Abstract
Stem rust caused by Puccinia graminis f. sp. tritici was among the most devastating diseases of barley in the northern Great Plains of the U.S. and Canada before the deployment of the stem rust-resistance gene Rpg1 in 1942. Since then, Rpg1 has provided durable protection against stem rust losses in widely grown barley cultivars (cvs.). Extensive efforts to clone Rpg1 by synteny with rice provided excellent flanking markers but failed to yield the gene because it does not seem to exist in rice. Here we report the map-based cloning and characterization of Rpg1. A high-resolution genetic map constructed with 8,518 gametes and a 330-kb bacterial artificial chromosome contig physical map positioned the gene between two crossovers ≈0.21 centimorgan and 110 kb apart. The region including Rpg1 was searched for potential candidate genes by sequencing low-copy probes. Two receptor kinase-like genes were identified. The candidate gene alleles were sequenced from resistant and susceptible cvs. Only one of the candidate genes showed a pattern of apparently functional gene structure in the resistant cvs. and defective gene structure in the susceptible cvs. identifying it as the Rpg1 gene. Rpg1 encodes a receptor kinase-like protein with two tandem protein kinase domains, a novel structure for a plant disease-resistance gene. Thus, it may represent a new class of plant resistance genes.
The barley stem rust-resistance gene Rpg1 confers resistance to many pathotypes of the stem rust fungus Puccinia graminis f. sp. tritici. North American barley cultivars (cvs.) with Rpg1 were first released in 1942 and since that time there have been no significant losses as a result of stem rust except for a minor epidemic caused by pathotype QCC in 1989–91 (1). The Rpg1 resistance is dominant and considered durable (2), because it has remained effective for a long time in widely grown cvs. across the stem rust-prone northern Great Plains (1). The earliest known source of stem rust resistance in the U.S. was derived from an unimproved cv. imported by the U.S. Department of Agriculture from Canton Lucerne, Switzerland, in 1914. Two selections from this seed lot became the cvs. Chevron and Peatland. Peatland has been the main source of stem rust resistance in most Canadian cvs. since 1947 (1). A potentially different source of resistance was discovered during the severe stem rust epidemic of 1935 by Sam Lykken, a farmer from Kindred, North Dakota, who identified a single healthy plant in a heavily rusted field of Wisconsin 37 barley (3). This plant was saved and gave rise to the cv. Kindred. Kindred and cvs. derived from it have dominated the barley hectarage in the northern Great Plains of the U.S. from 1942 to the present (1). Both sources of barley stem rust resistance are the results of the Rpg1 gene (4, 5).
The Rpg1 gene was mapped to the short arm of barley chromosome 1(7H) with morphological and molecular markers (6, 7). Our initial efforts at map-based cloning of the Rpg1 gene were focused on using rice as an intergenomic cloning vehicle (8, 9). Despite the excellent synteny between barley chromosome 1(7H) short arm and rice chromosome 6 short arm and the excellent alignment of flanking markers, the Rpg1 gene was not found in the syntenous position in rice (10). The development of a barley bacterial artificial chromosome (BAC) library allowed us to initiate a chromosome walk toward Rpg1 (11). A resistance gene analog (RGA), B9 (12), and barley homologues of the maize rust-resistance gene Rp1-D (pic20) (13) were proposed as the barley stem rust-resistance gene Rpg1. We mapped both B9 and the pic20 homologues at high resolution and found that they did not cosegregate with Rpg1. The pic20 probe, however, did identify a complex RGA locus near Rpg1 and helped to extend the BAC contig (14).
Numerous genes that confer resistance to a variety of plant pathogens have been cloned and characterized (15–17). The genes that seem to be involved in signal transduction and behave in a gene for gene manner (18) have been classified into four groups. The majority resembles intracellular receptors and contains a predicted nucleotide binding site/leucine-rich repeat (NBS/LRR) structure. Another group encodes an LRR putative extracellular receptor and a membrane anchor. Two additional groups contain serine/threonine kinase domain without any obvious ligand-binding domain (i.e., Pto) and receptor kinases with LRR extracellular ligand-binding, transmembrane, and serine/threonine kinase domains (i.e., Xa21) (16). A recently reported tomato Verticillium wilt-resistance gene Ve seems to encode a new group with cell-surface glycoprotein, signals for receptor-mediated endocytosis, and leucine zipper or PEST sequences [PEST, region rich in proline (P), glutamate (E), serine (S), and threonine (T); ref. 19].
Here we report map-based cloning and partial characterization of the barley stem rust-resistance gene Rpg1. The Rpg1 gene identity was confirmed by high-resolution genetic and physical mapping, sequencing of multiple alleles from resistant and susceptible lines, a fortuitous recombination that combined parts from the resistant and susceptible parents, and exclusion of other potential candidate genes by sequence analysis. The Rpg1 gene encodes a receptor kinase-like protein with two tandem protein kinase domains, which is a novel structure for a plant disease-resistance gene. It does not contain a strong membrane-targeting motif and known receptor sequences, suggesting a mechanism of action similar to the tomato Pto gene (20, 21). The barley Rpg1 gene may be useful for durable stem rust-resistance in other species.
Materials and Methods
Materials.
Barley crosses used to produce recombinant gametes are given in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Most of the crosses were produced by, and for, the North American Barley Genome Project and are described at http://www.css.orst.edu/barley/nabgmp/nabgmp.htm. The F2 materials were generated in this study. FN53 is a fast neutron-induced brachytic dwarf (brh1.ae) derived from cv. Steptoe (22). MD2 is a barley genetic stock carrying multiple dominant morphological markers (23). Resistant (24) and susceptible (25) barley lines used are described in supporting information on the PNAS web site. Libraries used include the cv. Morex BAC library (11) and cDNA libraries made from various tissues and with different biotic and abiotic stresses (26) (http://wheat.pw.usda.gov/ggpages/bgn/31/close.htm).
Molecular Marker Development.
Low-copy probes suitable for genetic mapping and hybridization-based BAC library searching were identified from small insert libraries developed from individual BAC clones. The BAC clone DNA was digested with four- or six-base recognition restriction enzymes and subcloned into a plasmid vector. The subclones were hybridized with 32P-labeled barley total genomic DNA probe. Subclones that did not show any or very little hybridization signal were selected and tested further by hybridization to Southern blots of HindIII-digested barley genomic DNA. The clones generated with six-base recognition enzymes were further digested with several restriction enzymes to develop small inserts. The digested DNA was electrophoresed, blotted, and transferred to filters for hybridization with barley total genomic DNA probe (27). DNA fragments that failed to hybridize were selected as low-copy probes.
Genetic Mapping.
Recombinants between the Rpg1 flanking markers ABG704 and ABG077, derived from 8,518 gametes (Table 1), were used for high-resolution mapping. The genetic distances are expressed in number of crossovers. The stem rust phenotype of progeny was determined as described and mapped in reference to the molecular markers (7).
BAC Contig Development.
BAC clones were isolated from the 6.3X cv. Morex BAC library (11) by hybridizing the arrayed BAC colony filters with 32P-labeled probes. BAC DNA was isolated by using the standard alkaline lysis procedure (28). Low-copy probes were restriction-enzyme mapped on the most-extending BAC clone, and the most-extending probes were hybridized to the BAC library to identify further extending BAC clones. The process was repeated as needed to complete the BAC contig.
Sequencing and Sequence Analysis.
DNA was sequenced with the BigDye terminator system on ABI Prizm 377 DNA sequencer (Applied Biosystems) at the Bioanalytical Center, Washington State University, Pullman. The region delimited by the crossovers flanking Rpg1 (Fig. 1) was screened for putative genes by sequencing low-copy clones. Small insert sublibraries were developed with four- and six-base restriction endonucleases essentially as described above for low-copy probe development except that more restriction enzymes were used and higher coverage was achieved. All of the low-copy clones were sequenced and analyzed with the blastx and blastn algorithms (29). Sequences without significant matches in blast searches were screened for ORFs and putative genes with the gene prediction software GENEMARK.HMM (http://opal.biology.gatech.edu/GeneMark/eukhmm.cgi).
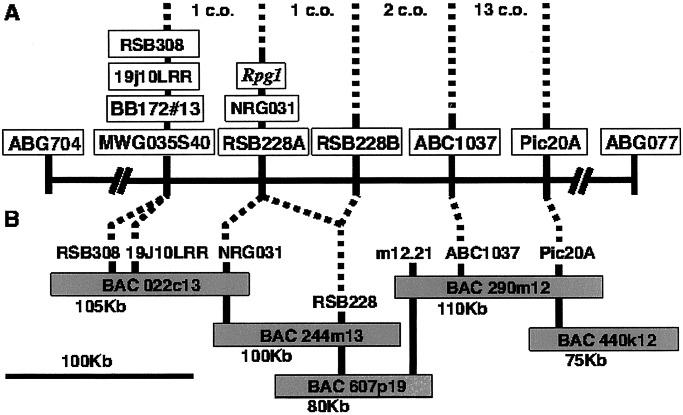
Figure 1.
A genetic and physical map of barley stem rust-resistance gene Rpg1 region on chromosome 1(7H). (A) high-resolution genetic map based on 8,518 gametes showing critical markers and the number of total crossovers (c.o.) separating them. (B) Physical map encompassing Rpg1 based on BAC clone contig. The distance from Pic20A to RSB308 was estimated to be ≈270 kb.
Genomic DNA was isolated with a modified CTAB method (30), and specific fragments were amplified by PCR. PCR reactions of 50 μl contained 100 ng of genomic DNA, 0.2 mM of each dNTP, 25 pmols of each primer, 2.5 μl of RedTaq DNA polymerase (Sigma), and 5 μl of 10× RedTaq reaction buffer. Amplification was performed in a PTC-100 programmable thermal controller (MJ Research, Cambridge, MA) at 95°C for 4 min; 35 cycles of 95°C for 1 min, 62°C for 1 min, and 72°C for 1 min; followed by 7 min at 72°C. The PCR fragments were electrophoresed on 1% agarose gels and eluted from the agarose. The PCR fragments were sequenced with the same forward and reverse primers used to amplify the fragments. Primers were designed from the Morex sequence to amplify about 1-kb overlapping fragments that covered the entire gene (Table 2, which is published as supporting information on the PNAS web site). Internal primers were designed to sequence across gaps and low-quality sequence. DNA fragments including sequence changes critical to the interpretation of the results were resequenced with different DNA preparations and different primer pairs. Sequences were assembled with the genetic computer group program (31) supported by the Visualization, Analysis, and Design in the Molecular Sciences center at Washington State University.
RNA Isolation and Reverse Transcription (RT)-PCR.
Total leaf RNA from 21-day-old Morex, Steptoe, and ASM170 plants was isolated with the Totally RNA kit (Ambion, Austin, TX). Integrity and quantity of the RNA samples were checked by formaldehyde denaturing agarose gel electrophoresis. Approximately 1 μg of the total RNA was used for RT-PCR reactions with SuperScript One-Step RT-PCR system (Invitrogen) under the manufacturer's recommended standard conditions. RT reactions were primed with gene-specific primers. Northern blots were done as recommended by Amersham Pharmacia Biotech (http://www.apbiotech.com/technical/documentation/dna_protein/tt159.htm).
Results
High Resolution Genetic Mapping of the Rpg1 Gene.
The Rpg1 locus was previously mapped to the telomeric end of barley chromosome 1(7H)S (9, 10). A high-resolution genetic map, based on 8,518 gametes (Table 1), was developed with recombinants from the region between the Rpg1 flanking markers ABG704 and ABG077. The interval between MWG035S40 (BB172#13) and the barley homologue of the maize Rp1-D gene (pic20A) delimited the Rpg1 region, contained 17 recombinants, and represented a genetic distance of ≈0.21 centimorgan (cM) (Fig. 1).
Physical Mapping of the Rpg1 Region.
A BAC clone contig was established for the Rpg1 region by chromosome walks from pic20A and 19j10LRR loci (Fig. 1). The probe 19j10LRR was obtained from L. D. Holappa (M. K. Walker-Simmons Laboratory, Pullman, WA). A blastx search revealed it to be “similar to a non-LTR [long terminal repeat] retroelement reverse transcriptase” (S = 170, 3e-41). Hybridization to barley polymorphism filters revealed many bands, some polymorphic, which were mapped. A 2.1-kb band, generated with the restriction enzyme NcoI, mapped one crossover distal to Rpg1 and cosegregated with MWG035S40 and other markers. The probe was used to isolate ≈2,000 BAC clones from the cv. Morex library. A BAC clone, 022c13, containing the mapped band was identified after digestion with NcoI and hybridization with 19j10LRR. A chromosome walk was initiated from this BAC clone and led to an overlap with BAC clones isolated from the pic20A walk.
Identification of Candidate Genes.
Three putative candidate genes were identified by draft sequencing in the region between the two closest Rpg1 flanking crossovers (Fig. 1). Two were receptor kinase-like genes and one was a truncated phophoenolpyruvate kinase-like gene that was not considered further. The first candidate gene was a 5-kb HindIII low-copy clone designated RSB228. This clone, when used as a probe, was polymorphic in all crosses, and in DraI restriction enzyme-digested Morex and Steptoe genomic DNA yielded three and two polymorphic bands, respectively (Fig. 4, which is published as supporting information on the PNAS web site). All bands cosegregated in all recombinants except one, AMS170, which contained one Morex and one Steptoe band and was susceptible to stem rust (rpg1). Sequence analysis of RSB228 revealed that it contained ORFs with high amino acid homology to several receptor kinase-like proteins from Arabidopsis and the SRK receptor kinase-like gene family. Thus, this gene was a strong candidate to be Rpg1, and subsequent sequence analysis of stem rust-resistant (Rpg1) and susceptible (rpg1) cv. alleles confirmed this conclusion (data presented below). A second receptor kinase-like gene, NRG031, was identified and sequenced from Morex, OSU6 (both resistant), and Steptoe (susceptible). Alleles from resistant and susceptible lines were very similar, excluding NRG031 from Rpg1 candidacy.
The candidate genes were identified by sequencing of the approximately 35 kb immediately distal to Rpg1. This sequenced region consisted primarily of long terminal repeat retrotransposons that are a common feature of the intergenic space in barley (32) and in other grass species, such as maize (33). For the about 70 kb remaining, sublibraries were constructed with the 4bp recognition endonucleases Sau3AI and TaqI and the 6-bp recognition restriction endonucleases HindIII and EagI. Approximately 18× coverage of this region was screened for low-copy clones, sequenced, and analyzed with the blastx and blastn algorithms. Sequences with putative ORFs were hybridized to Southern blots of HindIII-digested genomic DNA. Putative genes were completely sequenced from cv. Morex BAC DNA. In total, 80 kb of nonredundant putatively low-copy sequence was generated and examined with blast searches as well as with the gene prediction software GENEMARK.HMM. No other putative genes were identified.
Characterization of the Candidate Genes.
The Morex (resistant) allele of candidate gene 1 (RSB228) seemed to encode a functional gene. The Steptoe (susceptible) allele contained 40-bp differences, three of which introduced stop codons in the 4th, 8th, and 13th exons, indicating a defective gene. The Steptoe × Morex recombinant AMS170 allele had a crossover within the gene in the 436–524-aa region including parts of exons 7 and 8. The distal part of the AMS170 allele contained the two stop codons in the 8th and 13th exons from Steptoe, explaining the stem rust-susceptible phenotype (rpg1). The other part of the AMS170 gene had the Morex haplotype and eliminated all proximal genes from Rpg1 candidacy (Fig. 1).
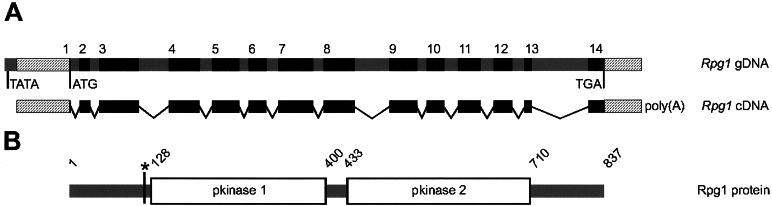
The genomic and cDNA sequence comparisons predicted the gene to contain 14 exons in a total sequence of 4,466 bp coding for an 837-aa (94.5 kDa) protein (Fig. 2). A blastp search of the nonredundant protein database showed the best homology to numerous hypothetical receptor kinase-like proteins. The best hit to characterized function proteins was to the SRK family of self-incompatibility S-receptor kinases. The highest homology was to the kinase domain regions. The N-terminal part of the protein sequence exhibited low homology to several predicted Arabidopsis proteins with unknown function. A blastn search with the Rpg1 coding sequence of the nonredundant database revealed no significant homologies. The deduced protein sequence contained two tandem pfam00069 protein kinase domains. Program SIGNALP VERSION 2.0 (http://www.cbs.dtu.dk/services/SignalP/) indicated that the predicted protein sequence does not contain a signal peptide. The predictprotein tool at the European Bioinformatics Institute (http://www.ebi.ac.uk/∼rost/predictprotein) identified a putative transmembrane domain from amino acid 321 to 331, although with low probability. Alternatively, the tmpred program at http://www.ch.embnet.org/software/TMPRED_form.html found 3 putative transmembrane domains, amino acids 23–47, amino acids 316–334, and amino acids 626–643, although only the 316–334-aa domain was predicted with a score higher than threshold. Future work will be required to determine whether the predicted RPG1 protein is membrane-bound or intracellular.
Figure 2.
Rpg1 gene, cDNA, and translated protein structure. (A) Genomic and cDNA structures with introns (gray) and exons (black) are shown to scale. The mRNA 5′ and 3′ untranslated regions are hatched. Exons are numbered above the drawing. Predicted TATA box, translation start site, and stop codon are shown below. The Rpg1 mRNA structure is shown below the gene. The mRNA was reconstructed with the sequence of 5′ truncated cDNA clone HVSMEf55g12 and sequences of the RT-PCR products from cv. Morex leaf total RNA. (B) Organization of the protein domains is shown to scale with predicted boundaries indicated above. pkinases 1 and 2 denote two tandem protein kinase domains. The asterisk indicates an asparagine residue within a putative N-glycosylation site NFSE.
Candidate gene 2 (NRG031) was homologous to putative receptor kinase-like genes. Eukaryotic GENEMARK.HMM version 2.2a (http://opal.biology.gatech.edu/GeneMark/eukhmm.cgi) gene prediction program predicted 2 exons in a total sequence of 981 bp coding for a 327-aa protein. Sequence analysis of alleles from resistant (Morex and OSU6) and susceptible (Steptoe) lines showed differences at only two amino acid positions among the three alleles sequenced. Morex and Steptoe had isoleucine, whereas OSU6 had threonine in position 4. At position 241, OSU6 and Steptoe had glycine, but Morex had alanine. Because all amino acid differences found in the susceptible allele were also present in one or the other of the resistant alleles, this gene was removed from further candidacy.
Analysis of Rpg1 cDNA.
Hybridization of the RSB228 probe to 30 arrayed cDNA filters representing 10 different tissue libraries and ≈500,000 clones identified four cDNA clones. Sequence analysis revealed three different expressed genes homologous to Rpg1. One partial cDNA clone, HVSMEf 055g12, was identical to the genomic sequence, whereas another came from a very similar member of the same gene family (ABC1037) located proximal to Rpg1 and excluded from candidacy by genetic mapping. The third clone came from a more distantly related member of the Rpg1 gene family and mapped to chromosome 7(5H). The HVSMEf 055g12 cDNA was used to predict the 3′ end of the Rpg1 gene and intron–exon structure for exons 9–14. The homologous, nearly full-length cDNA ABC1037 was used to predict a preliminary intron–exon structure of the 5′ part of the Rpg1 gene. Sequences of RT-PCR products generated from cv. Morex leaf total RNA with primers corresponding to predicted exons allowed for the construction of apparently the complete coding sequence of the Rpg1 gene. Even though the region from exon 1 to 3 forms an uninterrupted ORF, RT-PCR products indicated the presence of two introns. Translation of the reconstructed cDNA sequence suggested that the coding sequence was complete, because an in-frame stop codon was detected four codons upstream of the first predicted methionine. The gene prediction programs genscan (http://genes.mit.edu/GENSCAN.html) and fgenesh (http://www.softberry.com), as well as neural network promoter prediction (http://www.fruitfly.org/seq_tools/promoter.html) localized the putative transcription start site of the gene about 400 bp upstream of the translation start site.
Northern blots of Morex, Steptoe, and ASM170 total or poly(A) RNA failed to yield positive bands with Rpg1 probe although positive results were obtained with an actin probe (data not shown). This finding suggested that the gene was expressed at low levels.
Comparison of Rpg1 Alleles from Resistant and Susceptible Barley Lines.
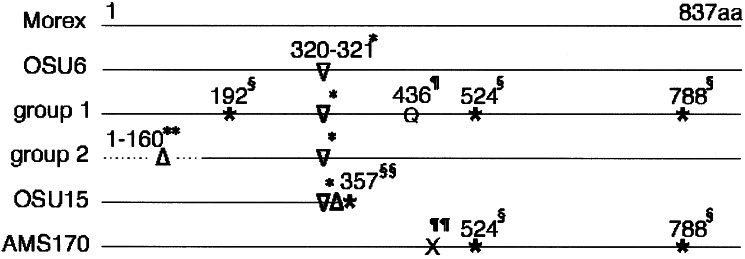
Sequence and RFLP analysis of 19 susceptible and 9 resistant barley lines confirmed that the RSB228 probe was the Rpg1 gene. The Morex gene sequence was compared with resistant cv. alleles from Kindred, Chevron, Peatland, Q21861, Leger, Bowman, 80-TT-29, and OSU6. All sequences, except OSU6, were identical, suggesting that the candidate gene was functional and probably from the same source (see Discussion). The resistant Hordeum vulgare subsp. spontaneum line OSU6 allele had a 3-bp insertion that resulted in a serine to arginine conversion and an insertion of a phenylalanine in the 6th exon at amino acid position 320 (Fig. 3 and Fig. 5, which are published as supporting information on the PNAS web site). This was the only nucleic acid difference between OSU6 and Morex that resulted in an amino acid difference. Other transitions occurred in the 7th exon and 8th and 13th introns, and a 30-bp deletion was observed in the 13th intron. Thus, the Rpg1 gene seemed to be very highly conserved and functional in all resistant lines analyzed.
Figure 3.
A diagram representation of the derived amino acid sequence alignment of Rpg1 alleles from resistant (Rpg1) and susceptible (rpg1) cvs. The differences from the cv. Morex sequence are indicated. “Morex” includes additional resistant cultivars Q21861, Kindred, Chevron, Peatland, Leger, Bowman, and 80-TT-29. “OSU6” is a resistant H. vulgare subsp. spontaneum line PBI004-7-0-015 (accession no. 8321). Group 1, Sequence from susceptible cultivars Steptoe, Lion, Wisconsin38, and MoravianIII. Group 2, Sequences from susceptible cultivars SM89010, Dicktoo, and Gobernadora. “OSU15” is a susceptible H. vulgaresubsp. spontaneum line Wadi Qilt 23–38. AMS170 is a doubled haploid line with a recombination in the Rpg1 gene. *, A 3 bp insertion that introduces a serine to arginine conversion and inserts a phenylalanine at amino acid positions 320 and 321. §, Base pair substitutions in group 1 cultivars which converts amino acid codons to stop codons at amino acid positions 192, 524, and 788. ¶, A single base pair substitution results in lysine to glutamine conversion. **, Highly diverged with multiple amino acid substitutions, a small deletion, and a large deletion. §§, A single base pair deletion introduces a frame shift that results in a stop codon. ¶¶, Crossover region in the recombinant AMS 170 was narrowed to a 295-bp region between the lysine at amino acid position 436 of Morex and the stop codon at amino acid position 524 of Steptoe.
In addition to Steptoe and AMS170 discussed above, the alleles from susceptible barley lines Lion, MoravianIII, Wisconson38, SM89010, Dicktoo, Gobernadora, and OSU15 were sequenced. The susceptible allele sequences formed three groups, all seeming to encode defective proteins (Figs. 3 and 5). Group I cvs. Lion, MoravianIII, Steptoe, and Wisconsin38 alleles, compared with Morex, had 42 bp differences translating into three stop codons and 8-aa changes. The 5′ end, including all of the first exon (6 bp), the first intron, and the first 38 bp of exon 2, of the alleles sequenced from Group II cvs. Dicktoo, Gobernadora, and SM89010 were very similar among themselves but were not homologous to the Morex allele. The sequence after this point to amino acid 160 is still quite diverged with several bp substitutions resulting in amino acid differences and a 96-bp deletion in exon 3. These differences indicated that the gene would not be functional. The allele from OSU15, the only representative of Group III, was very similar to the OSU6 sequence. However, it contained a single bp deletion in the 7th exon resulting in a frame shift that introduced a stop codon at amino acid position 358 and a predicted truncated protein.
Other susceptible barley lines (Baronesse, Betzes, Golden Promise, Harrington, Klages, Oderbrucker, OWB-D, OWB-R, and 80-tt-30) were analyzed and failed to hybridize with the RSB228 probe. PCR amplification, with the primers described previously, also failed. These lines seemed to have a highly diverged or deleted Rpg1 gene.
Synteny with Rice and Other Cereals.
Hybridization of the RSB228 probe with rice cvs. Nipponbare and M201 genomic DNA was negative. Searching two rice cv. Nipponbare BAC libraries representing 11X and 15X genomes yielded no positive clones, suggesting there are no homologues detectable by hybridization present in rice. A blastn search of the Chinese rice genome database (http://210.83.138.53/rice/) representing the rice subspecies indica genome sequence also failed to uncover homologous regions. A tblastx search of the “other” expressed sequence tag (EST) database yielded positive hits with wheat in addition to barley. Other than within the Triticeae, fair homology was observed with ESTs from Sorghum propinquum (S = 179, 4e-46) and Sorghum bicolor (S = 93, 9e-45), Medicago trancatula (S = 80, 3e-40), Lycopersicon esculentum (S = 75, 4e-37), and Zea mays (S = 99, 3e-35).
Discussion
A candidate gene for the barley stem rust-resistance gene Rpg1 was identified by map-based cloning. The Rpg1 gene identity was confirmed by high-resolution genetic and physical mapping, sequencing of multiple alleles from resistant and susceptible lines, a fortuitous recombination that combined parts from the resistant and susceptible parents, and exclusion of other potential candidate genes by sequence analysis. Conceptual translation of the sequence showed amino acid homology to the S receptor kinase gene SRK protein family. SRK (S locus receptor kinase) encodes a plasma membrane-spanning receptor serine/threonine kinase specific to the stigma epidermis (34, 35). It is presumed to be the determinant of self-incompatibility specificity in the stigma (36). Another interesting homology was to the maize kinase interacting kinase KIK1 gene predicted to encode a receptor kinase-like protein with an N-terminal signal peptide, an extracellular ligand-binding domain, a membrane-spanning domain, and a cytoplasmic serine/threonine protein kinase domain (37). The KIK1 protein was suggested to transfer a signal downstream by means of a kinase-associated protein phosphatase (KAPP) (37). Another protein with homology to the Rpg1 gene product was the GA-induced LRR receptor kinase-like protein OsTMK from rice, which was also suggested to interact with a KAPP protein (38).
Receptor kinase-like gene products are defined by the presence of a potential N-terminal hydrophobic signal peptide, an N-terminal domain with a transmembrane region, and a C-terminal intracellular protein kinase domain (39, 40). The two largest subclasses of plant receptor kinases are those whose extracellular domain contains LRRs or similarity to the S-locus glycoprotein. Some extracellular domains of plant receptor kinases are unique. The cloned rice Xa21 gene (41), which confers resistance to Xanthomonas oryzae pv. oryzae and belongs to a receptor kinase-like gene family, showed homology to Rpg1 in the kinase domain. However, the Xa21 gene contains the LRR extracellular domain whereas the Rpg1 gene has an N-terminal domain that does not resemble any previously described receptor. In that sense, Rpg1 might be more similar to the tomato Pto gene (20), which confers resistance to Pseudomonas syringae pv. tomato. The Pto gene, also showing similarity to Rpg1 in the kinase domain, is a serine/threonine kinase without an identifiable ligand-binding domain (21). However, the Pto gene product and the corresponding avirulence gene product of the pathogen, avrPto, have been shown to interact directly (42). The Pto threonine 204 residue implicated in interaction with avrPto (43) is conserved in both kinase domains of the RPG1 protein (positions 302 and 612) as it is in a number of other kinases including Xa21, KIK1, SRK, and OsTMK. Thus, the significance of this observation is not clear.
The Rpg1 gene product is unique because it contains two tandem protein kinase domains. This finding has not been previously reported in cloned plant pathogen-resistance genes and seems to be uncommon. Some Arabidopsis-predicted genes encode two kinase domains, e.g., AC007234_4 and AC069159_10. Examples of characterized genes encoding tandem kinase domains are the mammalian Janus kinases that, in addition to two kinase domains, also have plasma membrane-binding and SH2 domains (44). In the case of the mammalian ribosomal protein S6 kinase isoforms, which are involved in activation of mitogen-activated kinase cascade, both of the kinase domains are catalytically active although they exhibit different phosphotransferase activities (45). Analysis of the protein kinase catalytical domains from 65 different kinases found 9 invariant amino acid residues (46). The most conserved amino acid residues found in the core of the kinase catalytic domain were also conserved in RPG1 kinase domain 2 but not in kinase domain 1, indicating that this domain may not be active.
The origin of the Rpg1 gene bred into barley cvs. in the U.S. and Canada can be traced to two apparently different sources, one an introduction of an unimproved line from Switzerland, which yielded the resistant cvs. Chevron and Peatland, and another from a single resistant-plant selection made by a farmer from Wisconsin 37 barley that became the cv. Kindred. Our sequencing results suggested that both sources of Rpg1 are the same. Wisconsin 37 seed was no longer available, but analysis of Wisconsin 38, a sister line to Wisconsin 37, and the parents Oderbrucker (Wisconsin Pedigree 5) and Lion showed that even a rare recombination could not account for the resistant allele in Kindred, because Oderbrucker had an apparent deletion and Lion a susceptible allele nearly identical to the Steptoe allele. These results suggested that the resistant plant identified in the farmer's field of Wisconsin 37 was most probably the result of an admixture derived from Peatland or Chevron.
Other possible different sources of resistance were from the cv. Q21861 and the H. vulgare subsp. spontaneum line OSU6. The pedigree of Q21861 is not known, but the allele sequence was identical to Morex and thus, probably from the same source. The OSU6 line probably represented a unique source of the resistance gene, but still only had minor nucleotide substitutions and a 3-bp insertion. Interestingly, the 3-bp insertion is also seen in all of the susceptible alleles sequenced, suggesting that they arose from this type of resistance gene rather than from the one that gave rise to the Morex-type gene. Thus, the Rpg1 gene in all resistant lines seemed to be highly conserved. On the other hand, the rpg1 alleles from susceptible cvs. varied from highly conserved to highly diverged or even absent. Thus, the gene does not seem to be essential in absence of the disease.
Physical and genetic mapping of the Rpg1 region indicated a region of high recombination proximal to the Rpg1 gene and a region of low recombination distal to the gene (Fig. 1). Approximate calculations suggested that the region between markers pic20A and ABC1037 had a physical/genetic distance ratio of approximately 350 kb/cM similar to that calculated for the adjoining proximal pic20 region (14). The physical/genetic distance ratio for the ABC1037 to 19j10LRR region was ≈4 Mb/cM, the predicted barley genome average (30). The region defined by the markers MWG035S40, BB172#13, and 19j10LRR showed even higher physical/genetic distance ratio, because these three probes cosegregated in the 8,518 gamete population but did not form overlapping BAC clone contigs. Thus, the physical/genetic ratio cannot be calculated but must be in excess of 25 Mb/cM based on 1 recombinant and a minimum physical distance of 300 kb. Similar divergent ratios from 0.18 to 5 Mb/cM of physical to genetic distance were observed in the Mla cluster on chromosome 5(1H) (47).
We have suggested that map-based cloning in large grass genomes such as barley could be facilitated by using rice as an intergenomic cloning vehicle (9). Despite the excellent gene colinearity with the Rpg1 flanking markers, we did not find a candidate gene in rice (10). Screening of two rice cv. Nipponbare BAC libraries representing 11X and 15X genome coverage failed to find positive clones. A blastn search of the Chinese rice subspecies indica genome database was also negative. These results suggest that a homologous gene detectable by Southern hybridization does not exist in the rice genomes tested. Expressed sequence tags with good homology to the Rpg1 coding sequence were found in wheat and with fair homology in sorghum, maize, barrel medic, and tomato. These data suggest that Rpg1 homologues may have a function in other species.
In summary, we cloned the barley stem rust-resistance gene Rpg1 by map-based cloning. The gene seems to encode a receptor kinase-like protein with two tandem kinase domains but no recognizable receptor and membrane anchor domains. Tandem arrangement of kinase domains is a novel feature of plant disease-resistance genes. Structural similarity to tomato Pto protein supports the hypothesis that the RPG1 protein could potentially recognize the avirulence gene product of the Puccinia graminis f. sp. tritici pathotype MCC and function in a signal transduction pathway.
Supplementary Material
Acknowledgments
We are grateful to Tom Fetch, Jr., Yongliang Sun, and Chris Kavanaugh for technical assistance in determining the phenotypes for the disease reaction. This is Scientific Paper No. 0205-01 from the College of Agriculture and Home Economics Research Center, Washington State University, Project 0196. Research was supported by U.S. Department of Agriculture National Research Initiative Grant 9901325.
Abbreviations
- BAC
bacterial artificial chromosome
- cv.
cultivar
- LRR
leucine-rich repeat
- RT
reverse transcription
- cM
centimorgan
Note
Preliminary evidence indicates that the Rpg1 genomic fragment transformed into the susceptible cv. Golden Promise confers resistance to the barley stem rust pathotype MCC. Data were based on the analysis of 12 different transgenic plant families, 10 of which produced resistant progeny.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF509747 (Rpg1 cDNA); AF509748–AF509765 (Rpg1 genomic sequences from barley lines Morex, OSU6, Q21861, 80-TT, Bowman, Chevron, Leger, Kindred, Peatland, Steptoe, Lion, Wisconsin 38, Moravian III, OSU15, SM89010, Dicktoo, Gobernadora, and AMS170, respectively); AF509744–AF509746 (NRG031 genomic sequence from barley lines Morex, Steptoe, and OSU6, respectively); AF509766–AF509779 (genomic sequences from low copy probes isolated from the Rpg1 genomic region NRG019, NRG021, NRG026, NRG027, NRG046, NRG048, NRG050, RSB347, RSB409A, RSB409B, RSB410, RSB414, and RSB416, respectively)].
References
- 1.Steffenson B J. Euphytica. 1992;63:153–167. [Google Scholar]
- 2.Johnson R. Annu Rev Phytopathol. 1984;22:309–330. [Google Scholar]
- 3.Lejeune A J. In: Barley Improv. Conf. Rpt. Parker J H, editor. Minneapolis: Midwest Barley Improvement Association; 1951. [Google Scholar]
- 4.Powers L, Hines L. J Agric Res. 1933;46:1121–1129. [Google Scholar]
- 5.Miller J D, Lambert J W. Agron J. 1955;47:373–377. [Google Scholar]
- 6.Jin Y, Steffenson B J, Franckowiak J D. Crop Sci. 1993;33:642–643. [Google Scholar]
- 7.Kilian A, Steffenson B J, Maroof M A S, Kleinhofs A. Mol Plant–Microbe Interact. 1994;7:298–301. doi: 10.1094/mpmi-7-0298. [DOI] [PubMed] [Google Scholar]
- 8.Kilian A, Kudrna D A, Kleinhofs A, Yano M, Kurata N, Steffenson B, Sasaki T. Nucleic Acids Res. 1995;23:2729–2733. doi: 10.1093/nar/23.14.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilian A, Chen J, Han F, Steffenson B, Kleinhofs A. Plant Mol Biol. 1997;35:187–195. [PubMed] [Google Scholar]
- 10.Han F, Kilian A, Chen J P, Kudrna D, Steffenson B, Yamamoto K, Matsumoto T, Sasaki T, Kleinhofs A. Genome. 1999;42:1071–1076. doi: 10.1139/g99-060. [DOI] [PubMed] [Google Scholar]
- 11.Yu Y, Tomkins J P, Waugh R, Frisch D R, Kudrna D, Kleinhofs A, Brueggeman R S, Muehlbauer G J, Wise R P, Wing R A. Theor Appl Genet. 2000;101:1093–1099. [Google Scholar]
- 12.Leister D, Kurth J, Laurie D A, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P. Proc Natl Acad Sci USA. 1998;95:370–375. doi: 10.1073/pnas.95.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayliffe M A, Collins N C, Ellis J G, Pryor A. Theor Appl Genet. 2000;100:1144–1154. [Google Scholar]
- 14. Rostoks, N., Zale, J. M., Soule, J., Brueggeman, R., Druka, A., Kudrna, D., Steffenson, B. & Kleinhofs, A. (2002) Theor. Appl. Genet., 10.1007/s00122-002-0902-8. [DOI] [PubMed]
- 15.Hammond-Kosack K E, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 16.Richter T E, Ronald P C. Plant Mol Biol. 2000;42:195–204. [PubMed] [Google Scholar]
- 17.Hulbert S H, Webb C A, Smith S M, Sun Q. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 18.Flor H H. J Agric Res. 1947;74:241–262. [Google Scholar]
- 19.Kawchuck L M, Hachey J, Lynch D R, Kulcsar F, van Rooijen G, Waterer D R, Robertson A, Kokko E, Byers R, Howard R J, et al. Proc Natl Acad Sci USA. 2001;98:6511–6515. doi: 10.1073/pnas.091114198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin G M, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 21.Sessa G, Martin G B. Microbes Infect. 2000;2:1591–1597. doi: 10.1016/s1286-4579(00)01315-0. [DOI] [PubMed] [Google Scholar]
- 22.Franckowiak J D. Barley Genet Newslett. 1995;24:56–59. [Google Scholar]
- 23.Wolfe R I, Franckowiak J D. Barley Genet Newslett. 1990;20:117–121. [Google Scholar]
- 24.Steffenson B J, Miller J D, Jin Y. Plant Dis. 1993;77:626–629. [Google Scholar]
- 25.Vivar H. Rachis. 1987;6:17–19. [Google Scholar]
- 26.Close T J, Wing R, Kleinhofs A, Wise R. Barley Genet Newslett. 2001;31:29–30. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Manniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Birnboim H, Doly J D. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinhofs A, Kilian A, Maroof M A S, Biyashev R M, Hayes P, Chen F Q, Lapitan N, Fenwick A, Blake T K, Kanazin V, et al. Theor Appl Genet. 1993;86:705–712. doi: 10.1007/BF00222660. [DOI] [PubMed] [Google Scholar]
- 31.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rostoks N, Park Y-J, Ramakrishna W, Ma J, Druka A, Shiloff B A, SanMiguel P J, Jiang Z, Brueggeman R, Sandhu D, et al. Funct Integr Genomics. 2002;2:51–59. doi: 10.1007/s10142-002-0055-5. , 10.1007/s10142-002-0055-5. [DOI] [PubMed] [Google Scholar]
- 33.SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 34.Stein J C, Howlett B, Boyes D C, Nasrallah M E, Nasrallah J B. Proc Natl Acad Sci USA. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein J C, Dixit R, Nasrallah M E, Nasrallah J B. Plant Cell. 1996;8:429–445. doi: 10.1105/tpc.8.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. Nature (London) 2000;403:913–916. doi: 10.1038/35002628. [DOI] [PubMed] [Google Scholar]
- 37.Braun D M, Stone J M, Walker J C. Plant J. 1997;12:83–95. doi: 10.1046/j.1365-313x.1997.12010083.x. [DOI] [PubMed] [Google Scholar]
- 38.van der Knaap E, Song W-Y, Ruan D-L, Sauter M, Ronald P C, Kende H. Plant Physiol. 1999;120:559–569. doi: 10.1104/pp.120.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker J C. Plant Mol Biol. 1994;26:1599–1609. doi: 10.1007/BF00016492. [DOI] [PubMed] [Google Scholar]
- 40.Torji K U. Curr Opin Plant Biol. 2000;3:361–367. doi: 10.1016/s1369-5266(00)00097-2. [DOI] [PubMed] [Google Scholar]
- 41.Song W-Y, Wang G-L, Chen L-L, Kim H-S, Pi L-Y, Gardner J, Holsten T, Wang B, Zhai W-X, Zhu L-H, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 42.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:767–768. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 43.Frederick R D, Thilmony R L, Sessa G, Martin G B. Mol Cell. 1998;2:241–245. doi: 10.1016/s1097-2765(00)80134-3. [DOI] [PubMed] [Google Scholar]
- 44.Rane S G, Reddy E P. Oncogene. 1994;9:2415–2423. [PubMed] [Google Scholar]
- 45.Fisher T L, Blenis J. Mol Cell Biol. 1996;16:1212–1219. doi: 10.1128/mcb.16.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 47.Wei F, Gobelman-Werner K, Morroll S M, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise R P. Genetics. 1999;153:1929–1948. doi: 10.1093/genetics/153.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.