesis. In fact, despite the severe block of muscle growth, the synthesis of MyHC-slow and troponin I-slow induced by slow motor neuron activity is unaffected by rapamycin. Recent studies support the notion that mTOR is a downstream effector of the PI3K–PKB pathway both in insects (33) and mammals (34, 35). The results reported here, in agreement with those reported by Bodine et al. (6), indicate that mTOR is the major downstream effector of the PI3K–PKB pathway in the control of muscle fiber size.
Interpretation of the effect of rapamycin is complicated by the possibility that this drug has other cellular targets in addition to mTOR. Both FK506 and rapamycin bind to the peptidyl-prolyl cis-trans isomerase FKBP12 and dissociate this protein from the Ca2+ release channel of the sarcoplasmic reticulum, also known as ryanodine receptor. FKBP12 modulates the calcium release activity of the ryanodine receptor (36), and disruption of the interaction between FKBP12 and ryanodine receptor by rapamycin or FK506 leads to loss of cooperative interactions between subunits of the ryanodine receptor tetramer (37). However, it is unlikely that these effects are involved in the inhibition of muscle fiber growth, because rapamycin and FK506 have opposite effects on regenerating muscle. Rapamycin inhibits muscle fiber growth but not the nerve-dependent emergence of the slow phenotype, as determined by immunofluorescence and SDS/PAGE analyses of MyHC expression. In contrast, FK506 blocks the slow phenotype but not muscle growth. Therefore the differential effect of rapamycin and FK506 most likely results from the selective inhibition of mTOR kinase and calcineurin, respectively. Indeed, we have recently shown that two other inhibitors of calcineurin, cyclosporin A and cain/cabin-1, affect fiber type but not fiber growth in the regenerating muscle system (10).
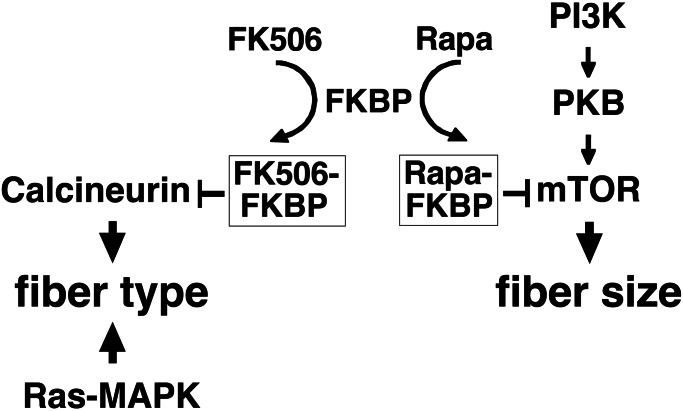
In conclusion, we show here that the effect of motor neuron activity on skeletal muscle growth in regenerating muscle depends on the PI3K–PKB–mTOR pathway. Taken together with our previous studies (10, 11), the present results indicate that fiber size and fiber type are regulated by nerve activity through different mechanisms in regenerating muscle (Fig. 5). Calcineurin and Ras–mitogen-activated protein kinase pathways control muscle fiber type differentiation, whereas PI3K–PKB–mTOR and other as-yet-unidentified pathways control muscle growth. It remains to be established whether the mechanisms responsible for muscle growth and fiber type specification during regeneration are identical to those involved in the hypertrophy of adult muscle fibers and the maintenance of the fast/slow fiber phenotype. A challenge for future studies is to determine how these transduction pathways are coupled to the electrical and mechanical signals that accompany nerve-dependent muscle activity and to identify the final effectors at the transcriptional and posttranscriptional level.
Figure 5.
Scheme of the signaling pathways involved in the regulation of fiber type and fiber size by nerve activity in regenerating skeletal muscle. The scheme is based on the results of the present study and previous works on the role of Ras–mitogen-activated protein kinase (MAPK) (11) and calcineurin (10) in regenerating rat soleus muscle between day 3 and day 10 after bupivacaine injury.
Acknowledgments
We are grateful to M. Murgia for initial experiments and S. Bortolotto for help with SDS/PAGE analysis of MyHCs. We thank E. Taparowsky for the RasV12C40 plasmid, B. A. Hemmings for the constitutively active PKB plasmid, and B. M. Burgering for the dominant negative PKB plasmid. We also thank Regeneron Pharmaceuticals (Tarrytown, NY) for rapamycin and Fujisawa Pharmaceutical (Osaka) for FK506. This work was supported by grants from the European Commission (Contracts ERBBIO4CT960216 and QLK6-2000-00530), the Giovanni Armenise-Harvard Foundation for Advanced Scientific Research, the Italian Space Agency (ASI), and the Italian Ministry of University and Scientific and Technological Research (MURST-COFIN).
Abbreviations
- PI3K
phosphoinositide 3-kinase
- PKB
protein kinase B
- mTOR
mammalian target of rapamycin
- MyHC
myosin heavy chain
- HA
hemagglutinin
- c.a.
constitutively active
- EDL
extensor digitorum longus.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Naya F J, Mercer B, Shelton J, Richardson J A, Williams R S, Olson E N. J Biol Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 2.Delling U, Tureckova J, Lim H W, De Windt L J, Rotwein P, Molkentin J D. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swoap S J, Hunter R B, Stevenson E J, Felton H M, Kansagra N V, Lang J M, Esser K A, Kandarian S C. Am J Physiol. 2000;279:C915–C924. doi: 10.1152/ajpcell.2000.279.4.C915. [DOI] [PubMed] [Google Scholar]
- 4.Chin E R, Olson E N, Richardson J A, Yang Q, Humphries C, Shelton J M, Wu H, Zhu W, Bassel-Duby R, Williams R S. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn S E, Burns J L, Michel R N. J Biol Chem. 1999;274:21908–21912. doi: 10.1074/jbc.274.31.21908. [DOI] [PubMed] [Google Scholar]
- 6.Bodine S C, Stitt T N, Gonzalez M, Kline W O, Stover G L, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence J C, Glass D J, Yancopoulos G D. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 7.Dunn S E, Chin E R, Michel R N. J Cell Biol. 2000;151:663–672. doi: 10.1083/jcb.151.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchell P O, Mills S T, Pavlath G K. Am J Physiol. 2002;282:C984–C992. doi: 10.1152/ajpcell.00483.2001. [DOI] [PubMed] [Google Scholar]
- 9.Dupont-Versteegden E E, Knox M, Gurley C M, Houle J D, Peterson C A. Am J Physiol. 2002;282:C1387–C1395. doi: 10.1152/ajpcell.00424.2001. [DOI] [PubMed] [Google Scholar]
- 10.Serrano A L, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lømo T, Schiaffino S. Proc Natl Acad Sci USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murgia M, Serrano A L, Calabria E, Pallafacchina G, Lømo T, Schiaffino S. Nat Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- 12.Vitadello M, Schiaffino M V, Picard A, Scarpa M, Schiaffino S. Hum Gene Ther. 1994;5:11–18. doi: 10.1089/hum.1994.5.1-11. [DOI] [PubMed] [Google Scholar]
- 13.Ramocki M B, Johnson S E, White M A, Ashendel C L, Konieczny S F, Taparowsky E J. Mol Cell Biol. 1997;17:3547–3555. doi: 10.1128/mcb.17.7.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andjelkovic M, Jakubowicz T, Cron P, Ming X F, Han J W, Hemmings B A. Proc Natl Acad Sci USA. 1996;93:5699–5704. doi: 10.1073/pnas.93.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Weeren P C, de Bruyn K M, de Vries-Smits A M, van Lint J, Burgering B M. J Biol Chem. 1998;273:13150–13156. doi: 10.1074/jbc.273.21.13150. [DOI] [PubMed] [Google Scholar]
- 16.Ausoni S, Gorza L, Schiaffino S, Gundersen K, Lømo T. J Neurosci. 1990;10:153–160. doi: 10.1523/JNEUROSCI.10-01-00153.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiaffino S, Gorza L, Sartore S, Saggin L, Ausoni S, Vianello M, Gundersen K, Lømo T. J Muscle Res Cell Motil. 1989;10:197–205. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- 18.Morisco C, Zebrowski D, Condorelli G, Tsichlis P, Vatner S F, Sadoshima J. J Biol Chem. 2000;275:14466–14475. doi: 10.1074/jbc.275.19.14466. [DOI] [PubMed] [Google Scholar]
- 19.Talmadge R J, Roy R R. J Appl Physiol. 1993;75:2337–2340. doi: 10.1152/jappl.1993.75.5.2337. [DOI] [PubMed] [Google Scholar]
- 20.Saggin L, Gorza L, Ausoni S, Schiaffino S. J Biol Chem. 1989;264:16299–16302. [PubMed] [Google Scholar]
- 21.Brazil D P, Hemmings B A. Trends Biochem Sci. 2001;26:657–664. doi: 10.1016/s0968-0004(01)01958-2. [DOI] [PubMed] [Google Scholar]
- 22.Datta S R, Brunet A, Greenberg M E. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 23.Stocker H, Hafen E. Curr Opin Genet Dev. 2000;10:529–535. doi: 10.1016/s0959-437x(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 24.Coolican S A, Samuel D S, Ewton D Z, McWade F J, Florini J R. J Biol Chem. 1997;272:6653–6662. doi: 10.1074/jbc.272.10.6653. [DOI] [PubMed] [Google Scholar]
- 25.Cuenda A, Cohen P. J Biol Chem. 1999;274:4341–4346. doi: 10.1074/jbc.274.7.4341. [DOI] [PubMed] [Google Scholar]
- 26.Jiang B H, Aoki M, Zheng J Z, Li J, Vogt P K. Proc Natl Acad Sci USA. 1999;96:2077–2081. doi: 10.1073/pnas.96.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang B H, Zheng J Z, Vogt P K. Proc Natl Acad Sci USA. 1998;95:14179–14183. doi: 10.1073/pnas.95.24.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandromme M, Rochat A, Meier R, Carnac G, Besser D, Hemmings B A, Fernandez A, Lamb N J. J Biol Chem. 2001;276:8173–8179. doi: 10.1074/jbc.M005587200. [DOI] [PubMed] [Google Scholar]
- 29.Rommel C, Bodine S C, Clarke B A, Rossman R, Nunez L, Stitt T N, Yancopoulos G D, Glass D J. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 30.Rommel C, Clarke B A, Zimmermann S, Nunez L, Rossman R, Reid K, Moelling K, Yancopoulos G D, Glass D J. Science. 1999;286:1738–1741. doi: 10.1126/science.286.5445.1738. [DOI] [PubMed] [Google Scholar]
- 31.Allen D L, Roy R R, Edgerton V R. Muscle Nerve. 1999;22:1350–1360. doi: 10.1002/(sici)1097-4598(199910)22:10<1350::aid-mus3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 32.Gingras A C, Raught B, Sonenberg N. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Stallock J P, Ng J C, Reinhard C, Neufeld T P. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aoki M, Blazek E, Vogt P K. Proc Natl Acad Sci USA. 2001;98:136–141. doi: 10.1073/pnas.011528498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neshat M S, Mellinghoff I K, Tran C, Stiles B, Thomas G, Petersen R, Frost P, Gibbons J J, Wu H, Sawyers C L. Proc Natl Acad Sci USA. 2001;98:10314–10319. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shou W, Aghdasi B, Armstrong D L, Guo Q, Bao S, Charng M J, Mathews L M, Schneider M D, Hamilton S L, Matzuk M M. Nature (London) 1998;391:489–492. doi: 10.1038/35146. [DOI] [PubMed] [Google Scholar]
- 37.Marx S O, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks A R. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]