Abstract
Maternal transmission of RNAs or proteins through the egg cytoplasm plays an important role in eukaryotic development. We show that the transposase activity encoded by the P transposable element of Drosophila melanogaster is transmitted through the oocytes of females heterozygous for this element even when these oocytes do not carry the element itself. However, this maternal transmission is abolished when the last of three introns is removed from the P element. These facts imply that maternal transmission of transposase activity involves the RNA transcribed from the P element rather than the polypeptide it encodes, and that to be transmitted maternally, this RNA must possess the last intron. Examination of the intron's sequence reveals that it contains a motif of nine nucleotides that has been implicated in the maternal transmission of developmentally significant RNAs. This same intron limits expression of the P transposase to the germ line of Drosophila. Thus, the last P intron has two important biological functions.
DNA sequencing projects have revealed a plethora of transposable elements in the genomes of different organisms (1). Among these transposons, the P elements of the fruit fly Drosophila melanogaster are among the best understood and technologically most useful (2). P elements have been widely used as insertional mutagens to tag genes for cloning, and as vectors for the genetic transformation of Drosophila. These applications have become paradigms for the use of transposons as genetic tools in other organisms.
P elements are found in natural populations, but not in long-standing laboratory stocks, apparently because they invaded the D. melanogaster genome sometime in the middle of the 20th century (3, 4). These elements were discovered through their involvement in a syndrome of germ-line abnormalities called hybrid dysgenesis (5). These abnormalities include high frequencies of mutation, chromosome breakage, and sterility—all caused by P element excision and transposition in the germ-line cells. The traits of hybrid dysgenesis occur in the offspring from crosses between males that carry P elements in their genomes and females that do not, but usually not in the offspring from the reciprocal cross. This difference between genetically identical offspring indicates that hybrid dysgenesis is repressed by a maternally inherited condition associated with the P elements. This condition, called the P cytotype (6), is thought to arise from some product(s) of the P elements themselves. In most models, these products are hypothesized to pass from mother to offspring through the egg cytoplasm (2, 7).
When P elements are introduced into laboratory stocks via crosses, they transpose in the germ line but not in the somatic tissues. This tissue-specific behavior is caused by the synthesis of a P-encoded enzyme, an 87-kDa transposase that catalyzes P excision and transposition, in germ-line cells only (8). In somatic cells, the last of the three introns in the transposase gene is not spliced out of the P element's RNA. Because this intron contains a stop codon, somatic P RNA's are translated into a 66-kDa polypeptide that does not have catalytic function. This polypeptide is also produced in the germ line, where it appears to repress P element activity (7, 9). Thus, the 66-kDa polypeptide has been postulated to contribute to the P cytotype (7).
The P transposase is encoded by P elements that are 2.9 kb long. In nature, many shorter P elements exist, most apparently derived from the 2.9-kb complete P elements by deletions of internal sequences (2). Some of these elements encode polypeptides that repress hybrid dysgenesis, albeit partially (10). Thus, they may also contribute to the P cytotype.
Although maternal inheritance is a key feature of the P cytotype, it has been difficult to ascertain which, if any, P element products are maternally transmitted (11). It is not even known if the P transposase can be passed from mother to offspring through the egg cytoplasm. To investigate this issue, we have used a genetic approach involving stable, transposase-producing transgenes that carry visible markers allowing them to be followed in a crossing scheme. Furthermore, rather than monitor the transposase biochemically, we have used a quantitative assay for transposase activity based on the mutability of a P-element-insertion allele that has an easily scored phenotype.
Materials and Methods
Stable Transposase-Producing Transgenes.
H(hsp/CP)2, a transgene inserted on an autosome by means of a hobo transposable element, contains a terminally truncated, but otherwise complete P element, including the native P promoter, fused to the Drosophila hsp70 (heat shock protein 70) promoter (9). H(hsp/CP)2 is marked with the mini-white eye color gene, and produces the P transposase in the germ line, even in the absence of heat shock. H(hsp/CP)3 is another autosomal insertion of this same transgene (9). P(ry+, Δ2-3)99B, a transgene inserted on an autosome by means of a P element, contains a P element lacking the last intron (between exons 2 and 3) of the transposase gene (12). It is marked with the wild-type rosy eye color gene and produces the P transposase in both the germ line and somatic tissues. However, because P(ry+, Δ2-3)99B has abnormal termini, it cannot be excised or transposed (13).
Mutability Assay for Transposase Activity.
Transposase activity was assayed by monitoring changes in a double-P element insertion mutation of the X-linked singed (sn) bristle gene. This mutation, called weak singed (snw), is a sensitive target for the P transposase (14, 15). When one of the incomplete P elements inserted in the snw allele is excised by transposase action in the germ line, flies with wild-type (sn(+)) or extreme mutant (sne) bristles are detected in the next generation (16). The frequency of these phenotypically different flies is an index of transposase activity in the previous generation. To assay for transposase activity in the male germ line, individual snw males were crossed to females with attached-X chromosomes, and their sons were scored for bristle phenotype. The snw mutability was calculated for each culture as the fraction of sne and sn(+) sons among those emerging within 17 days after starting the culture. An unweighted average of these mutabilities was obtained for each experimental group. Statistical differences between these averages were evaluated by z tests.
Genetic Scheme to Detect Maternal Transmission of Transposase Activity.
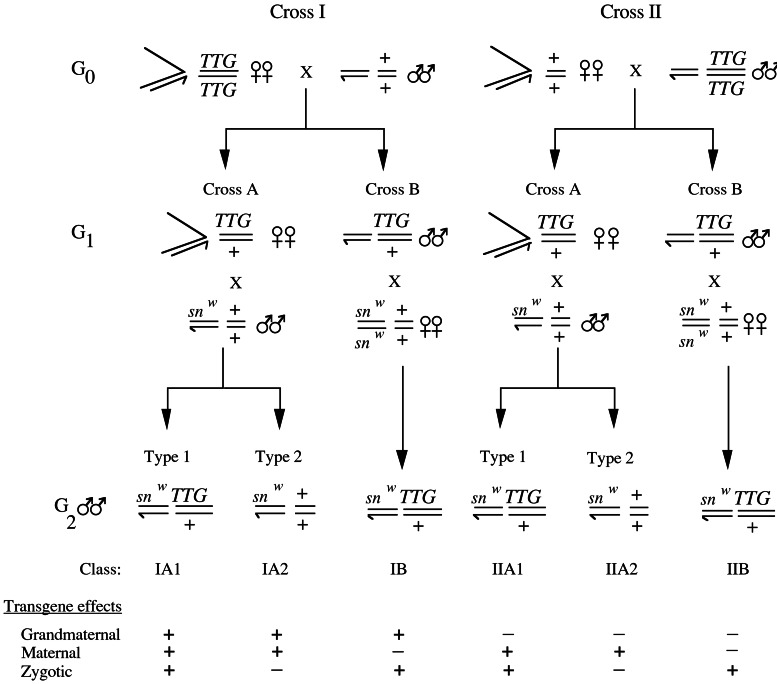
We used a crossing scheme (Fig. 1) in which the H(hsp/CP) and P(ry+, Δ2-3)99B transgenes could be followed because they conferred pigment in the eyes of flies that were mutant for an endogenous eye color gene. In cross I of the scheme, the transposase transgene was present in the grandmothers of the flies that were tested for transposase activity; in cross II, it was present in their grandfathers. From each cross, G1 flies that were heterozygous for the transgene were crossed to snw males (cross A) or snw females (cross B) to obtain snw G2 males, which were then tested individually for germ-line mutability of the snw allele. Attached-X chromosomes were used in cross A and in all of the mutability tests to achieve patroclinous transmission of the snw allele. Two types of males, distinguishable by their eye colors, were derived from cross A. Type 1, with pigmented eyes, was heterozygous for the transposase transgene and was therefore expected to exhibit a high level of germ-line snw mutability. Type 2, with mutant eyes, lacked this transgene and was therefore not expected to show germ-line snw mutability unless P transposase activity had been transmitted independently through the egg cytoplasm. From cross B, only males heterozygous for the transgene were tested for germ-line snw mutability. Altogether, six different classes of males from the various combinations of crosses (IA1, IA2, etc.) were studied. All crosses and snw mutability tests were incubated at 25°C without the administration of any heat shock treatments.
Figure 1.
Scheme to test for maternal transmission of P element transposase activity encoded by a transgene. The autosomal transposase-producing transgene (TTG) was either H(hsp/CP)2, H(hsp/CP)3, or P(ry+, Δ2-3)99B. In the experiments using H(hsp/CP)2 or H(hsp/CP)3, all of the flies were mutant for the endogenous white gene. In the experiment using P(ry+, Δ2-3)99B, all of the flies were mutant for the endogenous rosy gene. In all three experiments, the G0 and G1 females had attached-X chromosomes. To test for transposase activity, G2 males were mated individually to females with attached-X chromosomes and their sons were scored for the weak singed, extreme singed, and wild-type bristle phenotypes. The frequency of extreme singed and wild-type sons was used as a measure of transposase activity. In the experiment using P(ry+, Δ2-3)99B, the type 1 males were crossed to attached-X females from a P strain to repress somatic transposase activity in their offspring (17).
Results
Maternal Transmission of Transposase Activity.
The results of tests with the H(hsp/CP)2 transgene are given in the top of Table 1. The data from the IA2 and IIA2 males demonstrate that P transposase activity was transmitted independently of the transgene through the egg cytoplasm. The respective mutation rates for these males, 0.091 and 0.224, are 20% and 50% of the rates for males that inherited the transgene from their fathers (classes IB and IIB, with respective mutation rates of 0.468 and 0.441). Limited tests with H(hsp/CP)3, another insertion of this same transgene, corroborated the transmission of transposase activity through the egg cytoplasm (see middle of Table 1). For this insertion, IIA2 males had a mutation rate of 0.280. In all these tests to detect maternal transmission of transposase activity, the observed mutations were scattered among the test cultures, not confined to a small group.
Table 1.
Induction of snw mutability by transposase-producing transgenes in the germ lines of G2 males
| Class of G2 males | Transgene effects*
|
No. G2 males tested | G3
males
|
Mutation rate ± SE | |||||
|---|---|---|---|---|---|---|---|---|---|
| Gmtl | Mtl | Zyg | snw | sn+ | sne | Total | |||
| H(hsp/CP)2 transgene | |||||||||
| IA1 | + | + | + | 168 | 2,118 | 1,274 | 1,181 | 4,573 | 0.534 ± 0.013 |
| IIA1 | − | + | + | 170 | 1,810 | 1,233 | 1,175 | 4,218 | 0.575 ± 0.013 |
| IA2 | + | + | − | 244 | 6,205 | 356 | 296 | 6,857 | 0.091 ± 0.007 |
| IIA2 | − | + | − | 246 | 4,897 | 826 | 639 | 6,362 | 0.224 ± 0.011 |
| IB | + | − | + | 70 | 875 | 340 | 396 | 1,611 | 0.468 ± 0.018 |
| IIB | − | − | + | 63 | 756 | 266 | 289 | 1,318 | 0.441 ± 0.024 |
| H(hsp/CP)3 transgene | |||||||||
| IIA1 | − | + | + | 48 | 570 | 287 | 318 | 1,175 | 0.515 ± 0.018 |
| IIA2 | − | + | − | 98 | 1,733 | 400 | 275 | 2,408 | 0.280 ± 0.013 |
| P(ry+, Δ2-3)99B transgene | |||||||||
| IA1 | + | + | + | 49 | 204 | 641 | 436 | 1,281 | 0.841 ± 0.013 |
| IIA1 | − | + | + | 46 | 202 | 471 | 378 | 1,051 | 0.808 ± 0.016 |
| IA2 | + | + | − | 94 | 3,007 | 0 | 0 | 3,007 | 0.000 ± 0.000 |
| IIA2 | − | + | − | 81 | 2,125 | 0 | 0 | 2,125 | 0.000 ± 0.000 |
Gmtl, grandmaternal; Mtl, maternal; Zyg, zygotic.
The statistically significant difference between the mutation rates of the IA2 and IIA2 males in the experiment involving the H(hsp/CP)2 transgene suggests that maternal transmission of transposase activity is more effective when the mother inherits the transgene from her father than when she inherits it from her mother. Maternal transmission of transposase activity is therefore negatively influenced by a grandmaternal effect of the transgene. This effect might be caused by partial repression of the H(hsp/CP)2 transgene by the 66-kDa polypeptide produced by alternate splicing of P RNA in the grandmaternal germ line (9).
The combined effects of cytoplasmic transmission of transposase activity and chromosomal transmission of the H(hsp/CP)2 transgene are seen in the IA1 and IIA1 males. Their respective mutation rates of 0.534 and 0.575 are significantly greater than the rates observed for the IB and IIB males in which the transgene was paternally transmitted. In addition, the mutation rate of the IIA1 males is greater than that of the IA1 males, although not significantly so. This difference, which is in the same direction as that between the mutation rates for the IA2 and IIA2 males, may reflect the grandparental origin of the transgene.
To exclude the possibility that a source of the P transposase not associated with the H(hsp/CP)2 transgene was responsible for the observed snw mutability, snw males lacking the transgene were collected from the G3 flies and tested individually for snw mutability. No mutability was found (Table 2). Thus, the mutability detected in the experiment involving H(hsp/CP)2 was a bona fide result of transgene expression.
Table 2.
Stability of snw in the germ lines of G3 males lacking the H(hsp/CP)2 transgene
| Class of G3 males | No. G3 males tested | G4 males
|
|
|---|---|---|---|
| snw | Others | ||
| To exclude extraneous transposase sources | |||
| IA1 | 32 | 802 | 0 |
| IIA1 | 30 | 862 | 0 |
| IA2 | 81 | 2,116 | 0 |
| IIA2 | 97 | 2,390 | 0 |
| IB | 34 | 894 | 0 |
| IIB | 36 | 1,009 | 0 |
| To test for maternal transmission through two generations | |||
| IA3 | 48 | 1,272 | 0 |
| IIA3 | 49 | 1,379 | 0 |
An additional experiment was performed to determine if the transposase activity encoded by H(hsp/CP)2 could be transmitted maternally through two generations. G1 females from cross IA were mated to w-mutant males to obtain G2 females with mutant eyes (i.e., lacking the transgene). These females were then crossed to snw males, and their snw G3 sons (denoted as classes IA3 and IIA3) were tested for germ-line snw mutability. As the bottom of Table 2 shows, no such mutability was detected. Thus, transposase activity was not transmitted maternally through 2 generations.
An Impediment to Maternal Transmission of Transposase Activity.
The same type of reciprocal crossing scheme was used to investigate maternal transmission of the P transposase activity encoded by P(ry+, Δ2-3)99B, a transgene lacking the last intron in the P sequence. However, in this experiment only males derived from cross A were tested for transposase activity. As can be seen from the results in the bottom of Table 1, the two classes of males carrying the P(ry+, Δ2-3)99B transgene had high germ-line mutation rates, 0.840 (IA1) and 0.808 (IIA1). Moreover, in all these males the cuticle had patches of snw, sn(+), and sne bristles, indicating that the transposase encoded by P(ry+, Δ2-3)99B was somatically active. By contrast, none of the males lacking the P(ry+, Δ2-3)99B transgene (classes IA2 and IIA2) showed this somatic mosacism, and furthermore, none of them exhibited any germ-line snw mutability. This lack of snw mutability was unexpected given the results from the experiments involving H(hsp/CP)2 and H(hsp/CP)3. Thus, unlike the H(hsp/CP) transgene, the P(ry+, Δ2-3)99B transgene is unable to transmit transposase activity through the egg cytoplasm.
Discussion
H(hsp/CP)2, H(hsp/CP)3, and P(ry+, Δ2-3)99B all produce active P transposase in the female germ line (9, 18). What, therefore, might explain the inability of P(ry+, Δ2-3)99B to transmit this activity through the egg cytoplasm independently of the transgene itself? The obvious candidate is the missing intron in P(ry+, Δ2-3)99B. In nature this sequence of 190 nucleotides keeps the transposase out of the soma, where it could jeopardize the fly's life by causing P element excision and transposition. However, transmission of transposase activity through the oocyte poses a problem that does not seem to have been appreciated previously. If either the transposase or fully spliced P RNA passed into the oocyte, either could become localized in regions that would give rise to somatic tissues in the fly, thereby subverting the RNA processing mechanism that prevents transposase synthesis (and action) in the soma. Consequently, some mechanism must exclude the transposase and fully spliced P RNA from regions of the oocyte that will later form somatic tissues. The failure of P(ry+, Δ2-3)99B to transmit transposase activity through the oocyte suggests a simple hypothesis: the transposase protein cannot enter the oocyte, and the same intron that precludes transposase synthesis in the soma is required for RNA transport into the oocyte, or for its persistence there. On this hypothesis, only RNAs that possess this intron can enter or persist in the oocyte, and those that do so can be fully processed only if they become localized in the region that will become the primordial germ line in the embryo after fertilization. Thus, even within the oocyte, the P element would preserve a sense of the germ line/soma distinction.
Analysis of the last P intron reveals that it contains a motif of nine nucleotides (CTGTTTCTT, beginning at nucleotide 2089 in the P sequence) similar to sequences thought to be involved in the maternal transmission of bicoid and nanos RNAs, two of the factors that establish embryonic polarity in Drosophila (19). The bicoid sequence, TTGTTCCTG, differs from the sequence in the last P intron by three nucleotides; the nanos sequence, CTGTTTCTG, differs from it by only one nucleotide. Both the bicoid and nanos sequences are located in the 3′ untranslated regions of their respective mRNAs, rather than within an intron. During oogenesis, these mRNAs are transported from the nurse cells into the oocyte; bicoid mRNA becomes localized at the oocyte's anterior pole, whereas nanos mRNA becomes localized at the posterior pole. Later, the proteins translated from these mRNAs play key roles in creating the anterior–posterior axis of Drosophila embryos.
Based on an analysis of bicoid mRNA, Gottlieb (19) proposed that the consensus motif YTGTTYCTG is a generalized signal for RNA localization in Drosophila oocytes, and possibly in other organisms as well. The signals that localize RNA to specific positions within oocytes seem to be more complex (20). Deletion of the last intron in P(ry+, Δ2-3)99B may therefore have eliminated a key signal for the transport of P element RNA from the nurse cells into the oocyte.
It seems unlikely that other differences between the H(hsp/CP) and P(ry+, Δ2-3)99B transgenes can account for the ability of one but not the other to transmit transposase activity through the egg cytoplasm independently of the transgene itself. Both transgenes produce transposase activity in the female germ line, and H(hsp/CP) does so without induction by heat shock (9). In fact, judging from snw mutability experiments, P(ry+, Δ2-3)99B produces five times as much transposase activity in the female germ line as uninduced H(hsp/CP) (9, 18). Maternal transmission of transposase activity therefore does not seem to be caused by the presence of the hsp70 promoter in the H(hsp/CP) transgene. Nor does it seem to be caused by the genomic position of this transgene because two different insertions of it were able to transmit transposase activity through the egg cytoplasm. These observations argue that the lack of maternal transmission of transposase activity with P(ry+, Δ2-3)99B is not caused by something other than the missing last P intron.
These findings have important implications for analyses of P element regulation, which have assumed that P-encoded polypeptides are responsible for the P cytotype, the maternally inherited condition that naturally represses transposase activity in the germ line. Our results imply that P-encoded polypeptides cannot be the basis of the P cytotype unless they are synthesized in the oocyte or embryo from RNAs that were derived from the mother. Because these RNAs apparently must carry the last P intron, polypeptides encoded by P elements lacking this intron cannot contribute to the P cytotype, although they could contribute to other forms of P element regulation. The polypeptide encoded by KP, a geographically widespread element implicated in P regulation, is an example (21). Indeed, repression by a H(hsp/KP) transgene is not transmitted from mother to offspring independently of the transgene itself (10).
The 66-kDa polypeptide produced by alternate splicing from complete P elements has been hypothesized to form the basis of the P cytotype, and transgenes designed to express this polypeptide in the germ line have been found to have some regulatory ability (11). However, this ability was not maternally transmitted and the polypeptide encoded by these transgenes could not be detected in oocytes (11). The P sequence in these transgenes was truncated at nucleotide 2078 in the last intron, which deletes the nine nucleotide motif implicated in maternal transmission of developmentally significant Drosophila RNAs. Failure to detect the 66-kDa polypeptide in oocytes may therefore have been caused by loss of the signal for maternal transmission of the P RNA.
Our findings indicate that P elements are adapted to the Drosophila germ line because they possess an intron that simultaneously precludes transposase synthesis in the soma and excludes transposase mRNA from the oocyte. This bifunctional intron therefore plays an important role in P element regulation.
Acknowledgments
Technical help was provided by Bradley Morrison and John Raymond. Stanley J. P. Iydurai identified the nine nucleotide motif in the last P element intron. Jarad Niemi helped to prepare the manuscript, and Johng Lim provided comments on a draft. Financial support came from National Institutes of Health Grant R01 GM40263.
References
- 1.International Human Genome Sequencing Consortium. Nature (London) 2001;409:860–921. [Google Scholar]
- 2.Engels W R. In: Mobile DNA. Berg D E, Howe M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 437–484. [Google Scholar]
- 3.Kidwell M G. Proc Natl Acad Sci USA. 1983;80:1655–1659. doi: 10.1073/pnas.80.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engels W R. Genetics. 1997;145:11–15. doi: 10.1093/genetics/145.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidwell M G, Kidwell J F, Sved J A. Genetics. 1977;86:813–833. doi: 10.1093/genetics/86.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engels W R. Genet Res. 1979;33:219–236. doi: 10.1017/s0016672300018267. [DOI] [PubMed] [Google Scholar]
- 7.Roche S, Schiff M, Rio D C. Genes Dev. 1995;9:1278–1288. doi: 10.1101/gad.9.10.1278. [DOI] [PubMed] [Google Scholar]
- 8.Laski F A, Rio D C, Rubin G M. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 9.Simmons M J, Haley K J, Grimes C D, Raymond J D, Niemi J B. Genetics. 2002;161:195–204. doi: 10.1093/genetics/161.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons M J, Haley K J, Grimes C D, Raymond J D, Fong J C L. Genetics. 2002;161:205–215. doi: 10.1093/genetics/161.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misra S, Rio D C. Cell. 1990;62:269–284. doi: 10.1016/0092-8674(90)90365-l. [DOI] [PubMed] [Google Scholar]
- 12.Robertson H M, Preston C R, Phillis R W, Johnson-Schlitz D, Benz W K, Engels W R. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson H M. Dros Inform Service. 1996;77:99. [Google Scholar]
- 14.Engels W R. Proc Natl Acad Sci USA. 1979;76:4011–4015. doi: 10.1073/pnas.76.8.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engels W R. Science. 1984;226:1194–1196. doi: 10.1126/science.6095450. [DOI] [PubMed] [Google Scholar]
- 16.Roiha H, Rubin G M, O'Hare K. Genetics. 1988;119:75–83. doi: 10.1093/genetics/119.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson H M, Engels W R. Genetics. 1989;123:815–824. doi: 10.1093/genetics/123.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmusson K E, Raymond J D, Simmons M J. Genetics. 1993;33:605–622. doi: 10.1093/genetics/133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlieb E. Proc Natl Acad Sci USA. 1992;89:7164–7168. doi: 10.1073/pnas.89.15.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gavis R, Curtis D, Lehmann R. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 21.Black D M, Jackson M S, Kidwell M G, Dover G A. EMBO J. 1987;6:4125–4135. doi: 10.1002/j.1460-2075.1987.tb02758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]