Abstract
General transcription initiation factor IID (TFIID) plays a central and critical role in transcription initiation from both naked and chromatin templates. Although interaction between several DNA-binding proteins and TFIID were identified and well characterized, functional linkage between TFIID and chromatin factors has remained to be elucidated. Here we show the identification and characterization of human CIA/hASF1 (identified previously as a histone chaperone) as an interactor of two tandem bromodomain modules of human (h)TAFII250/CCG1, the largest subunit of TFIID. Although yeast (y)TAFII145, a homologue of hTAFII250/CCG1 in Saccharomyces cerevisiae, lacks bromodomains, glutathione S-transferase pull-down and immunoprecipitation assays revealed that Asf1p (antisilencing function 1), the counterpart of CIA in S. cerevisiae, interacts with Bdf1p (bromodomain factor 1), which is reported to serve as the missing bromodomain in yTAFII145. Furthermore, yeast strain lacking the BDF1 gene shows the Spt phenotype that is shown also by the ASF1 gene disruptant, and a double-knockout strain of both genes shows synthetic lethality, indicating that ASF1 genetically interacts with bromodomains associated with yTFIID. We also found that Asf1p coprecipitates with yTFIID subunits from yeast whole-cell extract, and overexpression of yTFIID subunits suppress the Spt phenotype caused by gene disruption of the ASF1. This study describes the functional linkage between TFIID and a histone chaperone.
Chromosomal DNA in eukaryotes is packaged into chromatin, and the nucleosome is the fundamental structural unit of chromatin (1). Nucleosomes act as general repressors of transcription by RNA polymerase II, which transcribes protein-coding genes in eukaryotic cells (2–4). Initiation of transcription by RNA polymerase II is a multistep reaction in which several general transcription initiation factors are involved (5). Transcription initiation factor IID (TFIID) is the only general transcription initiation factor that specifically binds the TATA box that is found within the promoter of many mRNA-encoding genes and required for specific transcription initiation both in vivo and in vitro. Therefore it is suggested that binding of TFIID to the promoter is a crucial step for eukaryotic transcriptional regulation (6–8). To date, numerous functional interactions between TFIID and DNA binding activators/repressors have been characterized (7–10). Functional interactions between TFIID and chromatin factors have remained to be elucidated, although they have been anticipated from studies showing that TFIID is not capable of binding to preassembled nucleosomes in vitro (11). Recently it was reported that histone acetyltransferase (HAT)-containing complexes and nucleosome remodeling factors affect the binding of TFIID to nucleosomal DNA templates (12, 13).
Biochemical studies have demonstrated that TFIID is a multisubunit complex comprised of TBP, the TATA box-binding protein, and several TBP-associated factors (TAFIIs; refs. 14–16). TAFII250/cell cycle gene 1 (CCG1), the largest subunit of TFIID (17, 18), directly binds to TBP via an N-terminal region that mimics the partially unwound minor groove surface of the TATA box (19). Binding of this ≈100-aa N-terminal portion of TAFII250/CCG1 regulates the association between TBP and the TATA box DNA element (20). Furthermore, TAFII250/CCG1 possesses HAT activity (21) and two tandem bromodomains, which are evolutionarily conserved domains found in both ATP-dependent nucleosome remodeling factors and various HAT complexes (22). Recent biochemical and structural studies indicate that the bromodomain binds with high affinity to the N-terminal tail of core histones through recognition of specifically acetylated lysine residues (22–26). Thus, because TAFII250/CCG1 interacts both with TBP/TATA DNA and histone tails, this TFIID subunit is likely a key player in the regulation of transcriptional initiation from nucleosomal DNA as well as from naked DNA.
In the yeast Saccharomyces cerevisiae, the ortholog of human (h)TAFII250/CCG1 is TAFII145. This protein, which is only half the length of its metazoan counterparts, lacks tandem bromodomains. It has been reported that bromodomain-containing proteins bromodomain factor 1 (Bdf1p) and Bdf2p associate with yeast (y)TFIID, and that these proteins might serve as the missing bromodomain region of hTAFII250/CCG1 (27). These data suggest that bromodomains are a conserved structural feature of TFIID from yeast to human. Therefore, factors that interact with TAFII250/CCG1 bromodomains would be expected to be involved in the regulation of transcription from chromatin in an evolutionarily conserved manner.
Here we report the characterization of human CCG1-interacting factor A (hCIA) as a protein capable of binding to the bromodomains of TAFII250/CCG1. The counterpart of CIA in S. cerevisiae, antisilencing function 1 (Asf1p), was isolated as a factor that disrupts telomeric silencing when overexpressed (28), suggesting that Asf1p is involved in alteration of chromatin structure. We previously reported that CIA/hASF1 directly binds to histones H3/H4 and possesses a histone-chaperone activity that regulates alteration of nucleosomal structure in an ATP-independent fashion (29, 30). In this report, biochemical and genetic interactions between ASF1 and BDF1 and also between ASF1 and yTFIID are revealed. Thus this report shows functional linkage between TFIID and a histone chaperone.
Materials and Methods
Plasmids.
To overexpress several factors by galactose induction in yeast cells, plasmids pMH521 and pMH524 (A. Kimura and M.H., unpublished data) were constructed. Briefly, the GAL1-10 promoter and a multicloning site were ligated into the single-copy plasmid pRS314 [LEU2 CEN ARS] (31). pMH521 expresses fusion protein carrying FLAG and His tags at the C terminus of the subcloned gene, whereas pMH524 expresses fusion proteins carrying a FLAG tag at the N terminus and a His tag at the C terminus. Gene encoding Asf1p was cloned into pMH521. TBP and TAFIIs were cloned into pMH524. pRS-ASF1 was constructed by inserting the ASF1 genomic region (from 773 bp upstream of the start codon to 879 bp downstream of the stop codon) into pRS316 [URA3 CEN ARS] (31). The disruption plasmid for the ASF1 gene was constructed by inserting URA3 between NdeI and BccI sites of ASF1 (32).
Yeast Strains and Media.
Disruption of ASF1 was performed by one-step gene replacement selecting for URA3. Disruption of BDF1 and BDF2 involved a PCR-mediated strategy selecting for Candida TRP1. Asf1p and TAFIIs were tagged at the C terminus with multiple hemagglutinin (HA) and Myc epitopes, respectively, at their original chromosomal loci by using a PCR-based epitope-tagging system (33). FY120 (MATa his4-912δ lys2-128δ ura3-52 leu2Δ1) was used as a parent strain for suppressor of Ty (Spt) assay (34). For synthetic lethality tests and preparation of whole-cell extract (WCE) for immunoprecipitation, W303 (MATa leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100) was used as a parent strain. Synthetic complete (SC) medium and SC lacking histidine (SC-His) contain 0.17% (wt/vol) yeast nitrogen base without amino acids, 0.5% (wt/vol) ammonium sulfate, 2% (wt/vol) glucose or galactose, and 2% (wt/vol) bacto agar, supplemented with appropriate amino acids.
Yeast Two-Hybrid Screen.
The isolation of hCIA with the two-hybrid screen was done as described (29). The HMG-like domain and bromodomains of hCCG1 correspond to amino acids 1,101–1,352 and 1,342–1,629, respectively. The β-galactosidase assay for detecting specific interactions also was done as described (35).
Synthetic Lethality Assay.
Diploid [asf1Δ/ASF1; bdf1Δ/BDF1] strains harboring pRS-ASF1 or pRS316 (31) were sporulated, and an asf1Δbdf1Δ strain harboring pRS-ASF1 and a bdf1Δ strain harboring pRS316 were isolated. Yeast cells were streaked on rich medium in the presence or absence of 1 mg/ml 5-fluoroorotic acid (5-FOA) and incubated at 30°C for 3 days.
Recombinant Proteins.
The recombinant hCIA and bromodomains of Bdf1p (amino acids 147–422) and Bdf2p (amino acids 132–427) were expressed as fusion proteins with glutathione S-transferase (GST) in the pGEX5X-2 vector (Amersham Pharmacia). The bromodomains of hCCG1 and yAsf1p were expressed as fusion proteins with hexa-His tags in the pET28c vector (Novagen). After induction of expression with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for 3 h, the fusion proteins were affinity-purified with glutathione-Sepharose beads (Amersham Pharmacia) or ProBond nickel resin (Invitrogen).
GST Pull-Down Assay.
The method for the GST pull-down assay between recombinant proteins was described previously (29). HeLa nuclear extract was prepared as described (36). Recombinant GST-hCIA or GST (0.5 μg) was mixed with 333 μl of nuclear extract and 467 μl of binding buffer (25 mM Hepes, pH 7.5/12.5 mM MgCl2/20% glycerol/0.1% Nonidet P-40/150 mM KCl/20 μM ZnCl2). Mixed fractions were incubated at 4°C for 2 h with rotating. The binding fraction was washed 3 times with 1 ml of NETN buffer (150 mM NaCl/1 mM EDTA/0.5% Nonidet P-40/20 mM Tris, pH 7.9/15% glycerol). Half the binding fraction and 1/20 of the nuclear extract were subjected to SDS/PAGE. CCG1/TAFII250 was detected by immunoblotting using anti-CCG1 antibody (15).
Immunoprecipitation.
Fifty A600 units of aliquots of exponentially growing yeast cells were harvested, washed with distilled water, and resuspended in 0.3 ml of lysis buffer (50 mM potassium phosphate, pH 7.6/150 mM potassium acetate/1 mM sodium pyrophosphate/1 mM sodium fluoride/1% Nonidet P-40/10% glycerol/2 mM magnesium chloride/1 mM EDTA, pH 8.0/4 mg/liter leupeptin/2 mg/liter pepstatin A/1 mM benzamidine/1.25 mg/liter chymostatin/1 mM phenylmethylsulfonyl fluoride). Glass beads (1 g) were added, and cells were lysed with a multibead shocker (model MB400U, YASUI KIKAI, Osaka) at −2°C. All subsequent steps were performed at 4°C. Cell debris was pelleted by centrifugation (Hitachi, 13,000 × g for 5 min). Immunoprecipitation reactions contained 1.5 mg of protein extract. The sample volumes were adjusted to 500 μl with lysis buffer, and Nonidet P-40 was added to a final concentration of 2%. Then the sample was incubated with 10 μl of anti-Myc agarose (Santa Cruz Biotechnology) and 250 units of DNase I (Takara Shuzo, Otsu, Japan) at 4°C for 3 h on a rotator. The beads were collected by brief centrifugation and washed three times with 0.5 ml of lysis buffer. The precipitates were resuspended in 10 μl of SDS/PAGE sample buffer (50 mM Tris⋅HCl, pH 6.5/2 mM EDTA/1% SDS/8% glycerol/0.25 mg/ml bromophenol blue), and half the total volume was subjected to SDS/PAGE and immunoblot analysis as the bound fraction.
Western Immunoblot Analysis.
For detection of Asf1p in the immunoprecipitation analysis, 2% of input and half the precipitate were subjected to SDS/PAGE as “input” and “bound,” respectively. Separated proteins were transferred to PROTRAN (Schleicher & Schuell) and detected by using horseradish peroxidase-conjugated anti-HA tag monoclonal antibody (Santa Cruz Biotechnology) and enhanced chemiluminescence detection kit (Amersham Pharmacia). For detection of His-tagged protein, anti-His polyclonal antibody (Santa Cruz Biotechnology) was used.
Results
Isolation of CIA/hASF1 as a TAFII250/CCG1-Interacting Factor.
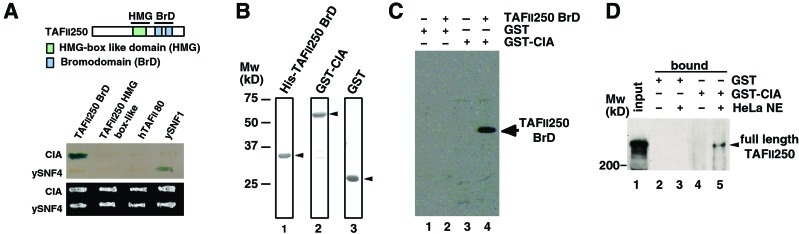
To isolate factors that interact with the bromodomain region of hTAFII250/CCG1, a yeast two-hybrid screen was performed by using GAL4-DNA binding domain-fused bromodomains as the bait. We isolated three independent clones from a human peripheral lymphocyte cDNA library. The determined sequence of full-length hCIA was strikingly similar to yAsf1p (28, 29). To confirm the specificity of this interaction between CIA/hASF1 and bromodomains of TAFII250/CCG1, we examined the interaction between CIA/hASF1 and the HMG box-like domain of TAFII250/CCG1 (Fig. 1A), another TFIID subunit (TAFII80), and yeast SNF1 by the yeast two-hybrid system. In these experiments the interaction between SNF1 and SNF4 served as a positive control (Fig. 1A; ref. 37). These assays show that CIA/hASF1 specifically interacts with the bromodomains of TAFII250/CCG1.
Figure 1.
Interaction between CIA/hASF1 and the two tandem bromodomain regions of TAFII250/CCG1. (A) Specific interaction between CIA/hASF1 and TAFII250/CCG1 bromodomains by yeast two-hybrid system. (Upper) Result of β-galactosidase assay. (Lower) Growth activities of yeast strains as a control. Combination of ySNF1-ySNF4 serves as a positive control. (B) Purified recombinant protein used in the GST pull-down assay. Approximately 0.5 μg of His-tagged TAFII250/CCG1 bromodomains (lane 1), GST-tagged CIA/hASF1 (lane 2), and GST (lane 3) were separated in SDS/PAGE and stained with Coomassie brilliant blue. Mw, molecular mass markers. (C) Direct interaction between CIA/hASF1 and TAFII250/CCG1 bromodomains by GST pull-down assay. Each fraction coprecipitated with GST or GST-CIA/hASF1 was separated by SDS/PAGE, and the bromodomains were detected by immunoblot analysis using anti-His-tag antibody. (D) Interaction between CIA/hASF1 and full-length TAFII250/CCG1 bromodomains by GST pull-down assay. GST (lanes 2 and 3) or GST-CIA/hASF1 (lanes 4 and 5) was mixed with HeLa nuclear extract (NE), and bound proteins were analyzed by using anti-CCG1 polyclonal antibody. The arrowhead indicates the full-length TAFII250 detected in HeLa nuclear extract (input) and bound (bound) fraction.
Next we examined whether this interaction is direct or indirect by GST pull-down assays using purified recombinant hexa-His-tagged TAFII250/CCG1 bromodomains and GST-tagged CIA/hASF1 (Fig. 1B). As shown in Fig. 1C, TAFII250/CCG1 bromodomains specifically bound (GST-) CIA/hASF1. From these results, we concluded that CIA/hASF1 directly interacts with the bromodomains of TAFII250/CCG1.
To elucidate whether CIA/hASF1 interacts with full-length TAFII250/CCG1, a pull-down experiment was performed (Fig. 1D). Purified recombinant GST-CIA/hASF1 was mixed with HeLa nuclear extract, and bound proteins were analyzed by immunoblotting using anti-TAFII250/CCG1 antibody. As shown in Fig. 1D, Full-length TAFII250/CCG1 was pulled down by GST-CIA/hASF1 but not by GST. This result indicates that CIA/hASF1 interacts with full-length TAFII250/CCG1.
Evolutionarily Conserved Interaction Between CIA/ASF1 and Bromodomains Associated with TFIID.
We next asked whether the interaction between CIA/ASF1 and the bromodomain is evolutionarily conserved. We previously reported that a counterpart of CIA/hASF1 in S. cerevisiae, Asf1p, binds directly to histone H3/H4 and possesses histone-chaperone activity similar to CIA/hASF1 (29, 30), indicating that the function of CIA/ASF1 is conserved from yeast to human. Although the counterpart of hTAFII250/CCG1 in yeast, TAFII145, lacks bromodomains, Bdf1p and Bdf2p associate with yTFIID and correspond to the missing bromodomain in yTFIID (27).
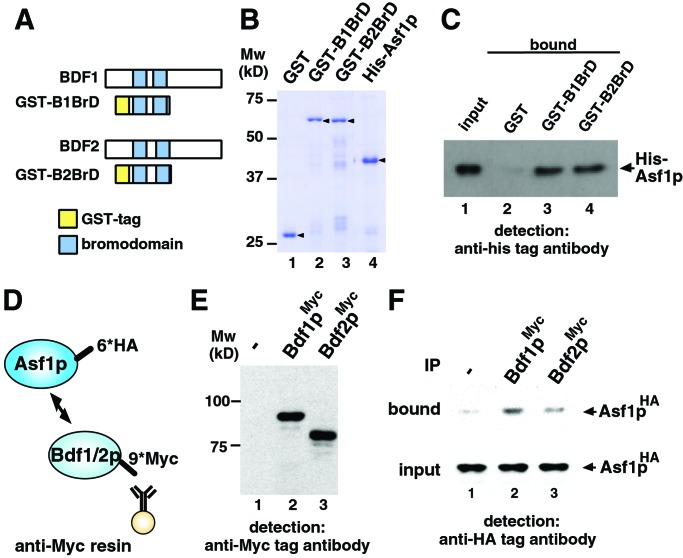
Thus, we reasoned that Asf1p should interact directly with bromodomains of Bdf1p and/or Bdf2p. To test this possibility, pull-down assays were conducted by using Bdf1p and Bdf2p bromodomains (B1BrD and B2BrD, respectively) fused to GST and His-tagged Asf1p (Fig. 2 A and B). As shown in Fig. 2C, Asf1p interacted with the bromodomains of both Bdf1p and Bdf2p (compare lanes 3 and 4 with lane 2). These data argue that Asf1p directly interacts with the Bdf1p and Bdf2p bromodomains.
Figure 2.
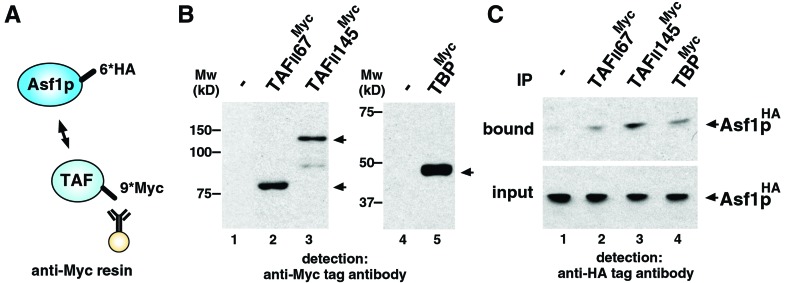
Interaction between Asf1p and Bdf1/2p bromodomains. (A) Representation of the bromodomains in Bdf1p and Bdf2p. Schematic drawings of GST-tagged bromodomains of Bdf1p and Bdf2p also are shown as GST-B1BrD and GST-B2BrD, respectively. (B) Purified recombinant proteins used in the GST pull-down assay (arrowhead). Approximately 0.2 μg of each protein was separated by SDS/PAGE and stained with Coomassie brilliant blue. Mw, molecular mass markers. (C) Direct interaction between Asf1p and bromodomains of Bdf1p and Bdf2p by GST pull-down assay. GST-tagged proteins (8.0 × 10−12 mol) and His-tagged Asf1p (2.5 × 10−11 mol) were mixed. One percent of the input (lane 1) and 30% of the pull-down material (lanes 2–4) were separated by SDS/PAGE, and His-tagged Asf1p was detected by immunoblot analysis using anti-His-tag antibody. (D) Schematic drawing of immunoprecipitation analysis. 6×HA epitope (6*HA) and 9×Myc epitope (9*Myc) are tagged at the C terminus of Asf1p and Bdf1p/Bdf2p, respectively. Myc-tagged Bdf1p or Bdf2p was immunoprecipitated with anti-Myc affinity resin. (E) Expression of Myc-tagged proteins. WCE of yeast strain expressing Myc-tagged Bdf1p (lane 2) or Bdf2p (lane 3) was separated by SDS/PAGE, and Myc-tagged proteins were detected by Western immunoblot analysis using anti-Myc antibody. No signal was detected in the WCE from control strain in which no protein is Myc-tagged (lane 1). (F) Interaction between Asf1p and Bdf1p/Bdf2p in yeast cells. HA-tagged Asf1p in bound fraction was detected by immunoblot analysis using anti-HA tag antibody (bound). A 4% volume of each WCE also was immunoblotted (input) to confirm that an equal amount of HA-tagged Asf1p is expressed in Myc-tagged Bdf1p- or Bdf2p-expressing (lanes 2 and 3, respectively) or control (lane 1) strains. IP, immunoprecipitation.
Next we performed coimmunoprecipitation experiments to examine whether Asf1p interacts with Bdf1p and Bdf2p in yeast cells. For this purpose we constructed yeast strains, the ASF1 gene and BDF1 or BDF2 genes of which were C-terminally HA-tagged and Myc-tagged, respectively (Fig. 2D). Note that tagged proteins are expressed from their native promoters. Expressions of tagged proteins in yeast cells are shown in Fig. 2 E and F (input). WCE was prepared from each strain, and immunoprecipitation was performed by using anti-Myc antibody (Fig. 2D). As shown in Fig. 2F, Asf1p coprecipitated with Myc-tagged Bdf1p and Bdf2p. Bdf1p precipitated Asf1p more efficiently than did Bdf2p.
Genetic Interaction Between ASF1 and BDF1.
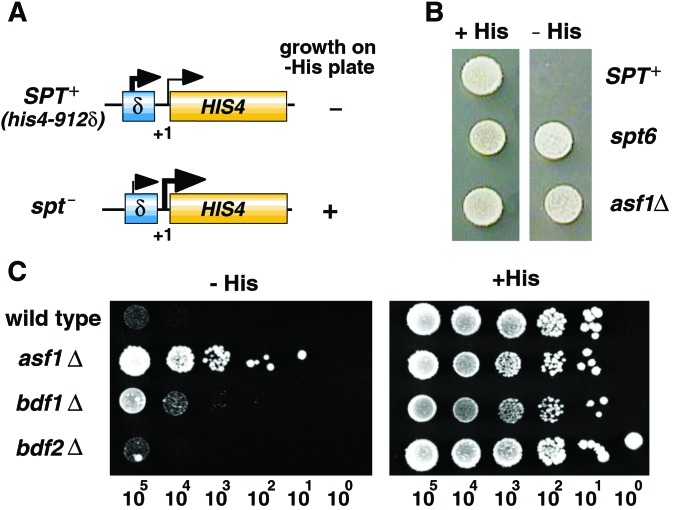
The evolutionarily conserved interaction between CIA/ASF1 and bromodomains associated with TFIID suggested the involvement of CIA/ASF1 in transcriptional regulation. Therefore, we next asked whether the disruption of genes encoding Asf1p (asf1Δ), Bdf1p (bdf1Δ), and Bdf2p (bdf2Δ) displayed similar phenotypes. It is known that mutations or disruption of several genes encoding factors involved in transcriptional regulation cause an Spt phenotype (38). After insertion of Ty or δ, transposable elements, into the promoter region of the HIS4 gene, wild-type SPT+ strains fail to grow on medium lacking histidine (−His plate) (39). However, mutations or disruption of any of several SPT genes allow cells to grow on −His plates (Fig. 3A). We assessed the Spt phenotype of asf1Δ cells by using isogenic yeast strains carrying the his4-912δ allele of HIS4 (Fig. 3B). They grew on the −His plate as well as the positive control, spt6 mutant strain, indicating that the loss of Asf1p function induces an Spt phenotype. Spt phenotypes of bdf1Δ and bdf2Δ strains also were assessed (Fig. 3C). Disruption of BDF1 but not BDF2 induced an Spt phenotype. Thus, both asf1Δ and bdf1Δ strains induced an Spt phenotype.
Figure 3.
Spt phenotype of asf1Δ and bdf1Δ strains. (A) Schematic drawing of the assay for Spt phenotype. Mutations in the SPT gene (spt−) suppresses transcriptional defect of the HIS4 gene in the SPT+ strain caused by insertion of the Ty element. The Spt phenotype is measured on the medium lacking histidine. (B) Spt phenotype of ASF1 disruptant (asf1Δ). The growth of cells carrying the asf1Δ allele on control plates (+His) and plates lacking histidine (−His) were measured. spt6 mutant strain serves as a positive control. (C) The Spt phenotype of BDF1 disruptant (bdf1Δ); the asf1Δ strain served as a positive control. The number of cells in each spot is indicated at the bottom.
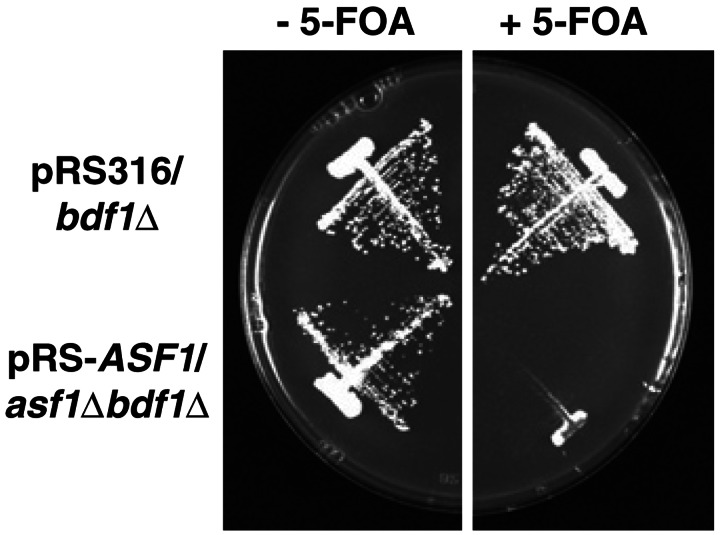
Moreover, as shown in Table 1, we found that the double-knockout strain carrying both asf1Δ and bdf1Δ shows synthetic lethality, whereas yeast carrying both asf1Δ and bdf2Δ did not. Next we asked whether synthetic lethality of asf1Δbdf1Δ strain is caused by defects in mitotic growth. Synthetic lethality of the asf1Δbdf1Δ strain is rescued by a single-copy plasmid, pRS-ASF1, that expresses ASF1 from its native promoter (Fig. 4, −5-FOA). Because pRS-ASF1 contains the URA3 gene as a selectable marker, the plasmid can be removed from yeast cells when the medium contains 5-FOA, which kills cells expressing URA3. As shown in Fig. 4 (Right, +5-FOA), the asf1Δbdf1Δ strain was not able to grow without pRS-ASF1, suggesting that gene disruption of both ASF1 and BDF1 causes loss of viability in mitotic growth. Together these data suggest the functional linkage between Asf1p and Bdf1p in vivo.
Table 1.
Synthetic lethality by double knockout of ASF1 and BDF1
|
asf1Δ/ASF1;
bdf1Δ/BDF1 diploid (n = 19
ascus)
|
asf1Δ/ASF1;
bdf2Δ/BDF2 diploid (n = 17 ascus)
|
||
|---|---|---|---|
| Genotype | Spore | Genotype | Spore |
| ASF1BDF1 | 16 | ASF1 BDF2 | 18 |
| asfΔ BDF1 | 20 | asf1Δ BDF2 | 17 |
| ASF1 bdf1Δ | 20 | ASF1 bdf2Δ | 16 |
| asf1Δ bdf1Δ | 0 | asf1Δ bdf2Δ | 17 |
Figure 4.
Synthetic lethality of asf1Δbdf1Δ double-knockout strain in mitotic growth. The synthetic lethality of asf1Δbdf1Δ is suppressed by a single-copy plasmid, pRS-ASF1, which expresses the ASF1 gene from its own promoter (pRS-ASF1/asf1Δbdf1Δ, −5-FOA). The viability of this strain without pRS-ASF1 was tested by the addition of 5-FOA into medium (+5-FOA). The pRS316/bdf1Δ strain, the ASF1 gene of which is coded on its chromosome, serves as a control to indicate that the bdf1Δ strain itself is viable in the presence of 5-FOA.
Association of Asf1p with yTFIID.
We showed here that ASF1 physically and genetically interacts with BDF1, which encodes a bromodomain protein that associates with TFIID. This observation suggested that Asf1p also interacts with TFIID in yeast cells. To test this hypothesis rigorously we performed coimmunoprecipitation experiments. We constructed three separate yeast strains, the ASF1 gene and TFIID-subunit-encoding genes (TAFII145, TAFII67, and TBP) of which were C-terminally HA-tagged and Myc-tagged, respectively (Fig. 5A). Immunoprecipitates were performed by using anti-Myc antibody and WCE prepared from each strain (Fig. 5A). Expression of tagged proteins in yeast cells were shown in Fig. 5 B and C (input). Asf1p coimmunoprecipitating with TAFIIs or TBP were detected by Western immunoblot analysis of precipitates using anti-HA antibodies (Fig. 5C). Associations between Asf1p and TBP, TAFII145, and TAFII67 were detected. We thus conclude that Asf1p associates with TFIID in yeast cells.
Figure 5.
Interaction between Asf1p and TFIID subunits in yeast cells. (A) Schematic drawing of immunoprecipitation analysis. 6×HA epitope (6*HA) and 9×Myc epitope (9*Myc) are tagged at the C terminus of Asf1p and TFIID subunits (TAFII67, TAFII145, and TBP), respectively. Myc-tagged TAFIIs or TBP were immunoprecipitated with anti-Myc affinity resin. (B) Expression of Myc-tagged proteins. WCE of yeast strain expressing Myc-tagged TAFII67, TAFII145, or TBP (lanes 2, 3, and 5, respectively) were separated by SDS/PAGE, and Myc-tagged proteins were detected by Western immunoblot analysis using anti-Myc antibody (arrowheads). No signal is detected in WCE from control strain in which no protein is Myc-tagged (lanes 1 and 4). Mw, molecular mass markers. (C) Interaction between Asf1p and TFIID subunits in yeast cells. HA-tagged Asf1p in bound fraction was detected by Western immunoblot analysis using anti-HA-tag antibody (bound). A 4% volume of each WCE also was immunoblotted (input) to confirm that an equal amount of HA-tagged Asf1p is expressed in tagged TAFII67-, TAFII145-, or TBP-expressing strains (lanes 2, 3, and 4, respectively) or a control strain (lane 1). IP, immunoprecipitation.
Genetic Interaction Between ASF1 and yTFIID.
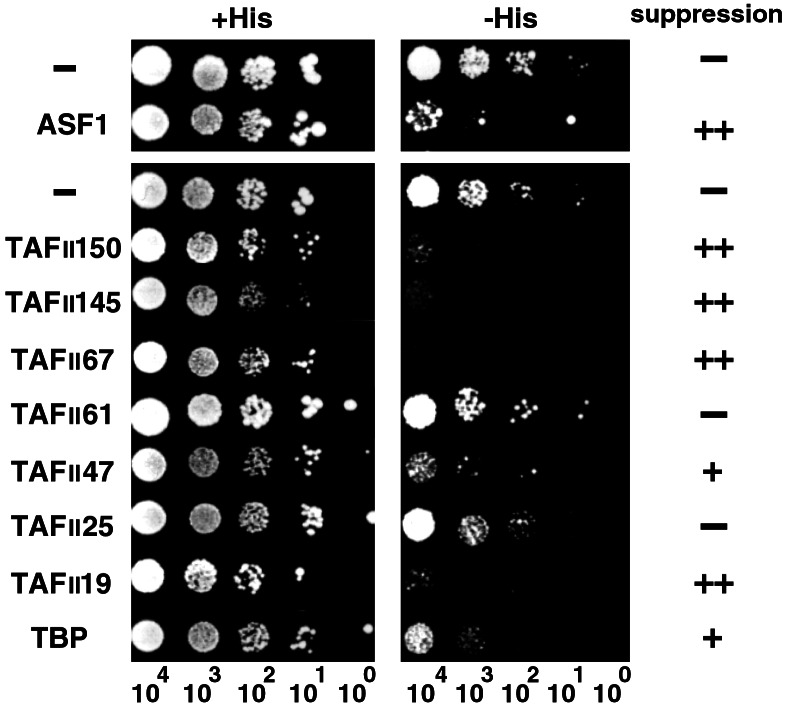
We next asked whether interaction between Asf1p and TFIID subunits is functional in transcriptional regulation in yeast cells. We reasoned that if Asf1p functions in the reaction where TFIID is involved, a transcriptional defect caused by deletion of the ASF1 gene likely would be suppressed by increased amounts of TFIID (subunits) in yeast cells. Therefore, we overexpressed subunits of yTFIID and tested whether the Spt phenotype caused by disruption of ASF1 is suppressed (Fig. 6). As a positive control, expression of the ASF1 gene suppressed growth of the asf1Δ strain on −His plates. Overexpression of yTAFIIs, including TAFII145, also suppressed the Spt phenotype of the asf1Δ. These data strongly suggested that ASF1 is involved in the process where TFIID plays critical roles in transcription.
Figure 6.
Genetic interactions between ASF1 and TFIID subunits. asf1Δ strains harboring either a plasmid for overexpression of Asf1p or TFIID subunits (Left) or a control plasmid (−) was diluted serially on control plates (+His) and plates lacking histidine (−His). The number of cells in each spot is indicated at the bottom. The efficiency of suppression is indicated (Right) with −, +, or ++ using suppression activity of ASF1 itself as a positive control (++).
Discussion
Interaction Between CIA/ASF1 and the Bromodomains of TFIID.
Core histones consist of two domains, a basic N-terminal tail domain and a histone-fold C-terminal domain. The histone-fold domain plays a central role in organization of the nucleosomal structure, and the N-terminal tail is the target for posttranslational modification enzymes including HATs, histone methyltransferases (HMTs), and kinases (40, 41). Specifically acetylated and methylated N-terminal tails are recognized by bromodomains and chromodomains, respectively (23–26, 42–44). Because these posttranslational modifications play critical roles in the organization and alteration of nucleosomal structure and higher order structure of chromatin (41), functional linkage between enzymes/factors that bind to the N-terminal tail of histones and enzymes/factors that bind to the histone-fold domain of histones seems likely.
We previously showed that CIA/hASF1 binds to the histone-fold domain of histone H3 and possesses histone-chaperone activity (29). Bromodomains of hTAFII250 bind to the specifically acetylated N terminus of histone H4 with high affinity, and it is suggested that TFIID is stabilized on the specifically acetylated nucleosome via bromodomains of TAFII250 (25). Therefore, CIA/ASF1 and the bromodomains of TFIID might bind to the histone-fold domain and specifically acetylated N-terminal region of core histones, respectively, to regulate nucleosome assembly/disassembly reactions and stability of TFIID on nucleosome template during the transcription initiation process. The synthetic lethality of asf1Δ and bdf1Δ that we observe here (Table 1 and Fig. 4) would support this notion.
A counterpart of CIA/ASF1 in Drosophila, which has been identified as a subunit of replication-coupled nucleosome assembly factor (RCAF), associates with histone H3/H4 proteins, the N-terminal lysine residues of which are specifically acetylated (45). In budding yeast, a deletion strain of the ASF1 gene shows sensitivity to DNA-damaging agents and inhibitors of DNA replication (28, 45). Under these stressed conditions, Asf1p binds to N-terminally acetylated histone H3/H4 (46); furthermore, the asf1Δ strain shows defects in transcriptional regulation (ref. 47 and T.C. and M.H., unpublished results). Because Rad53, an interactor of Asf1p (46, 48), is required in transcriptional regulation under these stressed conditions (49), it is possible that the interaction between Rad53 and Asf1p may play a critical role in transcriptional regulation. These considerations suggest that CIA/ASF1 might be a key component that mediates DNA replication- or DNA repair-coupled transcriptional regulation. The roles of interactions between CIA/ASF1 and bromodomains associated with TFIID under genotoxic stress or DNA replicational interference will have to be elucidated by further analyses.
Difference Between BDF1 and BDF2 in Interaction with ASF1.
We showed here that Asf1p physically interacts with Bdf1p and Bdf2p in vivo (Fig. 2F). In this assay, Bdf1p and Bdf2p interact with Asf1p with different affinities. Genetic analyses revealed that these two Bdfp-encoding genes also did not behave identically. For example, only Bdf1p interacts with Asf1p in Spt and synthetic lethality tests (Figs. 3 and 4 and Table 1). These genetic results seem to correlate with their affinities of interactions in vivo and might support the functional linkage between Asf1p and Bdf1p in vivo.
However, we also demonstrated that the bromodomains of Bdf2p directly interact with Asf1p in vitro to the same extent as that of Bdf1p (Fig. 2C), suggesting that regions other than bromodomains might modulate the interaction between bromodomains and Asf1p. It also is possible that additional factors regulate the association between Asf1p and Bdfs in vivo. Bdf2p might interact with Asf1p with higher affinity in certain specific stressed conditions.
Putative Roles of CIA/ASF1 in Eukaryotic Transcription Initiation.
We found that ASF1 interacts with TFIID subunits both physically and genetically in yeast cells, which shows that a histone chaperone interacts functionally with a general transcription initiation factor. Our genetic evidence that gene disruption of ASF1 induces an Spt phenotype that is suppressed by overexpression of specific TFIID subunits suggests that Asf1p plays an important role in modulation of the activities of specific TFIID subunits in transcriptional regulation in vivo. CIA/ASF1 may possibly regulate interaction between TFIID and nucleosomal DNA through association with nucleosomal histones. Alternatively, considering that TFIID contains several “histone-like TAFs” that contain domains similar to core histones (50–52) and that these histone-like TAFs interact with both themselves and core histones (35, 51), CIA/ASF1 also may modulate these interactions. Suppression of the Spt phenotype of the asf1Δ strain by TAFII19, a histone-like TAF (Fig. 6), might support this notion. It is an important issue to elucidate the roles of these specific TAFIIs in the alteration of nucleosomal structure together with Asf1p. Determination and classification of TFIID subunits that suppress the Spt phenotype of spt mutants other than asf1Δ might also be informative for understanding the functional linkages between TAFIIs and Spt factors in transcriptional regulation.
Alteration of nucleosomal structure is regulated by three major classes of enzymes and factors: HATs, ATP-dependent nucleosome remodeling factors, and histone chaperones (53, 54). To date, HATs and remodeling factors have been suggested to regulate the interaction between TFIID and nucleosomal DNA (12,13). In this present study, we suggest that CIA/ASF1 also participates in the regulation of the interaction between TFIID and nucleosomes. Furthermore, taking into consideration that several HATs and nucleosome remodeling factors contain bromodomains, CIA/ASF1 also might be involved in the regulatory process with HATs and/or remodeling factors via interactions with their bromodomains. These possibilities are an important issue for future analyses. In addition, we have isolated several TFIID-interacting factors such as Tip60 MYST-HAT that likely regulate the alteration of nucleosomal structure (55). To elucidate networks of TFIID and its interacting factors would be essential in understanding regulations in eukaryotic transcription initiation.
Acknowledgments
We thank Dr. F. Winston for gifts of FY120 (SPT+) and FY957 (spt6) yeast strains, Dr. T. Tanaka for gifts of plasmids for the epitope-tagging system, and Dr. P. A. Weil and M. Ohara for critical reading of the manuscript. We are grateful to A. Kimura, Dr. T. Suzuki, and all the members of the Laboratory of Developmental Biology at the University of Tokyo and the Horikoshi Gene Selector Project for comments on the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Technology, Sports, and Culture of Japan, and the Exploratory Research for Advanced Technology (ERATO) of the Japan Science and Technology Corporation (JST).
Abbreviations
- TFIID
transcription initiation factor IID
- HAT
histone acetyltransferase
- TBP
TATA box-binding protein
- TAFII
TBP-associated factor
- CCG1
cell cycle gene 1
- Bdf
bromodomain factor
- y
yeast
- CIA
CCG1-interacting factor A
- h
human
- ASF1
antisilencing function 1
- HA
hemagglutinin
- Spt
suppressor of Ty
- WCE
whole-cell extract
- 5-FOA
5-fluoroorotic acid
- GST
glutathione S-transferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kornberg R D. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Knezetic J A, Luse D S. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 3.Lorch Y, LaPointe J W, Kornberg R D. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 4.Owen-Hughes T, Workman J L. Crit Rev Eukaryotic Gene Expression. 1994;4:403–441. [PubMed] [Google Scholar]
- 5.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 6.Parker C S, Topol J. Cell. 1984;37:273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- 7.Horikoshi M, Carey M F, Kakidani H, Roeder R G. Cell. 1988;54:665–669. doi: 10.1016/s0092-8674(88)80011-4. [DOI] [PubMed] [Google Scholar]
- 8.Horikoshi M, Hai T, Lin Y S, Green M R, Roeder R G. Cell. 1988;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- 9.Kato H, Horikoshi M, Roeder R G. Science. 1991;251:1476–1479. doi: 10.1126/science.2006421. [DOI] [PubMed] [Google Scholar]
- 10.Verrijzer C P, Tjian R. Trends Biochem Sci. 1996;21:325–326. [PubMed] [Google Scholar]
- 11.Workman J L, Roeder R G. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 12.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 13.Lomvardas S, Thanos D. Cell. 2001;106:685–696. doi: 10.1016/s0092-8674(01)00490-1. [DOI] [PubMed] [Google Scholar]
- 14.Dynlacht B D, Hoey T, Tjian R. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 15.Takada R, Nakatani Y, Hoffmann A, Kokubo T, Hasegawa S, Roeder R G, Horikoshi M. Proc Natl Acad Sci USA. 1992;89:11809–11813. doi: 10.1073/pnas.89.24.11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders S L, Weil A P. J Biol Chem. 2000;275:13895–13900. doi: 10.1074/jbc.275.18.13895. [DOI] [PubMed] [Google Scholar]
- 17.Sekiguchi T, Nohiro Y, Nakamura Y, Hisamoto N, Nishimoto T. Mol Cell Biol. 1991;11:3317–3325. doi: 10.1128/mcb.11.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hisatake K, Hasegawa S, Takada R, Nakatani Y, Horikoshi M, Roeder R G. Nature (London) 1993;362:179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Ishima R, Tong K I, Bagby S, Kokubo T, Muhandiram D R, Kay L E, Nakatani Y, Ikura M. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 20.Kokubo T, Gong D W, Yamashita S, Horikoshi M, Roeder R G, Nakatani Y. Genes Dev. 1993;7:1033–1046. doi: 10.1101/gad.7.6.1033. [DOI] [PubMed] [Google Scholar]
- 21.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 22.Jeanmougin F, Wurtz J M, Le, Douarin B, Chambon P, Losson R. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 23.Ornaghi P, Ballario P, Lena A M, Gonzalez A, Filetici P. J Mol Biol. 1999;287:1–7. doi: 10.1006/jmbi.1999.2577. [DOI] [PubMed] [Google Scholar]
- 24.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M M. Nature (London) 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson R H, Ladurner A G, King D S, Tjian R. Science. 2000;288:1372–1373. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 26.Owen D J, Ornaghi P, Yang J C, Lowe N, Evans P R, Ballario P, Neuhaus D, Filetici P, Travers A A. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matangkasombut O, Buratowski R M, Swilling N W, Buratowski S. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 28.Le S, Davis C, Konopka J B, Sternglanz R. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 29.Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M. Genes Cells. 2000;5:221–233. doi: 10.1046/j.1365-2443.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- 30.Umehara T, Chimura T, Ichikawa N, Horikoshi M. Genes Cells. 2002;7:59–73. doi: 10.1046/j.1356-9597.2001.00493.x. [DOI] [PubMed] [Google Scholar]
- 31.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaki M, Umehara T, Chimura T, Horikoshi M. Genes Cells. 2001;6:1043–1054. doi: 10.1046/j.1365-2443.2001.00487.x. [DOI] [PubMed] [Google Scholar]
- 33.Knop M, Siegers K, Pareira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 34.Bortvin A, Winston F. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 35.Bando M, Ijuin S, Hasegawa S, Horikoshi M. J Biochem. 1997;121:591–597. doi: 10.1093/oxfordjournals.jbchem.a021626. [DOI] [PubMed] [Google Scholar]
- 36.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 38.Winston F, Sudarsanam P. Cold Spring Harbor Symp Quant Biol. 1998;63:553–561. doi: 10.1101/sqb.1998.63.553. [DOI] [PubMed] [Google Scholar]
- 39.Silverman S J, Fink G R. Mol Cell Biol. 1984;4:1246–1251. doi: 10.1128/mcb.4.7.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 41.Jenuwein T, Allis C D. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 42.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature (London) 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 43.Bannister A J, Zegerman P, Partridge J F, Miska E A, Thomas J O, Allshire R C, Kouzarides T. Nature (London) 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 44.Jacobs S A, Taverna S D, Zhang Y, Briggs S D, Li J, Eissenberg J C, Allis C D, Khorasanizadeh S. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tyler J K, Adams C R, Chen S R, Kobayashi R, Kamakaka R T, Kadonaga J T. Nature (London) 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 46.Emili A, Schieltz D M, Yates J R, 3rd, Hartwell L H. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 47.Sutton A, Bucaria J, Osley M A, Sternglanz R. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu F, Alcasabas A A, Elledge S J. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 50.Kokubo T, Gong D W, Wootton J C, Horikoshi M, Roeder R G, Nakatani Y. Nature (London) 1994;367:484–487. doi: 10.1038/367484a0. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann A, Chiang C M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R G. Nature (London) 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 52.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Nature (London) 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 53.Ito T, Tyler J K, Kadonaga J T. Genes Cells. 1997;2:593–600. doi: 10.1046/j.1365-2443.1997.1500348.x. [DOI] [PubMed] [Google Scholar]
- 54.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto T, Horikoshi M. J Biol Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]