Abstract
We followed adaptation in experimental microbial populations to inhibitory concentrations of an antimicrobial drug. The evolution of drug resistance was accompanied in all cases by changes in gene expression that persisted in the absence of the drug; the new patterns of gene expression were constitutive. The changes in gene expression occurred in four replicate populations of the pathogenic fungus Candida albicans during 330 generations of evolution in the presence of the antifungal drug fluconazole. Genome-wide expression profiling of over 5,000 ORFs identified 301 whose expression was significantly modulated. Cluster analysis identified three distinct patterns of gene expression underlying adaptation to the drug. One pattern was unique to one population and included up-regulation of the multidrug ATP-binding cassette transporter gene, CDR2. A second pattern occurred at a late stage of adaptation in three populations; for two of these populations profiled earlier in their evolution, a different pattern was observed at an early stage of adaptation. The succession of early- and late-stage patterns of gene expression, both of which include up-regulation of the multidrug major facilitator transporter gene, MDR1, must represent a common program of adaptation to this antifungal drug. The three patterns of gene expression were also identified in fluconazole-resistant clinical isolates, providing further evidence that these patterns represent common programs of adaptation to fluconazole.
The emergence of drug resistance is an evolutionary process common to all microorganisms exposed to antimicrobial drugs (1). The evolution of antifungal drug resistance now poses a growing public health problem because of the sharp increase in the incidence of opportunistic fungal infections in recent years (2, 3). Currently available antifungal drugs have a limited number of targets, primarily ergosterol and its biosynthesis, nucleic acid synthesis, and cell wall synthesis (4, 5). Resistance is documented for all of the antifungal drugs that have been widely deployed therapeutically (6). Resistance evolves because antimicrobial agents rarely achieve 100% mortality in the microbial population and natural selection operates on the survivors. Whenever the microbial pathogen population remains large enough through a course of treatment, the evolution of resistance is all but inevitable. The emergence of resistance mutations under natural selection can be observed in experimental populations evolving in the presence of inhibitory concentrations of a drug (7). How specific mutations conferring drug resistance are expressed in a phenotype depends on interactions with other genes (8). Genome-wide gene expression profiles of experimental populations can provide a finely resolved molecular phenotype that reflects the complex translation of genotype to phenotype (9). Starting with a very large number of possible changes in gene expression profiles, we found that these changes were actually channeled along only two paths—evidence that the number of adaptive solutions available through mutation is limited.
Initially isogenic experimental populations of the diploid pathogenic yeast Candida albicans were evolved with and without inhibitory concentrations of the antifungal drug fluconazole over 330 generations (10). The populations evolved with the drug diverged in their adaptive trajectories, reaching different levels of drug resistance associated with distinct molecular mechanisms of resistance (10) and with distinct fitness characteristics (11). In the present study, we measured changes in genome-wide gene expression in these evolved populations relative to their common ancestor by using DNA microarrays. In contrast to other microarray-based studies of gene expression in yeasts that have focused on the immediate effects of development (12), regulators (13), antifungal drugs (14, 15), and stressful conditions (16), our interest was in the changes in gene expression that became established during adaptation and persisted in the absence of the drug. The drug resistance phenotypes were stable during 50 generations in the absence of drug (10), reflecting genetic change rather than transient physiological response to exposure to the drug. We therefore profiled populations grown in a uniform environment without the drug. We sampled populations for transcriptional profiling (Table 1) to encompass the range of minimum inhibitory concentrations (MICs) of fluconazole, expression patterns of four genes involved in azole resistance, and fitness (10, 11). We measured gene expression profiles of four populations that evolved with the drug for 330 generations. For two of these populations we also assayed gene expression profiles at earlier time points. As a control for adaptation to the culture conditions, we measured the gene expression profile of one population that evolved without the drug.
Table 1.
Population samples and microarray experiments
| Population sample | Fluconazole MIC, μg/ml | Fitness with drug* | Fitness without drug | Microarray (no. ORFs) | Replicate hybridizations‡ |
|---|---|---|---|---|---|
| N4-330 | 0.25 | 0.10 ± 0.27 | 0.31 ± 0.12† | 4,651 | 6 |
| 3,609 | 4 | ||||
| D8-330 | 4.0 | 1.72 ± 0.27† | 0.46 ± 0.06† | 4,651 | 6 |
| 3,609 | 4 | ||||
| D9-165 | 4.0 | — | — | 5,669 | 6 |
| D9-330 | 64.0 | 0.62 ± 0.21† | −0.25 ± 0.11 | 4,651 | 6 |
| 3,609 | 4 | ||||
| D11-330 | 64.0 | −0.21 ± 0.14 | −0.27 ± 0.09 | 5,669 | 6 |
| D12-165 | 4.0 | — | — | 5,669 | 6 |
| D12-260 | 64.0 | −0.11 ± 0.07 | −0.35 ± 0.01† | 4,651 | 8 |
| 3,609 | 2 | ||||
| D12-330 | 4.0 | 0.38 ± 0.18 | −0.09 ± 0.12 | 5,669 | 6 |
In a previous study (11), fitness was measured relative to the genetically marked ancestor as the difference in the number of doublings of the two competitors (evolved population sample minus ancestor), standardized by the total number of doublings in the competition assay. Fitness measurements are the average from three replicates ± 95% confidence intervals. For competition experiments with the drug, the concentrations of fluconazole used were: 0.5 μg/ml for N4-330 and 128 μg/ml for the remaining population samples.
The difference in the number of doublings of the two competitors was significant (paired t test, P < 0.05).
The replicate hybridizations include an equal number of experiments with the paired samples reciprocally labeled. For each set of reciprocal hybridizations an independent RNA preparation was used, for both the evolved population sample and the ancestor.
We anticipated three possible outcomes from the comparison of genome-wide expression profiles. First, given the diversity of adaptive responses, all of the populations could have reached unique adaptive solutions with expression profiles of little or no similarity to one another. Second, if there are constraints on the evolution of drug resistance defining a limited number of adaptive solutions accessible by a small number of mutational steps, then the populations could have converged on several different adaptive solutions, forming distinct groups based on similarity of expression profiles. Third, if there is only one adaptive solution available to a genotype in a specific environment, then the populations could have converged on one adaptive solution with very similar expression profiles. The only other study characterizing genome-wide changes in gene expression associated with adaptation found the third outcome, with systematic changes in gene expression profiles in three yeast populations following evolution in a glucose-limited environment (17). We found the second outcome—the populations converged on two adaptive solutions by the end of the evolution experiment. These adaptive solutions must represent common pathways for the evolution of drug resistance, because the patterns of gene expression identified in the experimental populations also occurred in fluconazole-resistant, clinical isolates of C. albicans.
Methods
The Experimental Populations.
The evolutionary history of the experimental populations of C. albicans profiled in this study has been described (10). Briefly, 12 replicate populations were established from a single drug-sensitive cell and were serially propagated for 330 generations in RPMI 1640 medium (18) at 35°C with constant agitation. Population samples were archived in 1 ml of 40% (vol/vol) glycerol containing 3% (wt/vol) trisodium citrate at −70°C. Population N4 was one of six populations that were evolved without the drug. Because the six populations evolved without drug followed the same adaptive trajectories, one population was randomly selected for this study to control for adaptation to the culture conditions. Populations D8, D9, D11, and D12 were four of six populations that were evolved in the presence of twice their most recently measured MIC of fluconazole (Roerig-Pfizer, New York). These populations were selected for genome-wide expression profiling because they represented the diversity of adaptive trajectories, MICs, fitness profiles, and expression patterns of four genes known to be involved in azole resistance (10, 11). All of the populations included in this study were profiled at generation 330, the end of the evolution experiment. Populations D9 and D12 were also profiled at other time points in their evolutionary history to address how gene expression changed during the course of adaptation. Populations D9 and D12 were both profiled at generation 165, the midpoint of their evolutionary history and during their ascent in MIC of fluconazole. Population D12 was also profiled at generation 260, which was the generation that this population reached a peak MIC, before subsequently declining in MIC during continued evolution at the high drug concentration. MICs were determined by broth microdilution (18).
Clinical Isolates.
Thirty clinical isolates were screened for expression levels of the nine genes showing the largest change in expression in the microarray experiments (ADH4, MDR1, YPL88, YPX98, YPR127W, GRE99, YNL229C, HYR1, and HSP12) and for expression levels of CDR2. Twenty-nine isolates were from oral swabs from HIV-infected patients (19) and one (B1) was from a blood culture from a patient (20). These 30 isolates were selected because their MICs of fluconazole were ≥4 μg/ml.
Genome-Wide Gene Expression Analysis.
DNA microarrays with 51–86% coverage of the C. albicans genome (3,609–5,669 ORFs) were produced at the Biotechnology Research Institute, National Research Council, Montreal (http://dirac.bri.nrc.ca/microarraylab/) (21). For transcriptional profiling, independent replicates of each population sample (Table 1) were grown overnight from the frozen archives in 50 ml of YPD (1% yeast extract, 2% Bacto Peptone, and 2% d-glucose) at 30°C and 250 rpm. Each overnight culture was diluted to an optical density at 600 nm (OD600) of 0.05 in 100 ml of YPD and was grown until an OD600 of between 0.5 and 0.6. Cells were harvested by centrifugation for 2 min at 4,000 rpm at room temperature and cell pellets were frozen in liquid nitrogen and stored at −70°C. Total RNA was isolated using hot phenol, essentially as described (http://dirac.bri.nrc.ca/microarraylab/micro/candida_e.html). mRNA was purified using the Oligotex Spin-Column Protocol (Oligotex mRNA Maxi Kit, Qiagen, Valencia, CA). Each of the paired mRNA samples was reciprocally labeled with Cy3 and Cy5 (i.e., evolved-Cy3 vs. ancestor-Cy5 and evolved-Cy5 vs. ancestor-Cy3) for hybridization to two microarrays as described (http://dirac.bri.nrc.ca/microarraylab/micro/candida_e.html). Microarrays were scanned using ScanArray 5000 (Packard BioScience, Billerica, MA) and the intensity of spots was quantified with QuantArray. quantarray files were analyzed in excel spreadsheets (Microsoft, Redmond, WA). To be included in the normalization and analysis, each spot had to satisfy three quality control criteria: (i) the signal intensity had to be significantly greater than local background (the signal intensity minus half of the standard deviation had to be greater than the local background plus half of the standard deviation); (ii) the signal intensity had to be within the dynamic range of the photomultiplier tube; and (iii) the raw intensities of the duplicate spots for each gene had to be within 50% of one another. For spots that met these criteria, the ratio of intensity of the two channels was normalized by the median ratio for the entire subarray consisting of 400 spots. Finally, the log2 of the ratios for each duplicate spot was averaged. Statistical analysis and visualization were performed with genespring (Silicon Genetics, Redwood City, CA). Replication for each population sample was sufficient for the Student t test of genespring to measure the significance of changes in gene expression (see Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). Only ORFs that were significantly modulated (P ≤ 0.01) at least 1.5-fold in at least one evolved population sample relative to the ancestor were included in the analysis. The 1.5-fold cut-off, as opposed to the more common 2-fold cut-off, was possible because of the degree of replication in these microarray experiments.
Northern Analysis.
Three replicates of each of the population samples profiled in this study and one replicate of each of the clinical isolates were grown essentially as described above, except in 10 ml of YPD. RNA was prepared using glass beads (22) and Northern blots were prepared according to a standard protocol (23). The following primers were used for amplification of the ten probes from genomic DNA of the ancestor of the experimental populations: ADH4, 5′-ACATTTGGGTGGAGAAACAG-3′ and 5′-CCTTCAGTGACCAAAAATCT-3′; MDR1, 5′-TGCCCCAATAGCAATACATA-3′ and 5′-TGCTCTCAACTTTGGTCCGT-3′; YPL88, 5′-TCGTTTAGATCATGAAGTCA-3′ and 5′-ATCTTTGGCGTGATATGGTT-3′; YPX98, 5′-TTGCTTCATCAACAATTACA-3′ and 5′-GCAGCCAAAATATGCTTTCT-3′; YPR127W, 5′-TCGAAATATCGGGAAAGTTT-3′ and 5′-TCCTTCGAGAAATTGATTGT-3′; GRE99, 5′-ACAGTTTTCGTTTCTGGTGC-3′ and 5′-CGAATCGTCAATGGATTTTT-3′; YNL229C, 5′-TCCTTGAAAACCACTTTTCT-3′ and 5′-GACACCTTTGATAACAGCTG-3′; HYR1, 5′-TTTTTTTGCCTCCCTTCTCT-3′ and 5′-TCTTCAATCTTGGTACCGAT-3′; CDR2, 5′-CCAAGAGATAATGATCCAGA-3′ and 5′-ACCCCATCCTTATTTTTTCA-3′; and HSP12, 5′-CCGGAAGAAAAAACATTTCT-3′ and 5′-CGCCGGTTTTGGCACCTTCG-3′. Northern blots were probed sequentially. Blots were also probed with TEF3, which served as an internal control for loaded quantities of RNA (23). Membranes were analyzed with a PhosphorImager (Molecular Dynamics) for quantitative analysis of the signals. Expression values are relative to those of the drug-sensitive ancestor of the experimental populations. Clustering analysis of the population samples and the clinical isolates based on similarity of expression profiles of the ten genes was performed using genespring.
Results
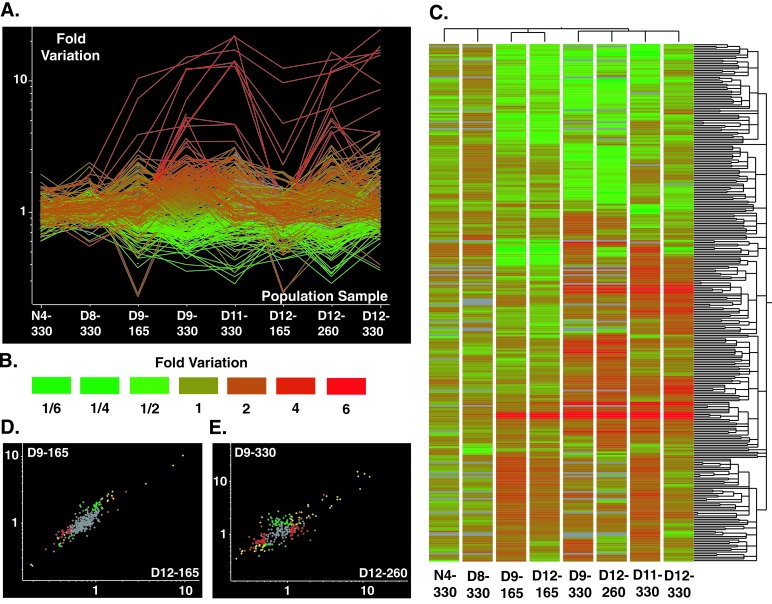
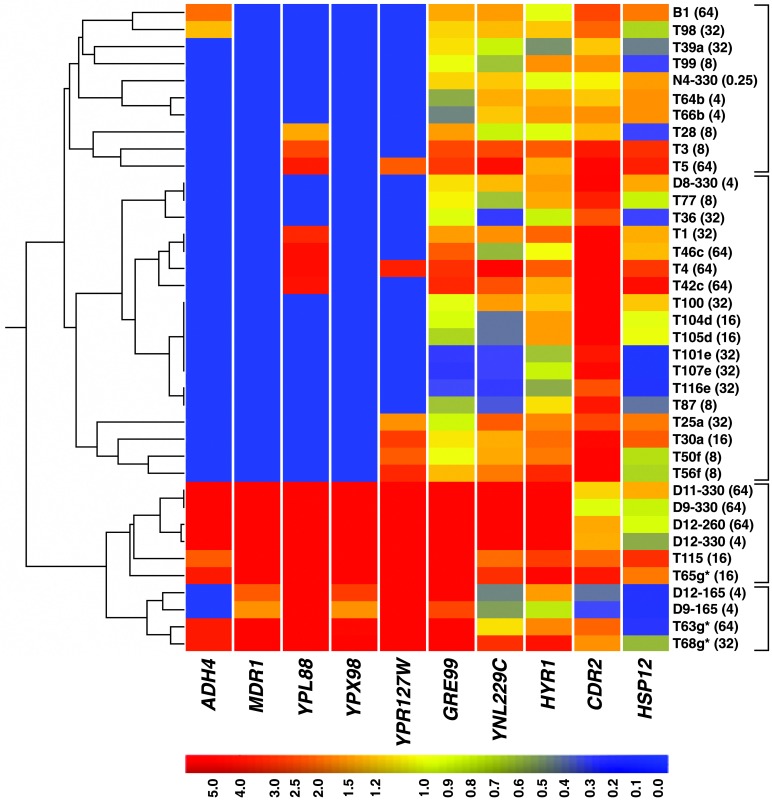
With replicated microarray experiments to monitor evolutionary change among eight population samples (Table 1), we have identified both parallelism and divergence in genome-wide expression profiles. There were 301 ORFs that were significantly modulated (P ≤ 0.01) at least 1.5-fold in at least one evolved population sample relative to the ancestor (Fig. 1A, and see Table 4, which is published as supporting information on the PNAS web site). In a previous study, results from northern analysis of mRNAs of four genes involved in azole resistance (the ATP-binding cassette transporter genes, CDR1 and CDR2, the gene encoding the target enzyme of the azoles, ERG11, and the major facilitator gene, MDR1) were consistent with results obtained with the microarrays (10). In the present study, results from northern analysis of ten genes were also consistent with results obtained with the microarrays (Fig. 2, and see Table 5, which is published as supporting information on the PNAS web site). Three genes were significantly modulated in the population evolved without fluconazole, N4 (EBP1, 2.0-fold, FAS1, 1.6-fold, and GRE2, 1.6-fold; see Table 6, which is published as supporting information on the PNAS web site). Cluster analysis based on similarity of expression profiles of the population samples identified three patterns of gene expression underlying adaptation to the drug (Fig. 1C). One pattern reflects the adaptive solution reached by only one population, D8. The other two patterns reflect an early stage and a late stage of the adaptive solution reached by populations D9, D11, and D12.
Figure 1.
Parallelism and divergence in genome-wide expression profiles. (A) Each line represents the change in expression of one of the 301 ORFs that were significantly modulated at least 1.5-fold (P ≤ 0.01) in at least one evolved population sample. The lines are colored according to the quantitative change in expression in D9-330 (see B). (B) Color scale for quantitative changes in gene expression shown in A and C. Increases in expression relative to the ancestor are shown as shades of red and decreases in expression are shown as shades of green. (C) Cluster analysis of the significantly modulated ORFs and of the experimental populations. Similarity of expression patterns of the 301 ORFs was analyzed using hierarchical clustering based on a matrix of Standard Correlations (not of distances) defined in GENESPRING and is shown as a dendrogram along the vertical axis. Similarity of expression profiles of the population samples was analyzed by the same method and is shown as a dendrogram along the horizontal axis. Data are graphically displayed with color to represent the quantitative changes in each population sample (see B). (D) Correlation in gene expression patterns between D9-165 and D12-165. The color of symbols indicate significance: red, ORFs significant in D9-165; green, ORFs significant in D12-165; yellow, ORFs significant in D9-165 and D12-165; gray, ORFs not significant in D9-165 or D12-165. (E) Correlation in gene expression patterns between D9-330 and D12-260. The color of symbols indicate significance: red, ORFs significant in D9-330; green, ORFs significant in D12-260; yellow, ORFs significant in D9-330 and D12-260; gray, ORFs not significant in D9-330 or D12-260.
Figure 2.
Cluster analysis of expression of ten genes in clinical isolates and samples of experimental populations. Similarity of expression patterns is shown as a dendrogram along the vertical axis. Data are graphically displayed with color to represent the quantitative changes in each sample relative to the ancestor of the experimental populations (see color bar). Designations of clinical isolates begin T or B. Isolates from the same patient share the same lowercase letter. Isolates with identical multilocus genotypes and DNA fingerprints (19) are indicated with an asterisk. MIC of fluconazole (μg/ml) for each isolate appears in parentheses. The brackets indicate the four clusters discussed in the text. The dendrogram was constructed from a matrix of Standard Correlations (GENESPRING), not of distances. Because the colors saturate at 5× expression, not all primary data are represented in the graphic (see Table 5).
The pattern of gene expression of population D8 at generation 330 (D8-330) included few changes relative to the ancestor (Fig. 1A). There were 11 ORFs that were significantly modulated in D8-330, with none showing greater than 2.4-fold change in expression [CDR2, 2.4-fold, YOR49, 2.2-fold, YKR3, 2.0-fold, YPL88, 1.9-fold, HSP12 (2148_0004), 1.8-fold, HSP12 (2734_0007), 1.8-fold, HSP12 (2734_0006), 1.7-fold, YLR63, 1.7-fold, 2690_0009, 1.5-fold, VMA8, 1.5-fold, and YLR336C, 0.6-fold; see Table 7, which is published as supporting information on the PNAS web site]. The gene with the largest change in expression was CDR2, an ATP-binding cassette transporter implicated in efflux of azoles from the cell (6). YLR63 was also overexpressed in D8-330 (see Table 7). This is consistent with the promoters of both CDR2 and YLR63 containing the same cis-acting element that mediates their up-regulation (D. Sanglard, personal communication). CDR1 was not present on the microarrays used for D8-330, although CDR1 was overexpressed in D8-330 based on northern analysis (10) and is often overexpressed in resistant isolates overexpressing CDR2 (23, 24). In population D8, this pattern of gene expression was associated with a low MIC, but very high fitness both in the presence and in the absence of the drug (Table 1).
The other two patterns of gene expression each evolved independently in different populations. The early-stage pattern was observed at generation 165 in D9 and D12, the two populations for which a temporal component was addressed in this study. D9-165 and D12-165 clustered together based on similarity of expression profiles (Fig. 1C), with 64 of the 301 ORFs significantly modulated (P ≤ 0.05) in the same direction in both population samples (Fig. 1D, and see Table 8, which is published as supporting information on the PNAS web site). In populations D9 and D12, the early-stage pattern was replaced by the late-stage pattern. In D9-330 and D12-260, 124 of the 301 ORFs were significantly modulated (P ≤ 0.05) in the same direction (Fig. 1E, and see Table 9, which is published as supporting information on the PNAS web site). The late-stage pattern was also detected in D12-330 and a third population, D11, sampled only at generation 330. The four population samples with the late-stage gene expression pattern (D9-330, D12-260, D12-330, and D11-330) all overexpressed MDR1, a major facilitator transporter implicated in efflux of fluconazole from the cell (6). MDR1 was also overexpressed, but to a lesser magnitude, in the early-stage pattern.
The three patterns of gene expression identified in the experimental populations evolved with fluconazole were also identified in fluconazole-resistant, clinical isolates. Clustering analysis of the population samples and the clinical isolates based on similarity of expression of the nine genes showing the largest changes on the microarrays and based on expression of CDR2 delineated the same three patterns as were identified with the microarrays (Fig. 2, and see Table 5). The early-stage pattern was shown by D9-165 and D12-165, as well as by two clinical isolates. The late-stage pattern was shown by D9-330, D11-330, D12-260, and D12-330, as well as by two clinical isolates. The two clinical isolates showing the early-stage pattern and one of the two isolates showing the late-stage pattern were from the same patient and had identical multilocus genotypes and DNA fingerprints (19). The pattern shown by D8-330 was also shown by 17 clinical isolates. There were nine clinical isolates that clustered with N4, the population evolved without drug.
Discussion
The three patterns of gene expression define two different adaptive solutions to the drug. The adaptive solution reached by D8 can be explained primarily by overexpression of a known drug-resistance determinant, CDR2. The more common adaptive solution reached by D9, D11, and D12 included changes in expression of numerous genes in addition to a known resistance determinant, MDR1. One gene that was overexpressed in all of the populations evolved with the drug, YPL88, belongs to the general class of dehydrogenases/oxidoreductases (Table 2). There were three other genes in this class (YPX98, YPR127W, and ADH4) that were among the most highly overexpressed in the experimental populations (Table 2). This class of proteins is thought to play an important protective role during oxidative stress (25) and may contribute to drug resistance because the azoles sensitize fungal cells to oxidative metabolites through inhibition of the target cytochrome P450 enzyme in the ergosterol biosynthesis pathway (6). Other genes that were highly overexpressed and are implicated in stress response include HYR1, GRE99, and YNL229C (Table 2). In addition to multidrug transporters, genes involved in lipid and cell wall metabolism have been implicated in pleiotropic drug resistance in yeast by altering permeability of cells to hydrophobic drugs (13). Genes in this class that were modulated in the experimental populations include YPL88, YPX98, and PDR16 (Table 2).
Table 2.
C. albicans ORFs showing greater than 3-fold change in expression in at least two population samples and ORFs with Saccharomyces cerevisiae homologues implicated in pleiotropic drug resistance
| ORF | Population sample*
|
Description | ||
|---|---|---|---|---|
| D8-330 | D12-165 | D12-330 | ||
| ADH4(2183_0001)† | 0.6 | 0.9 | 24.4 | Short-chain alcohol dehydrogenase‡ |
| ADH4(2499_0006)† | 1.1 | 1.1 | 18.4 | Short-chain alcohol dehydrogenase‡ |
| MDR1§ | 1.1 | 2.3 | 17.2 | Multidrug major facilitator transporter‡ |
| YPL88¶ | 1.9 | 12.4 | 16.4 | Putative aryl alcohol dehydrogenase‡/cell wall metabolism‖ |
| YPX98 | 1.2 | 9.6 | 15.6 | YPL88-like protein‡ |
| YPR127W | 1.0 | 2.8 | 9.5 | Similar to aryl alcohol dehydrogenase‖ |
| GRE99¶ | 1.2 | 4.7 | 8.6 | Involved in diamide tolerance and induced by osmotic stress‖ |
| YNL229C | 1.2 | 0.7 | 6.2 | Glutathione transferase-like protein‡ |
| HYR1 | 1.5 | 1.4 | 4.0 | Hyphal wall protein‡/glutathione peroxidase involved in oxidative stress‖ |
| PDR16¶ | 1.0 | 1.0 | 2.1 | Multidrug resistance‡‖/lipid synthesis‖ |
| EBP95§ | 1.0 | 1.1 | 1.8 | NADPH dehydrogenase isoform 3‖ |
| CDR2¶ | 2.4 | 0.8 | 1.3 | Multidrug ABC transporter‡ |
| 2913_0002 | 0.8 | 1.0 | 1.1 | ? |
| EBP1§ | 1.1 | 0.5 | 1.0 | Estrogen binding protein‡/NADPH dehydrogenase isoform 2‖ |
| 2866_0003 | 0.9 | 0.9 | 1.0 | ? |
| YOR49¶ | 2.2 | 0.9 | 1.0 | Putative transporter of unknown substrate‖ |
| HSP12(2148_0004)** | 1.8 | 0.5 | 0.7 | 12-kd heat-shock protein‡ |
| HSP12(2734_0007)** | 1.8 | 0.2 | 0.4 | 12-kd heat-shock protein‡ |
| HSP12(2734_0006)** | 1.7 | 0.2 | 0.4 | 12-kd heat-shock protein‡ |
Fold changes in expression of ORFs in one population sample representative of each of the three patterns of gene expression are shown here. For the complete list of 301 significantly modulated ORFs, including fold changes in all eight population samples, P values, S. cerevisiae homologues, Blast-E values, and annotation based on C. albicans (http://alces.med.umn.edu/candida/) and S. cerevisiae (http://www.proteome.com/databases/index.html), see Table 4.
The different versions of ADH4 may represent different portions of the same gene, different alleles of the same gene, or different copies of a duplicated gene.
Description based on C. albicans (http://alces.med.umn.edu/candida/).
ORFs that were significantly modulated and have S. cerevisiae homologues that are activated by YAP1 overexpression (25).
ORFs that were significantly modulated and have S. cerevisiae homologues that are activated by mutations in PDR1 and/or PDR3 (13).
Description based on S. cerevisiae (http://www.proteome.com/databases/index.html).
The different versions of HSP12 may represent different portions of the same gene, different alleles of the same gene, or different copies of a duplicated gene.
In contrast with other studies that have profiled the response of yeast cells to short-term exposure to azole drugs (14, 15), our results do not implicate genes in the ergosterol biosynthesis pathway as major players. The only genes in the ergosterol biosynthesis pathway that were significantly modulated in our experimental populations were ERG1, ERG3, ERG11, and ERG13; the gene with the largest change in expression was ERG1, which was repressed 2.9-fold in D12-260 (see Table 4). This highlights the importance of distinguishing genetically based adaptive change from transient responses to altered conditions.
The altered patterns of gene expression may include both changes that are related to drug resistance and changes that are collateral. For example, although D12-260 and D12-330 cluster together based on similarity of expression profiles, 120 ORFs that were significantly modulated at generation 260 (P ≤ 0.05) were no longer modulated at generation 330 (see Table 10, which is published as supporting information on the PNAS web site). In our previous study, we observed loss of the fitness cost of resistance in population D12 between generation 260 and generation 330 (Table 1). Although population D12 decreased in MIC of fluconazole between generation 260 and generation 330, it did not decrease in fitness in the presence of the drug. Genes that were altered in expression at generation 260, but not at generation 330, may have been responsible for the cost of resistance and may not be essential to the drug-resistant phenotype.
Given the short evolutionary time period, it is likely that a small number of mutations, used here in the broad sense to include base substitutions and intra- and interchromosomal rearrangements (10, 26), are responsible for the repetitive patterns of change in gene expression. Mutation in a few key transcription factors can directly regulate the expression of many genes. In S. cerevisiae, numerous determinants of both oxidative stress response and multidrug resistance are regulated by a complex interplay between the YAP and PDR networks of genes (27), including homologues of eight genes modulated in our experimental populations (Table 2). Suites of genes may have been co-regulated with the resistance determinant MDR1. FLR1 is the S. cerevisiae homologue of MDR1 and is regulated by overexpression of the transcription factor YAP1 (25). In C. albicans, a homologous transcription factor CAP1 is involved in oxidative stress response and multidrug resistance; CAP1 acts as a positive regulator of FLR1 when expressed in S. cerevisiae, but as a negative regulator of MDR1 in C. albicans (28). In our experimental populations, CAP1 was not significantly altered in expression and no mutations were detected in the DNA sequence (data not shown). In both yeasts, the interactions in the transcriptional networks regulating multidrug transporters have yet to be experimentally dissected (27, 29). In addition to transcriptional co-regulation, adaptive mutations could have indirect pleiotropic effects (30) on the expression of suites of genes through interactions among various gene products (8), signaling activity across different pathways (31), or changes in cell physiology (17).
The repetitive patterns of change in genome-wide expression profiles indicate that the evolution of drug resistance was canalized (8), or constrained, to two distinct paths of adaptation to the presence of the drug. Determining whether our populations might ultimately converge on one adaptive path would require further experimental evolution. Alternatively, this could be tested in S. cerevisiae by measuring the effect on fitness of combining different adaptive solutions in hybrid genotypes and their meiotic segregants. Although we do not yet know whether there are additional paths to drug resistance, that three of the four populations sampled followed one path suggests that this is a common adaptive solution. The three patterns of gene expression were also identified in fluconazole-resistant, clinical isolates, providing strong evidence that the two paths of adaptation to the drug are common. The pattern of gene expression unique to D8-330 among the experimental populations was the most frequently identified pattern among the clinical isolates. The clinical isolates that did not show any of the three patterns and clustered with N4–330 may have mutations in the target gene of the azoles, or may have yet other mechanisms of resistance. Genome-wide expression profiling is a sensitive tool for dissecting the genetic programs of adaptation. Teasing apart the functional contribution of different genes in the complex phenotypes defined by the genome-wide expression profiles may rely on further evolution experiments with C. albicans or complementary studies with the genetically tractable yeast, S. cerevisiae. Key genes that are necessary for the evolution of drug resistance would be ideal targets for companion drugs designed to minimize the evolution of resistance to existing antifungal drugs.
Supplementary Material
Acknowledgments
We thank Francois Benoit and Tracey Rigby for the construction of the microarrays. Fluconazole was provided by Pfizer Canada Inc. This is National Research Council publication no. 44795. This work was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to L.M.K. and J.B.A.), and by the National Research Council Genome Health Initiative. L.E.C. was supported by an NSERC postgraduate scholarship.
Abbreviation
- MIC
minimum inhibitory concentration
References
- 1.Levin B R, Lipsitch M, Bonhoeffer S. Science. 1999;283:806–809. doi: 10.1126/science.283.5403.806. [DOI] [PubMed] [Google Scholar]
- 2.Georgopapadakou N H, Walsh T J. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 3.Pfaller M A. Clin Infect Dis. 1998;27:1148–1150. doi: 10.1093/clinids/27.5.1148. [DOI] [PubMed] [Google Scholar]
- 4.DiDomenico B. Curr Opin Microbiol. 1999;2:509–515. doi: 10.1016/s1369-5274(99)00009-0. [DOI] [PubMed] [Google Scholar]
- 5.Georgopapadakou N H. Expert Opin Investig Drugs. 2001;10:269–280. doi: 10.1517/13543784.10.2.269. [DOI] [PubMed] [Google Scholar]
- 6.White T C, Marr K A, Bowden R A. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowen L E. FEMS Microbiol Lett. 2001;204:1–7. doi: 10.1111/j.1574-6968.2001.tb10853.x. [DOI] [PubMed] [Google Scholar]
- 8.Hartman J L, Garvik B, Hartwell L. Science. 2001;291:1001–1004. doi: 10.1126/science.291.5506.1001. [DOI] [PubMed] [Google Scholar]
- 9.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 10.Cowen L E, Sanglard D, Calabrese D, Sirjusingh C, Anderson J B, Kohn L M. J Bacteriol. 2000;182:1515–1522. doi: 10.1128/jb.182.6.1515-1522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowen L E, Kohn L M, Anderson J B. J Bacteriol. 2001;183:2971–2978. doi: 10.1128/JB.183.10.2971-2978.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 13.DeRisi J, van den Hazel B, Marc P, Balzi E, Brown P, Jacq C, Goffeau A. FEBS Lett. 2000;470:156–160. doi: 10.1016/s0014-5793(00)01294-1. [DOI] [PubMed] [Google Scholar]
- 14.Bammert G F, Fostel J M. Antimicrob Agents Chemother. 2000;44:1255–1265. doi: 10.1128/aac.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Backer M D, Ilyina T, Ma X J, Vandoninck S, Luyten W H, Vanden Bossche H. Antimicrob Agents Chemother. 2001;45:1660–1670. doi: 10.1128/AAC.45.6.1660-1670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasch A P, Spellman P T, Kao C M, Carmel H O, Eisen M B, Storz G, Botstein D, Brown P O. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferea T L, Botstein D, Brown P O, Rosenzweig R F. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Standard. National Committee for Clinical Laboratory Standards, Wayne, PA: document M27-A; 1997. [Google Scholar]
- 19.Cowen L E, Sirjusingh C, Summerbell R C, Walmsley S, Richardson S, Kohn L M, Anderson J B. Antimicrob Agents Chemother. 1999;43:2930–2938. doi: 10.1128/aac.43.12.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luu L N, Cowen L E, Sirjusingh C, Kohn L M, Anderson J B. J Clin Microbiol. 2001;39:1657–1660. doi: 10.1128/JCM.39.4.1657-1660.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessier D C, Thomas D Y, Brousseau R. In: Biotechnology. Rehm H-J, Stadler P, editors. 5b. Weinheim, Germany: Wiley-VCH; 2001. , Chapter 9. [Google Scholar]
- 22.Ausubel F M. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 2. Boston: Wiley; 1995. pp. 13.12.2–13.12.3. [Google Scholar]
- 23.Sanglard D, Ischer F, Monod M, Bille J. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 24.De Micheli M, Bille J, Schueller C, Sanglard D. Mol Microbiol. 2002;43:1197–1214. doi: 10.1046/j.1365-2958.2002.02814.x. [DOI] [PubMed] [Google Scholar]
- 25.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 26.Perepnikhatka V, Fischer F J, Niimi M, Baker R A, Cannon R D, Wang Y K, Sherman F, Rustchenko E. J Bacteriol. 1999;181:4041–4049. doi: 10.1128/jb.181.13.4041-4049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolaczkowska A, Goffeau A. Drug Resist Updat. 1999;2:403–414. doi: 10.1054/drup.1999.0113. [DOI] [PubMed] [Google Scholar]
- 28.Alarco A M, Raymond M. J Bacteriol. 1999;181:700–708. doi: 10.1128/jb.181.3.700-708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wirsching S, Michel S, Kohler G, Morschhauser J. J Bacteriol. 2000;182:400–404. doi: 10.1128/jb.182.2.400-404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waxman D, Peck J R. Science. 1998;279:1210–1213. [PubMed] [Google Scholar]
- 31.Roberts C J, Nelson B, Marton M J, Stoughton R, Meyer M R, Bennett H A, He Y D, Dai H, Walker W L, Hughes T R, et al. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.