Abstract
Filamentous fungi within the Fusarium graminearum species complex (Fg complex) are the primary etiological agents of Fusarium head blight (scab) of wheat and barley. Scab is an economically devastating plant disease that greatly limits grain yield and quality. In addition, scabby grain is often contaminated with trichothecene mycotoxins that act as virulence factors on some hosts, and pose a serious threat to animal health and food safety. Strain-specific differences in trichothecene metabolite profiles (chemotypes) are not well correlated with the Fg complex phylogeny based on genealogical concordance at six single-copy nuclear genes. To examine the basis for this discord between species and toxin evolution, a 19-kb region of the trichothecene gene cluster was sequenced in 39 strains chosen to represent the global genetic diversity of species in the Fg complex and four related species of Fusarium. Phylogenetic analyses demonstrated that polymorphism within these virulence-associated genes is transspecific and appears to have been maintained by balancing selection acting on chemotype differences that originated in the ancestor of this important group of plant pathogens. Chemotype-specific differences in selective constraint and evidence of adaptive evolution within trichothecene genes are also reported.
Fusarium head blight (scab) is an economically devastating disease of wheat and barley that reached epidemic proportions in North America during the last decade, causing nearly three billion dollars in losses to U.S. agriculture (1). Members of the Fusarium graminearum species complex (Fg complex) are the primary etiological agents of scab (2), and like other Fusarium species, they infect a broad range of plant hosts worldwide (3). Infection of cereals by Fusarium lowers grain yield and quality, and can result in contamination of cereal grains with trichothecene mycotoxins, potent inhibitors of eukaryotic protein synthesis that have been implicated in a number of human and animal mycotoxicoses (4). Trichothecenes also are acutely phytotoxic and act as virulence factors on sensitive host plants (5). The trichothecenes produced by Fusarium are divided into two broad categories based on the presence (B-trichothecenes) or absence (A-trichothecenes) of a keto group at the C-8 position of the trichothecene ring (4). Biochemical and genetic investigations of Fusarium sporotrichioides (an A-trichothecene producer) facilitated the identification of many of the genes involved in trichothecene biosynthesis (6). Homologous genes have since been identified in Fg complex species (B-trichothecene producers), and with the exception of a 3-O-acetyltransferase (TRI101) (7), all known trichothecene genes are localized within a gene cluster (6).
Three strain-specific profiles of trichothecene metabolites (chemotypes) have been identified within the B-trichothecene lineage of Fusarium: (i) nivalenol and acetylated derivatives (NIV chemotype), (ii) deoxynivalenol and 3-acetyldeoxynivalenol (3ADON chemotype), and (iii) deoxynivalenol and 15-acetyldeoxynivalenol (15ADON chemotype) (8). Only a hydroxyl group at C-4 in nivalenol distinguishes it from deoxynivalenol. However, these chemotype differences may have important fitness consequences, as differences in the pattern of oxygenation and acetylation can alter the bioactivity and toxicity of trichothecenes (7, 9).
On the basis of genealogical concordance at six single-copy nuclear genes, including TRI101, seven biogeographically distinct species have been identified within the Fg complex (2). However, trichothecene chemotypes do not correlate strongly with the Fg complex phylogeny (2), indicating that each of these chemotypes had multiple independent evolutionary origins or that the evolutionary history of these chemotypes was different from that predicted by the species phylogeny. To identify the basis for this discord between species and toxin evolution, a 19-kb region of the trichothecene gene cluster (TRI-cluster) was sequenced in 39 strains chosen to represent the global genetic diversity of species in the B-trichothecene lineage of Fusarium (2). Phylogenetic analyses of these sequences demonstrated that polymorphism within these virulence-associated genes has persisted through multiple speciation events and appears to have been maintained by balancing selection acting on chemotype differences that originated in the ancestor of extant species within the B-trichothecene lineage of Fusarium.
Materials and Methods
Biological Materials.
The strains selected for study are listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Thirty-seven of these strains were included in the description of seven biogeographically structured species within the Fg complex (2). Two additional strains, NRRL 29297 and NRRL 29306 from New Zealand, were included here because they represent an eighth species in the Fg complex. Hereafter, individual species within the Fg complex are referred to as Fg1–8.
Mycotoxin Analyses.
Mycotoxin analyses were conducted on wheat cultivar Norm. Wheat spikelets were excised 14 days after inoculation, weighed, extracted for trichothecenes in 1.5 ml acetonitrile/water (84:16 vol/vol) by shaking in glass vials for 24 h, and analyzed as described by using GC/MS (10). Strains were assigned to chemotype groups based on their major metabolite profiles (Table 1). Three strains that produced deoxynivalenol but no detectable acetylated derivatives were considered to have unknown chemotypes. In addition, Fg5 strains were considered to have unknown chemotypes because they either produced equal amounts of deoxynivalenol and nivalenol in a single trial or alternated between the production of these metabolites in replicate trials.
DNA Sequencing.
Primers were designed from published sequences (GenBank accession nos. U22462, U22463, AF364179, AF011355, AB024617, AB017495, and U22464) to amplify and sequence overlapping segments of an approximately 19-kb region of the TRI-cluster (Fig. 1) from the 39 strains listed in Table 1. Sequences for six single-copy nuclear genes (noncluster genes) described by O'Donnell et al (2) were obtained for Fg8 strains, and were aligned with previously published sequences (GenBank accession nos. AF212435–AF212825). Alignments were performed by using SEQUENCHER 4.1.2 (Gene Codes, Ann Arbor, MI) or WISCONSIN PACKAGE 10.2 (Genetics Computer Group, Madison, WI) and were improved manually.
Figure 1.
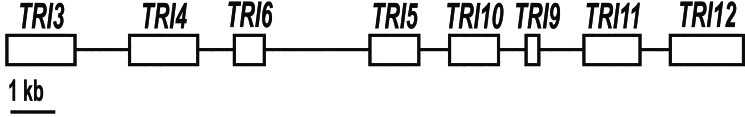
Organization of the trichothecene gene cluster in Fusarium. The genes sequenced include a 15-O-acetyltransferase (TRI3), two P450 oxygenases (TRI4 and TRI11), a transcription factor (TRI6), trichodiene synthase (TRI5), a regulatory gene (TRI10), a trichothecene efflux pump (TRI12), and a predicted protein of unknown function (TRI9) (9, 18).
Phylogenetic Analyses and Genetic Distance Estimation.
Maximum parsimony analyses were conducted by using the heuristic search algorithm of PAUP* V4.0b8 (12). Phylogenetic reconstructions based on variation at synonymous sites were performed by using the modified Nei–Gojobori method with a Jukes–Cantor correction as implemented in MEGA 2.1 (13). Relative support for individual nodes was assessed by nonparametric bootstrapping with 2,000 pseudoreplications of the data. TRI-cluster phylogenies were rooted with sequences from F. sporotrichioides (AF359360) or Myrothecium roridum (AF009417 and AF009416), and the combined noncluster phylogeny was rooted with TRI101 sequence from F. sporotrichioides (AF127176). A previously identified (2) recombination block within NRRL 28721 was removed before analyses involving noncluster genes. Kishino–Hasegawa tests performed with PAUP* V4.0b8 were used to assess concordance between gene trees and to test the fit of alternative phylogenetic hypotheses to various data partitions. Parsimony reconstructions of trichothecene chemotype evolution were obtained by using MACCLADE 4.0 (14). Genetic distance estimates were obtained as described for phylogenetic analyses using MEGA 2.1, with standard errors estimated by using the bootstrap method and 500 pseudoreplications of the data. The significance of differences in genetic distance estimates was assessed by using one-tailed t tests and infinite degrees of freedom.
Southern Hybridization and Chemotype-Specific PCR.
Genomic DNA from representatives of Fg6, Fg7, Fusarium lunulosporum, Fusarium cerealis, Fusarium culmorum, and Fusarium pseudograminearum was digested with HindIII and hybridized with a digoxigenin-labeled probe produced by using primers designed to amplify a 351-bp region of TRI12.
Sets of primers specific to individual chemotypes were designed from TRI3 and TRI12 sequences and used in noncompetitive PCR.
Analysis of Recombination.
Evidence of a history of recombination within the TRI-cluster sequences was assessed by using the informative-sites test implemented in PIST 1.0 (15), with maximum likelihood estimates of tree topologies and model parameters (HKY85+dΓ4) generated with PAUP* V4.0b8. Identification of individual recombination blocks was conducted by using RDP 1.07 (16).
Analysis of Selective Pressure.
The impact of selection on molecular evolution (selective constraint) was investigated for seven TRI-cluster genes. TRI9 was not examined because of its small size (132 bp). Before these analyses, recombination blocks identified by using RDP 1.07 were removed from sequence alignments. Likelihood ratio tests employing codon models of molecular evolution (17) were used to test for heterogeneous selective pressure among amino acid sites and adaptive evolution. The two-cluster test implemented in lintree (18) was used to identify significant differences in rates of amino acid substitution along branches leading to different chemotype lineages in individual TRI-cluster gene trees. Maximum likelihood methods (19) also were used to determine whether adaptive evolution contributed to increased amino acid substitution rates in these branches. Maximum likelihood analyses were performed by using the codeml program of the PAML package (20), with topologies estimated with the Neighbor Joining algorithm (HKY85) implemented in PAUP* V4.0b8.
Results
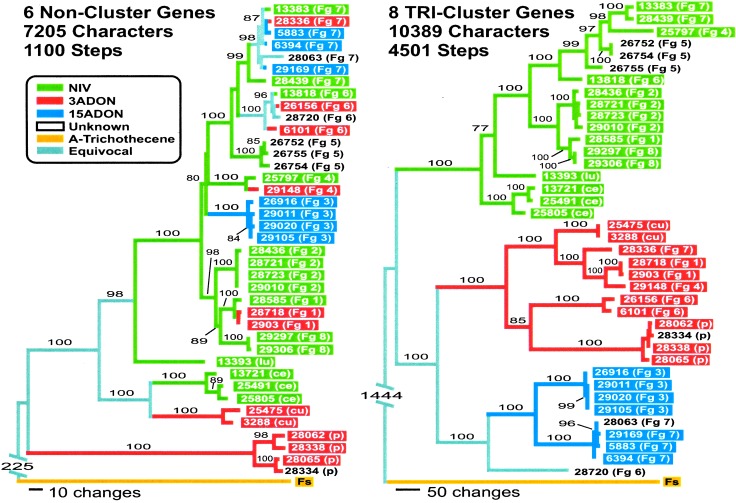
Significant incongruence (P < 0.001) was evident between combined trees for noncluster and TRI-cluster genes respectively (Fig. 2). In the combined noncluster tree, the monophyly of each of the species examined, and of the Fg complex, was strongly supported (bootstrap scores ≥99%) and was congruent with well-resolved clades in individual noncluster gene trees. However, the pattern of trichothecene chemotype evolution inferred from this noncluster tree suggested that either NIV, 3ADON, and 15ADON chemotypes had multiple independent evolutionary origins or the evolutionary history of these chemotypes was different from that predicted by the species phylogeny (Fig. 2). Phylogenetic trees derived by using topological constraints that forced strains into reciprocally monophyletic groups based on chemotype were significantly worse than unconstrained most parsimonious trees (242 additional steps; P < 0.001). In contrast, the Fg complex and five of the eight species within the Fg complex were nonmonophyletic in the combined TRI-cluster gene tree, whereas reciprocally monophyletic lineages corresponding to each of the B-trichothecene chemotypes were recovered in 100% of bootstrap replicates from the combined TRI-cluster gene data (Fig. 2). Constraints that forced the monophyly of the Fg complex onto the combined TRI-cluster gene data were significantly worse than the unconstrained most parsimonious trees (445 additional steps; P < 0.001), and at least 3,944 additional steps were required to produce a tree consistent with any of the four most parsimonious trees recovered from a combined analysis of noncluster genes (P < 0.001). Despite the exceptional differences between the combined noncluster and TRI-cluster trees, evolutionary relationships among strains within each of the chemotype lineages recovered in the combined TRI-cluster gene tree were largely consistent with species limits indicated by the combined noncluster tree (Fig. 2).
Figure 2.
One of four most parsimonious phylograms (differing only in the position of the Fg4 and Fg3 clades) inferred from a combined analysis of six noncluster genes, and one of three most parsimonious phylograms (differing only in the relationships within Fg2) inferred from a combined analysis of eight TRI-cluster genes. Colored blocks indicate the trichothecene chemotype of each strain, and colored branches reflect parsimony-based reconstructions of trichothecene chemotype evolution. In-group strains are identified by their Agricultural Research Service Culture Collection (NRRL) number followed by an abbreviation in parentheses indicating species: Fg complex (Fg 1–8), F. lunulosporum (lu), F. cerealis (ce), F. culmorum (cu), and F. pseudograminearum (p). F. sporotrichioides is abbreviated as Fs. The frequency (%) with which a given branch was recovered in 2,000 bootstrap replications is shown above all branches recovered in more than 70% of bootstrap replicates.
Phylogenetic analysis of sequences from paralogous genes can distort gene trees relative to species phylogenies. However, the results of Southern hybridizations using a probe derived from TRI12 were consistent with expectations for a single-copy gene. In addition, two sets of primers specific to the individual chemotype lineages were designed from TRI3 and TRI12 sequences. For all 39 strains examined, only one amplicon was produced for each set of chemotype-specific primers. The results of chemotype-specific amplifications from TRI3 and TRI12 were consistent within a strain, and were also consistent with the position of individual strains within the TRI-cluster phylogeny, indicating that the strains examined do not harbor heterogeneous populations of TRI-clusters belonging to different chemotype lineages. In addition, phylogenetic trees derived from variation at synonymous sites (not shown) were nearly identical to those shown in Fig. 2, indicating that differences between the combined TRI-cluster gene tree and the species phylogeny are not the result of convergent evolution.
The demonstration that TRI-cluster haplotypes group according to chemotype rather than by species indicated that NIV, 3ADON, and 15ADON chemotypes each have a single evolutionary origin (Fig. 2). Unfortunately, it was not possible to reliably calibrate a molecular clock for the Fusarium species examined. However, TRI4 and TRI5 gene trees, rooted with homologous sequences from M. roridum, were constructed (not shown) to assess the relative age of the trichothecene chemotype polymorphism. Monophyly of the B-trichothecene lineage was strongly supported (bootstrap score = 100%) relative to the position of F. sporotrichioides, indicating that TRI-cluster haplotype polymorphisms observed in the B-trichothecene lineage originated after the divergence of A- and B-trichothecene-producing species. In comparisons of F. sporotrichioides with the B-trichothecene producers listed in Table 1, average divergence at synonymous sites within TRI101 (47.8% ± 4.5%), a noncluster gene, was not significantly different from that for TRI-cluster genes (42.9% ± 1.6%; P > 0.15), indicating that comparisons between TRI-cluster genes and TRI101 could be used to estimate the age of the chemotype polymorphism relative to speciation events. Average genetic distances between chemotype lineages at synonymous sites within the TRI-cluster ranged from 13.1% ± 0.5% to 15.5% ± 0.7%, and were significantly greater than synonymous site distances (estimated from TRI101) between any of the B-trichothecene producing species (range = 0.5% ± 0.3% to 9.3%± 1.6%) (P < 0.05). Taken together, these results indicate that NIV, 3ADON, and 15ADON chemotypes originated in the ancestor of extant species within the B-trichothecene lineage of Fusarium, and that chemotype polymorphisms have been maintained through multiple speciation events.
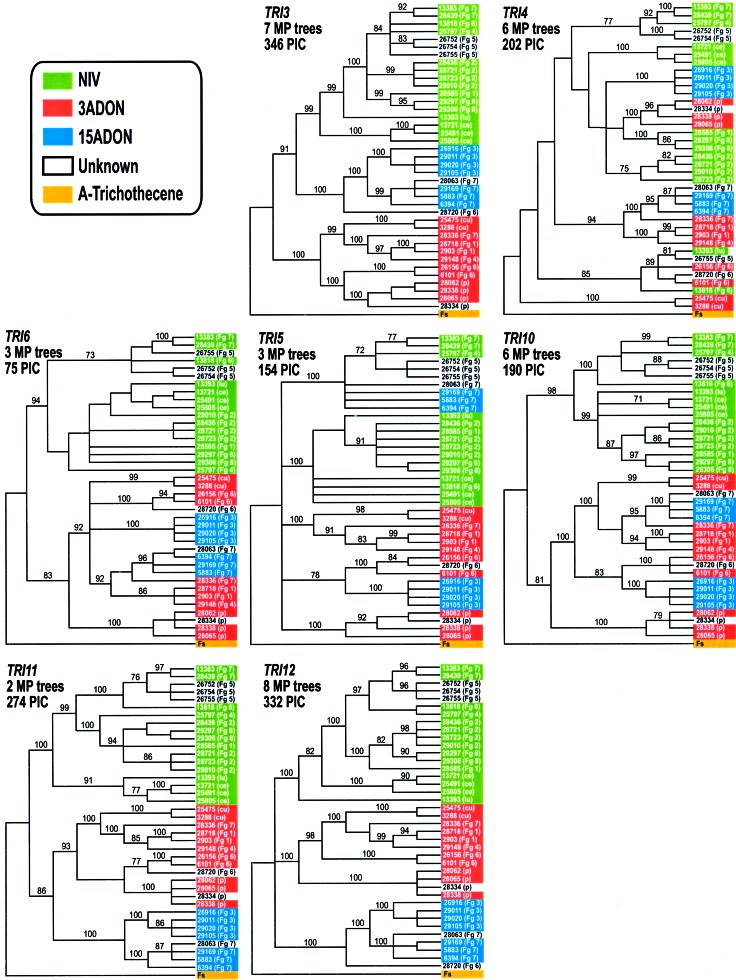
Despite strong bootstrap support for virtually every node in the combined TRI-cluster gene tree (Fig. 2), multiple differences were evident between phylogenies derived from individual TRI-cluster genes (Fig. 3). Differences were also observed between phylogenies derived from individual intergenic regions of the TRI-cluster (not shown). However, none of the individual TRI-cluster phylogenies were congruent with the combined noncluster tree (Fig. 2) or with the monophyly of the Fg complex (P < 0.001). Results of the informative-sites test indicated a significant history of recombination within the TRI-cluster (P < 0.001). When RDP 1.07 (window size = 100 sites, P < 0.05) was used, 92 possible recombination events were inferred from the TRI-cluster sequence alignment. Although many of these events were obviously not independent, the large number and overlapping nature of potential recombination blocks made it impossible to reconstruct the true pattern of recombination across the entire TRI-cluster. These results indicate that TRI-cluster haplotypes are mosaics of multiple evolutionary histories brought together by recombination. However, reciprocally monophyletic groups, corresponding to each of the B-trichothecene chemotypes, were strongly supported (bootstrap scores ≥93%) in TRI3, TRI11, and TRI12 gene trees (Fig. 3), and in the most parsimonious trees derived from the intergenic region between TRI11 and TRI12, indicating that recombination between chemotype lineages has been limited at either end of the sequenced region.
Figure 3.
Strict consensus cladograms derived from maximum parsimony (MP) analyses of individual TRI-cluster genes. Individual analyses of TRI9 were not performed because this gene contained only seven parsimony-informative characters (PIC). Colored blocks indicate the trichothecene chemotype of each strain. Individual strains are labeled as indicated in Fig. 2. The frequency (%) with which a given branch was recovered in 2,000 bootstrap replications is shown above all branches recovered in more than 70% of bootstrap replicates.
The majority of sites in each of the TRI-cluster genes were dominated by strong purifying selection, with the ratio of nonsynonymous to synonymous substitution (ω) < 0.08 (ω = 1 when all amino acid changes are neutral). However, a small proportion (f) of sites also were found to be evolving under positive selection (P < 0.05) within TRI4 and TRI10 (ω = 11.6 and 2.3, f = 1% and 3% respectively). In addition, the rate of amino acid substitution in TRI3 and TRI11 was found to be significantly (P < 0.05) greater in the branch leading to the 3ADON lineage (3ADON branch) than in branches leading to the NIV or 15ADON lineages (Fig. 3). TRI3 and TRI11 were analyzed with a codon model allowing changes in selective constraints at predefined branches (19). Results suggested that positive selection (ω = 7.59) at a small fraction (7%) of sites contributed to the increased rate of amino acid substitution along the 3ADON branch in TRI11, and that a relaxation of functional constraint along the 3ADON branch (ω < 1) contributed to the increased rate of amino acid substitution in TRI3.
Discussion
The results presented here extend the previously published observation that trichothecene chemotype differences are not well correlated with evolutionary relationships within the Fg clade (2), and demonstrate that these chemotypes have a polyphyletic distribution within the B-trichothecene lineage of Fusarium (Fig. 2). We used molecular phylogenetic methods to examine the basis for this discord between species and toxin evolution, and demonstrated that each of the B-trichothecene chemotypes had a single evolutionary origin in the ancestor of extant species within the B-trichothecene lineage of Fusarium (P < 0.05), and that polymorphism within these virulence-associated genes has persisted through multiple speciation events in this important group of plant pathogens (Fig. 2). The results of Southern hybridizations, chemotype-specific PCR amplifications, and phylogenetic analyses of variation at synonymous sites indicated that the discord between TRI-cluster and noncluster phylogenies is not caused by paralogy or convergence. Nonvertical transmission of trichothecene haplotypes by means of introgressive hybridization or horizontal gene transfer could also distort phylogenies derived from TRI-cluster genes. However, explanations based on nonvertical transmission between the examined species fail to account for the observation that TRI-cluster haplotype polymorphisms predate the divergence of extant species in the B-trichothecene lineage of Fusarium. Therefore, the results presented here indicate that the polyphyletic distribution of trichothecene chemotypes relative to the species phylogeny (Fig. 2) is the result of nonphylogenetic sorting of ancestral polymorphism into descendant species and the sharing of ancestral polymorphism among extant species (transspecies polymorphism), collectively referred to here as transspecies evolution.
Neutral polymorphisms have relatively short residence times in populations and are unlikely to persist through multiple speciation events (21). Large effective population sizes and rapid speciation may promote transspecies evolution for neutral variants. However, the impact of these historical circumstances should be evident across the genome, and patterns of transspecies evolution were not observed within the six noncluster genes examined. Therefore, the demonstration of transspecies evolution for TRI-cluster genes (Fig. 2) indicates that polymorphism within the TRI-cluster has been maintained through multiple speciation events by balancing selection. Although it is also possible that the maintenance of polymorphism within the TRI-cluster is an indirect result of selection acting on a tightly linked locus that is not involved in trichothecene biosynthesis, the recovery of reciprocally monophyletic TRI-cluster lineages that correspond to chemotype differences and also reflect transspecies evolution (Fig. 2) strongly suggests that balancing selection has acted directly on chemotype differences.
The joint effects of selection and linkage can result in extended residence times for neutral polymorphisms closely linked to a site under balancing selection (22). This hitchhiking effect will decrease with increasing distance from the selected site as recombination uncouples the evolutionary dynamics of neutral polymorphisms from those under selection. However, with balancing selection acting on combinations of linked sites, the impact of hitchhiking can be extended over a much greater chromosomal region than predicted by a single-locus model, and extended residence times for neutral polymorphisms located between selected loci can be expected even with relatively high recombination rates (22). Despite evidence of recombination (informative sites test, P < 0.001), patterns of transspecies evolution were identified in gene trees from across a 19-kb region of the TRI-cluster (Fig. 3). In addition, reciprocally monophyletic lineages corresponding to each of the B-trichothecene chemotypes were recovered from either end of the sequenced region, whereas phylogenies from the center of this region were less well correlated with chemotype differences (Fig. 3). These results are inconsistent with a single-locus model of balancing selection, which predicts that reciprocally monophyletic chemotype lineages should be recovered from across the entire TRI-cluster (with little or no recombination) or from a single region of the TRI-cluster surrounding the selected site (with higher levels of recombination), and suggest that the evolutionary dynamics of the TRI-cluster have been impacted by the presence of interacting balanced polymorphisms located within or near both ends of the sequenced region.
Evidence of chemotype-specific shifts in functional constraint at sites within TRI3 and TRI11 is interesting because such shifts may be associated with changes in protein function (23), and could indicate that variation within these genes either has a direct impact on chemotype or contributes to the observed pattern of transspecies evolution through epistatic interactions with polymorphisms that are directly responsible for chemotype differences. Although a more thorough discussion awaits additional information on the functional significance of variation within TRI-cluster genes and a detailed characterization of the late pathway steps differentiating NIV, 3ADON, and 15ADON biosynthesis, it is unlikely that variation within identified trichothecene genes is entirely responsible for the observed chemotype polymorphism. For instance, hydroxylation at C-4 distinguishes nivalenol from deoxynivalenol, but is not accounted for by the putative activities of any of the trichothecene genes identified to date (6). However, the recovery of reciprocally monophyletic chemotype lineages in the combined TRI-cluster phylogeny (Fig. 2), and evidence from genetic linkage analyses (24) indicate that the genetic variation responsible for hydroxylation at C-4 is closely linked to the sequenced region of the TRI-cluster. Therefore, the sequence data reported here are currently being extended to identify the boundaries of the TRI-cluster and to characterize the distribution of genetic variation within all TRI-cluster genes relative to species limits and trichothecene chemotype differences.
The results of this study provide the first evidence of balancing selection and transspecies evolution identified from analyses of fungal genes involved in virulence or secondary metabolism. Previous instances where balancing selection has resulted in transspecies evolution include loci with roles in self-recognition and immune response such as the vertebrate major histocompatibility complex (25), the self-incompatibility locus in plants (26), and a fungal heterokaryon-incompatibility locus (27), where overdominant (heterozygote advantage) or frequency-dependent selection are thought to be responsible for the maintenance of polymorphism. Environmental heterogeneity can also create distinct selective regimes that promote stable polymorphism (28). The present data do not address the mechanism responsible for balancing selection within the TRI-cluster. However, over-dominant selection is unlikely because the species examined are haploid except for a transient diploid stage during sexual reproduction, suggesting that some form of spatial or temporal heterogeneity in selective pressure exists such that the selectively optimal trichothecene chemotype varies by environment or changes over time. Host-specific differences in susceptibility to specific trichothecenes have been reported (29). However, systematic investigations of the impact of chemotype differences on host range and fitness in different environments are needed.
In conclusion, polymorphism within the TRI-cluster is transspecific and appears to have been maintained by balancing selection acting on chemotype differences. As a result, species within the Fg complex, the primary etiological agents of scab, exhibit unexpectedly high levels of genetic variation within these virulence-associated genes.
These findings have important implications for the control of scab and the reduction of mycotoxin contamination of cereals worldwide. First, they suggest that both species limits and chemotype differences need to be considered in plant breeding efforts to increase the likelihood that broad-based resistance, effective against all scab pathogens, is achieved. Second, the demonstration that divergent TRI-cluster haplotypes, corresponding to different trichothecene chemotypes, are segregating in at least four of the eight species within the Fg complex (Fig. 2), indicates that disease control and plant quarantine programs need to exercise increased vigilance to ensure that international trade in agricultural commodities does not result in the global transposition of strains with novel toxigenic potential. In that regard, the chemotype-specific PCR tests developed in the present study provide a rapid and direct genetic method for distinguishing among NIV-, 3ADON-, and 15ADON-producing strains, and could be incorporated into global monitoring programs to evaluate contamination of cereals and characterize biogeographic or host-specific differences in chemotype distributions. Lastly, the evolutionary framework developed in the present study provides the necessary context for additional comparative studies urgently needed to understand the ecology, virulence, and population dynamics of cereal scab pathogens.
Supplementary Material
Acknowledgments
We thank those individuals who made strains available through various culture collections. We also thank Steve Prather for preparing the figures, David Posada for advice on recombination analyses, Cletus Kurtzman for helpful criticisms of the manuscript, and Weiping Xie and the trichothecene diagnostic services of the University of Minnesota supported by the U.S. Wheat and Barley Scab Initiative. J.P.B. was supported by Biotechnology and Biological Sciences Research Council Grant 31/G10434.
Abbreviations
- Fg complex
Fusarium graminearum species complex
- NIV
nivalenol and acetylated derivatives
- 3ADON
3-acetyldeoxynivalenol
- 15ADON
15-acetyldeoxynivalenol
- TRI-cluster
trichothecene gene cluster
Note Added in Proof.
Lee et al. (30) have identified a trichothecene C–4 hydroxylase (TRI13) less than 1 kb away from TRI12.
Footnotes
References
- 1.Windels C E. Phytopathology. 2000;90:17–21. doi: 10.1094/PHYTO.2000.90.1.17. [DOI] [PubMed] [Google Scholar]
- 2.O'Donnell K, Kistler H C, Tacke B K, Casper H H. Proc Natl Acad Sci USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook R J. In: Fusarium: Diseases, Biology and Taxonomy. Nelson P E, Toussoun T A, Cook R J, editors. University Park: Pennsylvania State Univ. Press; 1981. pp. 39–52. [Google Scholar]
- 4.Ueno Y, Nakajima M, Sakai K, Ishii K, Sato N, Shimada N. J Biochem. 1973;74:285–296. [PubMed] [Google Scholar]
- 5.Proctor R H, Hohn T M, McCormick S. Mol Plant–Microbe Interact. 1995;8:593–601. doi: 10.1094/mpmi-8-0593. [DOI] [PubMed] [Google Scholar]
- 6.Brown D W, McCormick S, Alexander N J, Proctor R H, Desjardins A E. Fungal Genet Biol. 2001;32:121–133. doi: 10.1006/fgbi.2001.1256. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M, Kaneko I, Komiyama M, Takatsuki A, Koshino H, Yoneyama K, Yamaguchi I. J Biol Chem. 1998;273:1654–1661. doi: 10.1074/jbc.273.3.1654. [DOI] [PubMed] [Google Scholar]
- 8.Miller J D, Greenhalgh R, Wang Y, Lu M. Mycologia. 1991;83:121–130. [Google Scholar]
- 9.Alexander N J, Hohn T M, McCormick S P. Appl Environ Microbiol. 1998;64:221–225. doi: 10.1128/aem.64.1.221-225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirocha C J. J Agric Food Chem. 1998;46:1414–1418. [Google Scholar]
- 11.Tag A G, Garifullina G F, Peplow A W, Ake C, Phillips T D, Hohn T M, Beremand M N. Appl Environ Microbiol. 2001;67:5294–5302. doi: 10.1128/AEM.67.11.5294-5302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swofford D L. paup* (Sinauer Associates, Sunderland, MA), Version 4.0b8. 2001. [Google Scholar]
- 13.Kumar S, Tamura K, Jakobsen I B, Nei M. mega (Arizona State Univ., Tempe), Version 2.1. 2001. [Google Scholar]
- 14.Maddison D R, Maddison W P. macclade (Sinauer Associates, Sunderland, MA), Version 4.0. 2000. [Google Scholar]
- 15.Worobey M. Mol Biol Evol. 2001;18:1425–1434. doi: 10.1093/oxfordjournals.molbev.a003928. [DOI] [PubMed] [Google Scholar]
- 16.Martin D, Rybicki E. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Nielsen R, Goldman N, Pederson A-M K. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takezaki N, Rzhetsky A, Nei M. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Nielsen R. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 21.Takahata N, Nei M. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly J K, Wade M J. J Theor Biol. 2000;204:83–101. doi: 10.1006/jtbi.2000.2003. [DOI] [PubMed] [Google Scholar]
- 23.Knudesen B, Miyamoto M M. Proc Natl Acad Sci USA. 2001;98:14512–14517. doi: 10.1073/pnas.251526398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurgenson J E, Bowden R L, Zeller K A, Leslie J F, Alexander N J, Plattner R D. Genetics. 2002;160:1451–1460. doi: 10.1093/genetics/160.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 26.Ioerger T R, Clark A G, Kao T-H. Proc Natl Acad Sci USA. 1990;87:9732–9735. doi: 10.1073/pnas.87.24.9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J, Saupe S J, Glass N L. Proc Natl Acad Sci USA. 1998;95:12398–12403. doi: 10.1073/pnas.95.21.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt P S, Bertness M D, Rand D M. Proc R Soc London B. 2000;267:379–384. doi: 10.1098/rspb.2000.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhitch M J, McCormick S P, Alexander N J, Hohn T M. Plant Sci. 2000;157:201–207. doi: 10.1016/s0168-9452(00)00282-x. [DOI] [PubMed] [Google Scholar]
- 30.Lee T, Han Y-K, Kim K-H, Yun S-H, Lee Y-W. Appl Environ Microbiol. 2002;68:2148–2154. doi: 10.1128/AEM.68.5.2148-2154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.