Abstract
Several dinoflagellate species have plastids that more closely resemble those of an unrelated algal group, the haptophytes, suggesting these plastids have been obtained by tertiary endosymbiosis. Because both groups are photosynthetic, all of the genes for nuclear-encoded plastid proteins might be supplied by the dinoflagellate host or some of them might have been replaced by haptophyte genes. Sequences of the conserved nuclear psbO gene were obtained from the haptophyte Isochrysis galbana, the peridinin-containing dinoflagellate Heterocapsa triquetra, and the 19′hexanoyloxy-fucoxanthin-containing dinoflagellate Karenia brevis. Phylogenetic analysis of the oxygen-evolving-enhancer (PsbO) proteins confirmed that in K. brevis the original peridinin-type plastid was replaced by that of a haptophyte, an alga which had previously acquired a red algal chloroplast by secondary endosymbiosis. It showed clearly that during this tertiary symbiogenesis the original psbO gene in the dinoflagellate nucleus was replaced by a psbO gene from the haptophyte nucleus. The phylogenetic analysis also confirmed that the origin of the peridinin-type dinoflagellate plastid was indeed a red alga.
Dinoflagellates are enigmatic protists. About half of them are nonphotosynthetic and the other half have some sort of plastid and rely entirely or partially on photosynthesis. Most photosynthetic dinoflagellates have plastids that are surrounded by three membranes and contain chlorophylls (Chl)-a and -c and peridinin as the major photosynthetic pigments. However, a small fraction of the photosynthetic species has plastids that do not contain peridinin but have pigment compositions characteristic of other algal groups (1–4). These anomalously pigmented plastids are considered to have been acquired from other algal groups by means of secondary (4) or tertiary endosymbioses (5, 6).
Among the anomalously pigmented dinoflagellates, there is a monophyletic group that consists of species containing 19′-hexanoyloxy-fucoxanthin and/or 19′-butanoyloxy-fucoxanthin instead of peridinin (7). This group includes Karenia brevis ( = Gymnodinium breve), Karenia mikimotoi ( = Gymnodinium mikimotoi), Karlodinium micrum ( = Gymnodinium galatheanum), and a few others (7). Because these two carotenoids are otherwise found only in haptophytes, it has been suggested that these plastids came from a haptophyte alga (2). This suggestion was supported by two molecular phylogenetic analyses of plastid small-subunit rRNA genes (6, 8), although no peridinin-containing species was included. These anomalous dinoflagellates also resemble haptophytes in containing a plastid-encoded form-I rubisco (9), in contrast to the nuclear-encoded form-II rubisco of peridinin-containing dinoflagellates (10). In contrast, nuclear gene phylogenies with large- and small-subunit rRNA sequences put all of the 19′-hexanoyloxy-fucoxanthin-containing species in a single clade within the peridinin-containing dinoflagellate lineage (7, 11). These observations suggest that the common ancestor of the 19′-hexanoyloxy-fucoxanthin-containing species had a peridinin-type plastid but lost it and acquired a new one by engulfing a haptophyte alga (6, 11). The engulfed haptophyte was itself a secondary-endosymbiosis-derived alga, making this a tertiary endosymbiosis.
If this is so, it raises a question about the fate of nuclear-encoded genes for plastid proteins, because both host and symbiont would have had many genes for proteins that are synthesized on cytoplasmic ribosomes and targeted to the plastids. If the dinoflagellate host lost its original plastid a relatively short time before acquiring the haptophyte endosymbiont (or even after), it would have retained many of these genes, and their products might have been imported successfully into the new haptophyte plastid. On the other hand, genes that had been lost or whose products were incompatible with the new plastid would have to have been replaced by genes transferred from the haptophyte endosymbiont as its nucleus disintegrated.
PsbO (also called OEC33 or 33-kDa oxygen-enhancer 1 protein) is one of those nuclear-encoded plastid proteins. It is localized in the thylakoid lumen and serves to protect the tetra-manganese cluster and ionic environment, which are essential for the water-splitting reaction of photosystem II (12, 13). Although this protein has never been used for phylogenetic analysis, it has some qualities that make it a good candidate for elucidating the evolutionary relationship among plastids. It is unique to cyanobacteria and plastids that carry out oxygenic photosynthesis, therefore its phylogeny should represent that of photosynthetic organisms. The psbO gene is nuclear-encoded in all photosynthetic eukaryotes (14), thus its evolution should not have been influenced by the difference in evolutionary rates between nuclear and plastid genomes (15). It is a very conserved protein that makes it suitable for studying relationships over large evolutionary distances. In fact, the PsbO proteins of cyanobacteria, red algae, and green plants can be successfully reconstituted with each others' photosystem II cores (16).
Only PsbO sequences from cyanobacteria and Chl-a/b-containing organisms (“green line”) are available in the databases. In this article, we report previously uncharacterized PsbO sequences from haptophytes, peridinin-containing dinoflagellates, and 19′hexanoyloxy-fucoxanthin-containing dinoflagellates. Analysis of targeting sequences as well as phylogenetic analysis of PsbO protein sequences gives new insights into the origin of peridinin-type dinoflagellate plastids, and the fate of the psbO genes of both host and endosymbiont during the tertiary endosymbiotic replacement of the plastid in 19′hexanoyloxy-fucoxanthin-containing dinoflagellates.
Materials and Methods
Algal Cultures.
Unialgal cultures of a haptophyte, Isochrysis galbana (CCMP1323), and a 19′-hexanoyloxy-fucoxanthin-containing dinoflagellate, K. brevis (syn. G. breve) (CCMP718) were obtained from the National Center for Culture of Marine Phytoplankton (CCMP), Bigelow Laboratory for Ocean Sciences, U.S. Both strains were maintained in f/2-Si medium (http://ccmp.bigelow.org/CI/CI_01e.html) at 19°C under a 11-h/13-h light/dark cycle.
Total RNA Extraction, Reverse Transcription–PCR, and Rapid Amplification of cDNA Ends (RACE) for 5′ and 3′ cDNA Ends.
Total RNA was extracted from 100 ml of culture of each strain during the middle of the light period with the StrataPrep Total RNA Miniprep kit (Stratagene). First-strand cDNA synthesis was carried out with Avian Enhanced Reverse Transcriptase (Sigma); products were purified with a QIAquick PCR purification kit (Qiagen, Chatsworth, CA). Partial psbO cDNA fragments were amplified by PCR with degenerate primers based on known sequences. Bands of the expected size were cloned into pCR2.1-TOPO cloning vector (Invitrogen) for sequencing. Both ends of each psbO cDNA sequence were amplified with the RACE technique. For 5′ end RACE, first-strand cDNAs tailed with poly(A) at 3′ ends, using terminal deoxynucleotidyl transferase (Promega), were used as templates. Full-length PsbO precursor sequences were obtained by making contigs of the sequences obtained from the PCR and RACE reactions with the GAP4 program in the Staden et al. (17) package and translating them with the universal genetic code.
Phylogenetic Analyses.
PsbO protein sequences (21 taxa) used for phylogenetic analyses are listed in Table 1. The characterization of Heterosigma akashiwo PsbO will be published elsewhere. For Porphyra yezoensis, four sequences were downloaded from the web site for the P. yezoensis expressed sequence tag (EST) project (ref. 18; http://www.kazusa.or.jp/en/plant/porphyra/EST/) and assembled into contigs. An alignment data set of the PsbO protein sequences was created and edited manually with MACCLADE 4 (19); phylogenetic analyses used 231 sites (86.2% of the mature protein sites).
Table 1.
Sequences used in the present phylogenetic analyses
| Species | Accession nos. |
|---|---|
| Synechocystis sp. PCC6803 | P10549 |
| Synechococcus elongatus | P55221 |
| Synechococcus sp. PCC7942 | P11472 |
| Cyanothece sp. ATCC51142 | Q9R6W6 |
| Nostoc sp. PCC7120 | P13907 |
| P. yezoensis | AV432153, AV432421, AV432896, AV436570 |
| H. triquetra | This study |
| H. akashiwo | K.-i.I., unpublished data |
| K. brevis | This study |
| I. galbana | This study |
| Euglena gracilis | BAA03529 |
| Volvox carteri | Q9SBN6 |
| Chlamydomonas reinhardtii | P12853 |
| Pisum sativum | P14226 |
| Arabidopsis thaliana | P23321 |
| Spinacia oleracea | P12359 |
| Fritillaria agrestis | O49079 |
| Nicotiana tabacum | Q40459 |
| Solanum tuberosum | P26320 |
| Bruguiera gymnorrhiza | BAA96365 |
| Triticum aestivum | P27665 |
Two distance matrices were created by TREE-PUZZLE 5.0 (20) with JTT (21) and WAG (22) amino acid substitution models with gamma correction (8 + 1 categories). The estimated shape parameters (alpha) for gamma distribution in both models were 1.01 (JTT model) and 1.25 (WAG model). Neighbor-joining trees were constructed from both of the distance matrices with the neighbor program in the PHYLIP 3.572c package (23) with the -j (jumble) option. Fitch trees were constructed with the fitch program with -j (jumble) and -g (global rearrangement) options. For bootstrap analyses, 100 data sets were created by the seqboot program in the phylip package. Distances for each data set were calculated with puzzleboot script (by M. Holder and A. Roger, available at http://www.tree-puzzle.de/#puzzleboot). The consensus bootstrap tree was obtained with the consense program of phylip. For maximum likelihood tree analyses, two different programs were used: the proml program in PHYLIP 3.6a2.1 (24) and the protml program in MOLPHY 2.3 (25). Analysis with the proml program was performed with the JTT amino acid substitution model with gamma correction (8 + 1 categories). The parameters obtained from the tree-puzzle distance analysis (with the JTT model) were used for this gamma correction. For analysis with the protml program, a neighbor-joining tree topology was obtained first with the protml (with –D option) and njdist programs in the molphy package with the JTT-f substitution model. Based on this topology, a maximum likelihood tree was constructed with the protml with the RELL method (-R option) (26).
A maximum parsimony tree analysis was performed with heuristic search strategy with random sequence addition (100 times) in the PAUP 4.0 package (27). Gaps were treated as a 21st amino acid. This analysis gave five trees from which a majority-rule consensus tree was produced. A bootstrap analysis was also carried out with the same program and options.
Sequence Characterization.
The cleavage sites for the endoplasmic reticulum (ER)-targeting domain and the thylakoid-targeting domain (TTD) were predicted by the signal p program (ref. 28; http://www.cbs.dtu.dk/services/SignalP/). The stroma-targeting domain (STD) was predicted to be the region between the ER- and TTDs, taking into consideration the hydropathy plot, the few known cleavage sites for heterokont proteins (29), and the results from the chlorop program (ref. 30; http://www.cbs.dtu.dk/services/ChloroP).
Results
PsbO Precursor Sequences Obtained in This Study.
PsbO protein sequences of three Chl-c-containing algae, the haptophyte I. galbana and the dinoflagellates H. triquetra and K. brevis, were deduced from cDNA sequences obtained by reverse transcription–PCR and RACE. These are previously uncharacterized PsbO sequences reported from Chl-c-containing algae. In addition, an almost-complete red algal PsbO sequence was obtained by assembling contigs from the P. yezoensis expressed sequence tag database (18). It gave two separate contigs that turned out to be the N- and C-terminal halves of the molecule. Judged by an alignment with other PsbO proteins, it seemed that one amino acid was missing between the two contigs, and the C-terminal end of the molecule was slightly truncated (data not shown). For all of the species except K. brevis, some of the 5′ untranslated region was obtained, giving the entire N-terminal leader sequence (targeting sequence) of the precursor proteins. In the case of K. brevis, the alignment of the leader sequences (Fig. 1) suggests that only a few amino acids are missing at the N terminus. In comparing the sequences of the mature proteins from I. galbana, H. triquetra, K. brevis, and P. yezoensis with the cyanobacterial and green line sequences (not shown), it was apparent that this protein is generally very conserved.
Figure 1.
Alignment of PsbO N-terminal leader sequences. Sources are given in Table 1. Hydrophobic domains are underlined and italicized. Bold K and R residues, positively charged Lys and Arg preceding the hydrophobic domains; bold A, Ala in the TTD AXA motif. Names of species in red-line clade are bolded. ETD, ER-targeting domain.
N-Terminal Leader Sequences of PsbO Precursors in Chl-c-Containing Algae.
The leader sequences of I. galbana, K. brevis, and H. triquetra PsbO precursors clearly consist of three domains: two hydrophobic domains and a variable-length domain between them (Fig. 1). The second hydrophobic domains have the typical characteristics of a TTD: an Ala/Leu-rich hydrophobic stretch of 15–19 amino acids (Fig. 1, underlined) preceded by a single positively charged amino acid (Fig. 1, bolded) and followed by the typical thylakoidal-processing motif AXA (31). Our prediction of the start of the mature protein, which is located in the thylakoid lumen, is based on sequence alignment and the presence of this TTD shared by all of the sequences (Fig. 1).
It has now been well established that precursors for nuclear-encoded plastid proteins in algae that acquired their plastids secondarily from eukaryotic algal endosymbionts are synthesized and inserted into the rough ER as the first step of their journey into the plastid (32–34). The first hydrophobic domains of the I. galbana and the H. triquetra leader sequences (21 and 20 amino acids, respectively) were predicted as eukaryotic signal sequences (ETD, ER-targeting domain) by the signal p program (28). The first hydrophobic domain of the K. brevis leader sequence was not identified as an ETD by signal p, probably because some N-terminal residues are missing, but because the C- and S-scores were typical of signal sequences the first 16 amino acids were provisionally assigned to an ETD (Fig. 1). The P. yezoensis PsbO precursor does not have an ETD, because the red algal chloroplast has only two envelope membranes like that of a green plant: its first 43 amino acids are therefore a transit peptide or STD (Fig. 1).
The middle domain of the presequence should be an STD analogous to the transit peptides of higher plants. The STDs of Chl-c-containing algae are not well characterized and do not seem to be processed by higher plant-processing peptidases (ref. 29 and B. K. Chaal and B.R.G., unpublished data). Processing sites predicted by the chlorop program (30), which is trained on green plant sequences, may therefore not be valid. However, for the K. brevis and the H. triquetra sequences, the chlorop-predicted processing sites agreed well with the start of their TTD prediction, and their STDs were therefore assigned as in Fig. 1. The H. akashiwo and I. galbana cleavage sites were tentatively assigned to follow a methionine in agreement with the limited data available for heterokont algal STD cleavage sites (29). If these predictions are correct, the STDs of H. akashiwo (10 residues), I. galbana (about 9 residues), and K. brevis (about 11 residues) are very short, in agreement with the STDs of some nuclear-encoded heterokont proteins such as the fucoxanthin Chl-a/c protein (32, 34). In contrast, the predicted length of the H. triquetra STD is about 34 residues, closer in size to those of the red alga P. yezoensis and higher plants, which are usually more than 30 residues (35).
Phylogenetic Analysis of PsbO Sequences.
Phylogenetic analyses of all PsbO protein sequences currently available (Table 1) were performed with three commonly used methods: maximum parsimony, distance, and maximum likelihood. For distance analyses, two amino acid substitution models, JTT (21) and WAG (22), were used, and rate variation among sites was corrected with gamma distribution. For the maximum likelihood analyses, the JTT model was used with or without the gamma correction, and local bootstrap probabilities were also estimated by the RELL method (26) for the tree with no gamma correction.
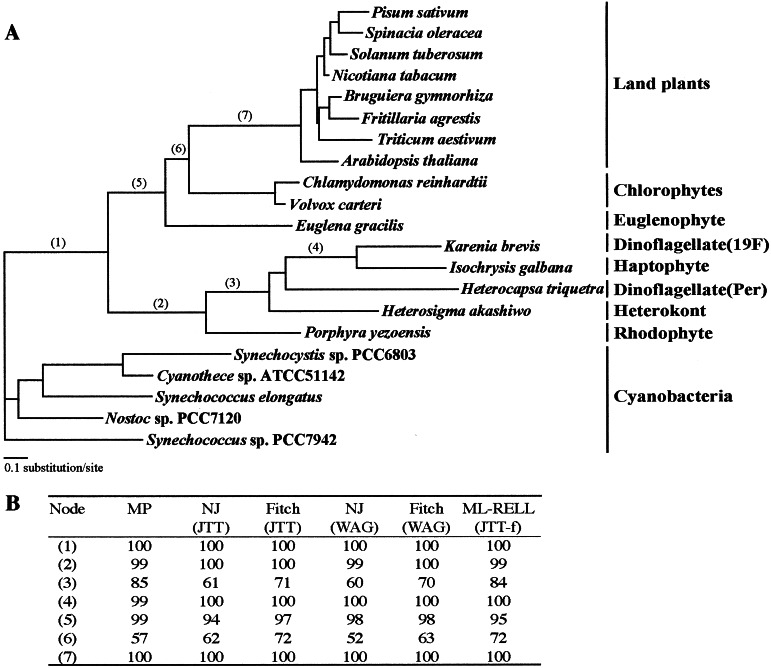
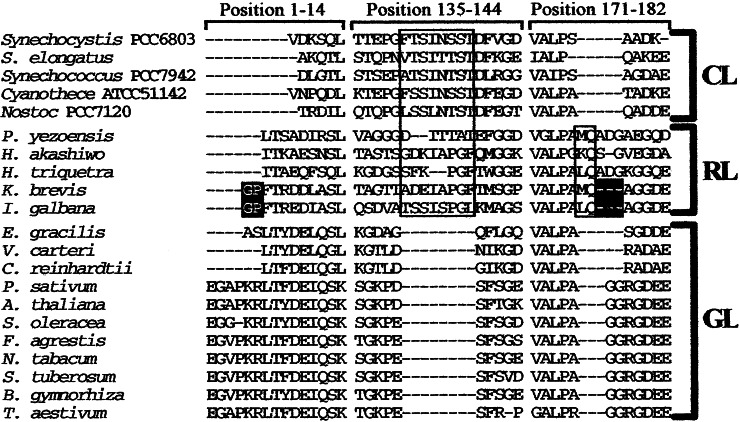
All PsbO protein trees gave nearly identical topologies (Fig. 2A). Three major clades, a cyanobacterial clade, a rhodophyte/Chl-c-containing algal clade (red-line clade), and a euglenophyte/chlorophyte/land plants clade (green line clade), were recognized in all trees. The red-line clade included sequences from the red alga P. yezoensis, the heterokont H. akashiwo, the haptophyte I. galbana, the peridinin-containing dinoflagellate H. triquetra, and the 19′-hexanoyloxy-fucoxanthin-containing dinoflagellate K. brevis. It was supported by very high bootstrap probabilities in all analyses (Fig. 2B). Besides the phylogenetic trees, there is other supporting evidence that the H. triquetra branch is placed within the red-line clade. The H. triquetra sequence does not have an 8-aa deletion that is specific to the Chl-a/b-containing organisms, making it unlikely that the plastid of H. triquetra originated from a green plastid, and it has a 2-aa insertion present in other sequences of the red-line clade (Fig. 3), indicating that this plastid shares a common ancestor with other members of the red-line clade. Taken together, all of the evidence demonstrate that the peridinin-containing plastids of dinoflagellates originated from a red algal plastid, not from a green algal plastid or a cyanobacterium.
Figure 2.
Phylogenetic trees of PsbO protein sequences. (A) Gamma-corrected (8 + 1 categories) maximum likelihood tree constructed by the PROML program with the JTT model. (B) Bootstrap values given by various tree-constructing methods for numbered nodes.
Figure 3.
Lineage-specific insertions/deletions (indels) in mature PsbO protein sequences. The large box in the second column indicates an 8-aa insertion specific to cyanobacteria and the red-line clade. The small box in the third column indicates a 2-aa insertion specific to the red-line clade. Indels shared only by I. galbana and K. brevis are highlighted. Numbers at the top of each column indicate amino acid positions based on the spinach PsbO sequence. CL, cyanobacteria-line; RL, red-line; GL, green-line.
The second major point shown by our PsbO trees is that the 19′-hexanoyloxy-fucoxanthin-containing dinoflagellate K. brevis clustered with the haptophyte alga I. galbana with 100% bootstrap probability, rather than with the peridinin-containing dinoflagellate H. triquetra (Fig. 2A). The presence of two insertions/deletions that are shared only by K. brevis and I. galbana (highlighted in Fig. 3) supports this relationship. The primer pairs used to PCR the Heterocapsa psbO sequence did not give any product with Karenia cDNA and vice versa. It is still possible that Karenia retains a copy of the original peridinien dinoflagellate psbO gene, but the lack of a product suggests that its sequence would have diverged, as would be expected for a nonfunctional or pseudogene.
Discussion
Replacement of a Nuclear-Encoded Chloroplast Protein Gene Along with a Tertiary Endosymbiotic Plastid Acquisition.
Our PsbO trees show conclusively that the nuclear-encoded PsbO sequence of the 19′-hexanoyloxy-fucoxanthin-containing dinoflagellate K. brevis is much more closely related to that of the haptophyte I. galbana than it is to that of the peridinin-containing dinoflagellate H. triquetra. This finding confirms the haptophyte origin of the 19′-hexanoyloxy-fucoxanthin-type plastids suggested on the basis of pigment content (2) and the phylogenetic analyses of two plastid-encoded molecules (6, 8, 9). That these dinoflagellates once had peridinin-type plastids was suggested by nuclear-encoded small- and large-subunit rRNA gene trees, in which the 19′-hexanoyloxy-fucoxanthin-containing dinoflagellates were placed within the peridinin-containing dinoflagellate clade (6, 7, 11, 36). We therefore conclude that the peridinin-type plastid was replaced by a haptophyte plastid by means of a tertiary endosymbiosis in the ancestor of the 19′-hexanoyloxy-fucoxanthin-containing dinoflagellates (Fig. 4).
Figure 4.
Hypothetical evolutionary scheme for the 19′-hexanoyloxy-fucoxanthin-containing dinoflagellates. (I) Acquisition of a plastid by a secondary symbiogenesis in which a red algal cell was engulfed by a colorless ancestor. The red algal nuclear psbO gene was transferred to the dinoflagellate nucleus (dashed arrow) and the sequence subsequently diverged. (II) Loss of the peridinin-type plastid and the nuclear psbO gene for the plastid. (III) Acquisition of a new plastid by a tertiary symbiogenesis in which a haptophyte alga was engulfed by a dinoflagellate that may or may not have already lost its original peridinin-type plastid. psbO-R, psbO-P, and psbO-H indicate the psbO genes of the red algal secondary endosymbiont, the peridinin-containing dinoflagellate, and the haptophyte tertiary endosymbiont, respectively.
To our knowledge, this is the first example of the replacement of a host nuclear gene with a gene originating in the nucleus of a tertiary endosymbiont. It has been argued that dinoflagellates can easily integrate tertiary plastids, because the preexisting genes for plastid-targeted proteins can be recycled to facilitate their integration into the cell (6). However, this is not the case for the psbO gene of K. brevis, because the original psbO gene for the peridinin-type plastid was superseded by the haptophyte gene instead of being recycled for the new plastid during the tertiary symbiogenesis (Fig. 4). This observation suggests that the systems for expressing the psbO gene and targeting its product from the cytoplasm to the new plastid were newly established during the plastid-replacement event. It will not be clear whether the relationship between the host and the new endosymbiont was completely rebuilt until origins of other genes for plastid-targeted proteins in these dinoflagellates are investigated. One possible explanation for why the psbO gene was replaced would be that the loss of the peridinin-type plastid occurred long before the host cell acquired the haptophyte endosymbiont, so that the preexisting psbO gene was no longer functional. Another possibility would be that the mechanism for targeting proteins into the peridinin-type plastids did not function for the new endosymbiont; therefore, the host cell had to give up the preexisting psbO gene.
Origin of the Standard Peridinin-Type Plastid.
One of the major unsolved problems in algal evolution is the origin of the typical peridinin-type dinoflagellate plastid. When molecules that are normally encoded by plastid genomes were used for tree construction, the positions of dinoflagellate sequences were obscured by their very long branches (37–39), even when a tree was constructed from a concatenated data set of nine different plastid proteins (38). A further complication is that in at least some of the peridinin-containing dinoflagellates, genes for the plastid proteins and rRNAs are encoded by single-gene minicircular chromosomes, which may be subjected to different evolutionary pressures (38, 39).
Detailed phylogenetic analysis has been carried out on two nuclear-encoded, plastid-targeted proteins of peridinin-containing dinoflagellates: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (40) and the large subunit of rubisco (41, 42). However, neither of them was directly derived from the original plastid (the primary cyanobacterial endosymbiont): GAPDH is from a duplicated cytoplasmic (gapC-type) counterpart (40) and the large subunit of rubisco is from an anaerobic bacterial form-II rubisco (41, 42). In contrast, PsbO did originate from the cyanobacterial ancestor of all plastids. In our PsbO tree, the peridinin-containing dinoflagellate branch (H. triquetra) is not particularly long and shows conclusively that this type of plastid originated from a red alga by means of a secondary endosymbiosis. This is direct phylogenetic evidence of the red algal origin of the peridinin-type dinoflagellate plastids.
Acknowledgments
We thank Dr. Z. Zhang for H. triquetra DNA, S. Quayle and G. Rossi for technical assistance, G. Huang for preliminary work on prediction of targeting sequences, and Dr. P. J. Keeling for helpful discussions. This research was supported by research grants from the Natural Science and Engineering Research Council (NSERC) of Canada and by a grant-in-aid for Creative Basic Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.
Abbreviations
- ER
endoplasmic reticulum
- STD
stroma-targeting domain
- TTD
thylakoid-targeting domain
- RACE
rapid amplification of cDNA ends
- Chl
chlorophyll
Footnotes
References
- 1.Mandelli E F. J Phycol. 1968;4:347–348. doi: 10.1111/j.1529-8817.1968.tb04706.x. [DOI] [PubMed] [Google Scholar]
- 2.Tangen K, Björnland T. J Plankton Res. 1981;3:389–401. [Google Scholar]
- 3.Hewes C D, Mitchell B G, Moisan T A, Vernet M, Reid F M H. J Phycol. 1998;34:945–951. [Google Scholar]
- 4.Watanabe M M, Suda S, Inouye I, Sawaguchi T, Chihara M. J Phycol. 1990;26:741–751. [Google Scholar]
- 5.Chesnick J M, Kooistra W H, Wellbrock U, Medlin L K. J Eukaryotic Microbiol. 1997;44:314–320. doi: 10.1111/j.1550-7408.1997.tb05672.x. [DOI] [PubMed] [Google Scholar]
- 6.Tengs T, Dahlberg O J, Shalchian-Tabrizi K, Klaveness D, Rudi K, Delwiche C F, Jakobsen K S. Mol Biol Evol. 2000;17:718–729. doi: 10.1093/oxfordjournals.molbev.a026350. [DOI] [PubMed] [Google Scholar]
- 7.Hansen G, Daugbjerg N, Henriksen P. J Phycol. 2000;36:394–410. [Google Scholar]
- 8.Takishita K, Nakano K, Uchida A. Phycol Res. 1999;47:257–262. [Google Scholar]
- 9.Takishita K, Nakano K, Uchida A. Phycol Res. 2000;48:85–89. [Google Scholar]
- 10.Morse D, Salois P, Markovic P, Hastings W J. Science. 1995;268:1622–1624. doi: 10.1126/science.7777861. [DOI] [PubMed] [Google Scholar]
- 11.Saldarriaga J F, Taylor F J R, Keeling P J, Cavalier-Smith T. J Mol Evol. 2001;53:204–213. doi: 10.1007/s002390010210. [DOI] [PubMed] [Google Scholar]
- 12.Seidler A. Biochim Biophys Acta. 1996;1277:35–60. doi: 10.1016/s0005-2728(96)00102-8. [DOI] [PubMed] [Google Scholar]
- 13.Bricker T M, Frankel L M. Photosynth Res. 1998;56:157–173. [Google Scholar]
- 14.Shibata M, Kashino Y, Satoh K, Koike H. Plant Cell Physiol. 2001;42:733–741. doi: 10.1093/pcp/pce092. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe K H, Li W-H, Sharp P M. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enami I, Yoshihara S, Tohrii A, Okumura A, Ohta H, Shen J-R. Plant Cell Physiol. 2000;41:1354–1364. doi: 10.1093/pcp/pcd069. [DOI] [PubMed] [Google Scholar]
- 17.Staden R, Beal K F, Bonfield J K. In: Computer Methods in Molecular Biology. Misener S, Krawetz S A, editors. Vol. 132. Totowa, NJ: Humana; 1998. pp. 115–130. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido I, Asamizu E, Nakajima M, Nakamura Y, Saga N, Tabita S. DNA Res. 2000;7:223–227. doi: 10.1093/dnares/7.3.223. [DOI] [PubMed] [Google Scholar]
- 19.Maddison D R, Maddison W P. MACCLADE 4, Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 2000. , Version 4.0. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt H A, Strimmer K, Vingron M, von Haeseler A. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 21.Jones D T, Taylor W R, Thornton J M. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 22.Whelan S, Goldman N. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 23.Felsenstein J. PHYLIP, Phylogeny Inference Package. Univ. of Washington, Seattle: Department of Genetics; 1995. , Version 3.57c. [Google Scholar]
- 24.Felsenstein J. PHYLIP, Phylogeny Inference Package. Univ. of Washington, Seattle: Department of Genetics; 2001. , Version 3.6a2. [Google Scholar]
- 25.Adachi J, Hasegawa M. Comp. Sci. Monographs 23. Tokyo: Inst. Stat. Math.; 1996. pp. 1–150. [Google Scholar]
- 26.Hasegawa M, Kishino H. Mol Biol Evol. 1994;11:142–145. [Google Scholar]
- 27.Swofford D L. PAUP*, Phylogenetic Analysis Using Parsimony (and Other Methods) Sunderland, MA: Sinauer; 1999. , Version 4.0 (test version). [Google Scholar]
- 28.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Lang M, Apt K E, Kroth P G. J Biol Chem. 1998;273:30973–30978. doi: 10.1074/jbc.273.47.30973. [DOI] [PubMed] [Google Scholar]
- 30.Emanuelsson O, Nielsen H, von Heijne G. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peltier J-B, Friso G, Kalume D E, Roepstorff P, Nilsson F, Adamska I, van Wijk L J. Plant Cell. 2000;12:319–341. doi: 10.1105/tpc.12.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhaya D, Grossman A. Mol Gen Genet. 1991;229:400–404. doi: 10.1007/BF00267462. [DOI] [PubMed] [Google Scholar]
- 33.McFadden G I. J Eukaryotic Microbiol. 1999;46:339–346. doi: 10.1111/j.1550-7408.1999.tb04613.x. [DOI] [PubMed] [Google Scholar]
- 34.Ishida K, Cavalier-Smith T, Green B R. J Phycol. 2000;36:1135–1144. [Google Scholar]
- 35.Cline K, Henry R. Annu Rev Cell Dev Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Daugbjerg N, Hansen G, Lasen J, Lasen, Moestrup Ø. Phycologia. 2000;39:302–317. [Google Scholar]
- 37.Takishita K, Uchida A. Phycol Res. 1999;47:207–216. [Google Scholar]
- 38.Zhang Z, Green B R, Cavalier-Smith T. Nature (London) 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Green B R, Cavalier-Smith T. J Mol Evol. 2000;51:26–40. doi: 10.1007/s002390010064. [DOI] [PubMed] [Google Scholar]
- 40.Fast N M, Kissinger J C, Roos D S, Keeling P J. Mol Biol Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- 41.Ueda K, Shibuya H. In: Endocytobiology V (5th International Colloquium on Endocytobiology and Symbiosis, Uji-Kyoto, June 23–27, 1992) Sato S, Ishida M, Ishikawa H, editors. Tübingen, Germany: Tübingen Univ. Press; 1993. pp. 369–376. [Google Scholar]
- 42.Delwiche C F, Palmer J D. Plant Syst Evol. 1997;11,Suppl.:53–86. [Google Scholar]