Abstract
We have previously identified an antigen (Ag) recognized on a human large cell carcinoma of the lung by a tumor-specific cytotoxic T lymphocyte clone derived from autologous tumor infiltrating lymphocytes (TILs). The antigenic peptide is presented by HLA-A2 molecules and is encoded by a mutated α-actinin-4 (ACTN4) gene. In the present report, we have isolated two anti-α-actinin-4 T cell clones from the same patient TIL and from his peripheral blood lymphocytes (PBLs) by using tetramers of soluble HLA-A2 molecules loaded with the mutated peptide. Although all of the clones displayed similar tetramer labeling, those isolated from PBL showed lower avidity of Ag recognition and killed the specific target much less efficiently, indicating that tetramer staining does not correlate with clone avidity/tumor reactivity. T cell receptor (TCR) analysis revealed that α-actinin-4-reactive clones used distinct α and β chain rearrangements, demonstrating TCR repertoire diversity. Interestingly, TCRβ chain gene usage indicated that only Ag-specific clones with high functional avidity were expanded at the tumor site, whereas a low-avidity clone was exclusively amplified in patient peripheral blood. Our results point to the existence of distinct but overlapping antitumor TCR repertoires in TIL and PBL and suggest a selective in situ expansion of tumor-specific cytotoxic T lymphocyte with high avidity/tumor reactivity.
The majority of tumor antigens (Ags) described so far have been identified from malignant melanoma by using specific autologous cytotoxic T lymphocytes (CTLs) (1). These Ags opened up new possibilities for immunotherapy of cancer, and several vaccination trials are now under way aimed at inducing a strong CTL response against defined tumor Ag (2–3). With respect to other human solid tumors, in particular lung carcinomas, much less is known about tumor-specific Ag recognized by CTL. About 80% of human lung tumors belong to the categories of non-small cell lung cancer, including squamous cell carcinomas, adenocarcinomas, and large-cell carcinomas (LCCs). These tumors are often infiltrated by T lymphocytes, most of which are T cell receptor (TCR)α/β+, CD8+, and CD28 − (4). The role and target Ag of these T cells are not very well known.
So far, only a few Ags have been identified in non-small cell lung cancer because of the difficulty of establishing in vitro tumor cell lines. Some of these tumor cells could be used to stimulate autologous peripheral blood lymphocytes (PBLs) or tumor-infiltrating lymphocytes (TILs) and to derive tumor-specific CTLs (5–7). Two tumor-specific Ags have been previously described. One of them, identified with HLA-A2-restricted CTL derived from TIL, is encoded by HER2/neu, which is overexpressed in many tumors (8). The other antigenic peptide was found to be recognized by HLA-A68.2-restricted CTL and is encoded by a mutated elongation factor 2 gene (9). Furthermore, two antigenic peptides presented by HLA-A24 molecules and recognized by tumor-specific CTLs have been reported. One of them was found to be encoded by the cyclophilin B gene, which is expressed ubiquitously (10), and the second was derived from an endoplasmic reticulum-resident protein (11). More recently, an antigenic peptide, presented by HLA-A2 and encoded by a mutated malic enzyme gene, was identified by autologous CTL on a squamous cell carcinomas (12).
The introduction of the HLA–tetramer method of identifying and phenotypically characterizing Ag-specific T lymphocytes (13) has rapidly advanced our understanding of antitumor immune response (14, 15). These reagents have been successfully used for detecting and isolating low-frequency tumor-specific CTLs (14). Tetramer-guided characterization of circulating T cells specific for tumor Ag has been performed, in particular in melanoma patients (16). These studies revealed various surface phenotypes and differential expression frequency of Ag-specific T cells, which may use a large TCRβ chain repertoire (17, 18). However, no data are available concerning TCR repertoire distribution of tetramer+ cells in TIL and PBL of tumor-bearing patients. We have previously reported that the antigenic peptide recognized by a HLA-A2.1-restricted CTL clone isolated from lung carcinoma TIL was encoded by a mutated α-actinin-4 (ACTN4) gene (19). ACTN4 is expressed ubiquitously. Its role is not clearly defined, but it was suggested that it might function as a tumor suppressor gene (20). To further investigate the antitumor CTL response in non-small cell lung cancer, we have identified, from the same patient TIL and from his PBL by using soluble HLA tetramers, several anti-mutated α-actinin-4 T cell clones. the tetramer staining and effector functions of these clones, as measured by cytokine secretion and specific lytic activity, were determined, and their distribution at the tumor site and peripheral blood was compared.
Materials and Methods
Derivation and Culture of CTL Clones Heu127 and Heu171.
The IGR-Heu cell line was derived from a LCC biopsy and maintained in culture as described (21). Heu127 and Heu171 TIL clones were derived as described (4). Briefly, viable TIL were seeded at 104 cells/well and stimulated by irradiated autologous tumor and Epstein–Barr virus (EBV)-transformed B cells in complete medium supplemented with IL-2 (4). After 3 weeks, the resulting cell line was cloned by limiting dilution, and CTL clones were isolated and restimulated every other week with the same protocol.
Isolation of Tetramer-Reactive T Cell Clones and Tetramer Staining.
Soluble phycoerythrin (PE)-labeled HLA-A2/mutated α-actinin-4 peptide tetramers were produced as described (19). Patient peripheral blood mononuclear cells (PBMCs) (1–2 × 106) were thawed and stimulated either in conditions similar to TIL with autologous IGR-Heu tumor cell line or with the mutated peptide. For peptide stimulation, PBMCs were incubated for 2 weeks with the mutated α-actinin-4 peptide (20 μM) as described (19). They were then collected, washed, and tetramer-stained. Positive cells were sorted and seeded at one cell/well in round-bottom microplates by using FACS-VANTAGE (Becton Dickinson). They were restimulated in similar conditions, and the resulting clonal populations were labeled with tetramers. Some of them, chosen randomly, including clones H2307 and H23018, were selected. For tetramer labeling, cells were incubated for the indicated time period and temperature conditions with tetramer-PE. Ab (anti-TCR or -CD8 coupled to FITC, Immunotech, Marseille, France) was then added for a further 30 min at 4°C (19).
Functional Assays.
Lytic activity was measured with a conventional 51Cr-release assay of 4 h by using 3,000 target cells/well. Tumor necrosis factor (TNF)α release was detected by measuring the cytotoxicity of the culture medium on the TNF-sensitive WEHI-164c13 cells (22), with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide colorimetric assay (23).
TCR Gene Sequence and Complementarity-Determining Region 3 (CDR3) Size Analysis of TCRVβ Transcripts.
TCRVα and -Vβ gene segment usage by CTL clones was determined by reverse transcription–PCR by using a panel of primers specific for the 29 Vα and 24 Vβ subfamilies (24). The PCR products were purified and sequenced as described (4). The Vβ13.2 primer sequence was 5′-ATG GGC ACC AGG CTC TTC TTC TAT-3′.
CDR3 size analysis of TCRVβ gene segments expressed by CTL clones in T cell lines, PBL, or tumor fragments was performed as described (4, 25–26). Briefly, cDNA, synthesized from 5 μg of RNA, were amplified in 30 or 40 cycles of PCR by using Vβ/Cβ primer pairs in 50-μl final volume, and aliquots (2 μl) were copied in 1–5 cycle runoff reactions primed with fluorescent (Applied Biosystems fluorophore 6-Fam)-labeled oligonucleotides specific for Cβ, Jβ, or clonotypic primers (Table 1). Runoff products were then subjected to electrophoresis on an Applied Biosystems sequencer in the presence of fluorescent size markers and analyzed by automatic fluorescence quantification and size determination by using the computer program GENESCAN 672 (Applied Biosystems). The sequences of the clonotypic primers hybridizing with the CDR3 region of the selected clones were: 5′-CATGGGGCTCTTCACTGCTGGCA-3′ for clone Heu127; 5′-GCTGGTCCGTCCCAAAACTGCT-3′ for clone Heu171; and 5′-GTCCCCCAGGGCGCTGCT-3′ for clone H23018. Quantification of selected T cell clones was performed as described (18). Briefly, cDNA samples were amplified with the appropriate Vβ and Cβ primers, and the aliquots of the PCR products were subjected to an elongation cycle with either the 6-Fam-labeled and nested Cβ or clonotypic primers. An equal amount of the two runoff products was size-fractionated in the automated sequencer. The proportion of the specific sequence in the total corresponding Vβ mRNA population was calculated by dividing the area under the curve obtained with the clonotypic primer by the sum of the area under the curve obtained with the Cβ primer. This method determines the respective proportion (%) of a unique recurrent transcript in the total VβX mRNAs (18).
Table 1.
TCRα and -β chain V-(D)-J amino acid sequences of α-actinin-4-specific T cell clones
| Heu127
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Vβ22
|
CDR3 | Jβ1.4
|
|||||||
| GCC | AGC | AGT | GAA | GAG | CCC | C | AT | GAA | AAA |
| A | S | S | E | E | P | H | E | K | |
| Vα2.1 | Jα24 | ||||||||
| CTC | TGT | GCC | GTG | ACA | ACT | GAC | |||
| L | C | A | V | T | T | D | |||
| Heu171 | |||||||||
| Vβ8 | CDR3 | Jβ1.5 | |||||||
| AGC | AGT | TT | T | GGG | ACG | GAC | CAG | CCC | CAG |
| S | S | F | G | T | D | Q | P | Q | |
| Vα12 | Jα8 | ||||||||
| TGT | GCT | CTG | AAC | GCC | AGG | GGC | TTT | CAG | |
| C | A | L | N | A | R | G | F | Q | |
| H23018 | |||||||||
| Vβ13.2 | CDR3 | Jβ2.7 | |||||||
| GCC | AGC | AGC | GCC | CTG | GGG | G | AC | GAG | CAG |
| A | S | S | A | L | G | D | E | Q | |
| Vα16 | Jα17.11 | ||||||||
| TTC | TGT | GCT | GGT | GGT | GCT | ||||
| F | C | A | G | G | A | ||||
Results
Establishment of Mutated α-Actinin-4-Reactive TIL Clones.
The cell line IGR-Heu was derived from a LCC patient tumor (21). Mononuclear cells that infiltrated the primary tumor were isolated and stimulated with irradiated IGR-Heu and autologous EBV-transformed B cells and IL-2. The resulting TIL cell line (17% Vβ22 and 3% Vβ8) was cloned by limiting dilution, and 14 CD3+/CD8+/CD28 − tumor-specific CTL clones were isolated and included five Vβ22+ represented by clone Heu127 and two Vβ8+ T cell clones represented by Heu171. Heu127 and Heu171 clones lysed the autologous tumor cell line in a HLA-A2.1-restricted manner but not the autologous EBV-B cell line (Fig. 1).
Figure 1.
Cytotoxic activity of T cell clones derived from patient TIL and PBL. Cytotoxicity was determined by a conventional 4-h 51Cr-release assay at indicated E:T ratios. Target cells included the autologous LCC (IGR-Heu) and EBV-transformed (Heu-EBV) cell lines.
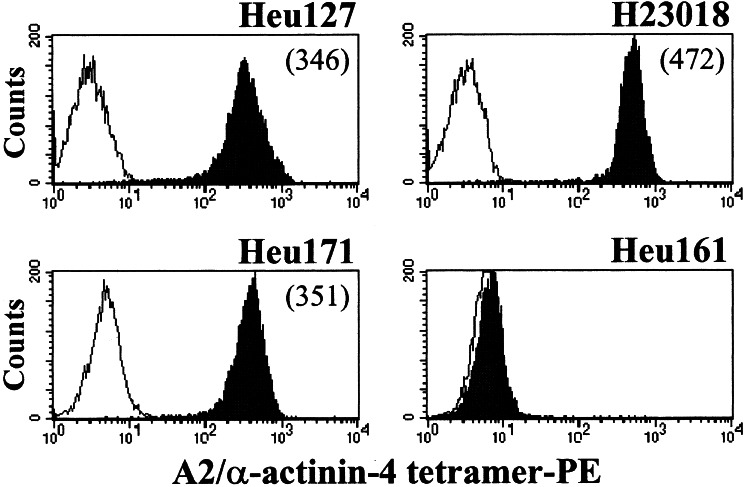
The antigenic peptide recognized by Heu127 was encoded by a mutated α-actinin-4 gene and corresponded to the FIASNGVKLV decamer, with the substitution (Asn instead of Lys) located at position 5 (19). Using soluble HLA-A2/mutated α-actinin-4-phycoerythrin tetramers, we demonstrated that Heu171 recognized the same tumor epitope as Heu127. Indeed, labeling of the CTL clones with HLA multimers showed similar fluorescence intensities (Fig. 2). In contrast, tetramers failed to stain a HLA-A2-restricted autologous TIL clone, Heu161 (4), used as control (Fig. 2). The specificity of Heu171 for the HLA-A2/mutated α-actinin-4 complex was confirmed by a TNFα secretion assay performed after CTL clone activation by an allogeneic HLA-A2-transfected melanoma cell line loaded with the antigenic peptide (data not shown).
Figure 2.
Labeling of anti-mutated α-actinin-4 T cell clones with HLA tetramers. CTL clones Heu127 and Heu171 derived from TIL and H23018 derived from PBL were incubated for 1 h at RT with HLA tetramers. An Heu161 irrelevant clone (4) was included as negative control. The cells were washed, fixed with paraformaldehyde, and analyzed by flow cytometry. Negative controls are shown as white histograms. Numbers in parenthesis correspend to mean fluorescence intensity.
Isolation of Anti-Mutated α-Actinin-4 PBL T Cell Clones with HLA Tetramers.
Patient PBMCs, including less than 0.01% of tetramer+/CD8+ T cells, were stimulated either with the antigenic α-actinin-4 peptide and IL-2, -4, and -7 or in similar conditions to TIL with the autologous IGR-Heu tumor cell line. After two or three stimulations, T cell lines were analyzed for multimer staining. Tumor Ag-specific cells could be detected with a frequency of 56% of the CD8+ cells after stimulation with the mutated decamer. Tetramer+ cells were seeded at one cell per well, and 17 T cell clones, including H23018 and H2307, were randomly selected. These clones (CD8+/CD28 −) showed similar multimer staining to Heu127 and Heu171 TIL clones (Fig. 2) and expressed Vβ13.2-Jβ2.7 rearranged gene segments with identical junctional region, indicating they are likely to be derived from the same precursor. In contrast, patient PBLs stimulated with autologous tumor cells (with probably lower epitope density than stimulation conditions with synthetic peptide) expressed up to 4.2% CD8+/tetramer+ cells, as determined by conventional staining conditions [1 h at room temperature (RT)], including Vβ13.2+ and Vβ8+ T lymphocytes. Interestingly, no Vβ22+/tetramer+ T cells were detected in PBLs after stimulations with either mutated peptide or autologous tumor cells.
Comparison of H23018 TCRβ chain amino acid junctional sequence to those of Heu127 and Heu171, expressing, respectively, Vβ22-Jβ1.4 and Vβ8-Jβ1.5 rearrangements, revealed no sequence homology. Furthermore, TCRα chain sequences indicated that the three clones used distinct TCRα rearrangements with no junctional region similarity (Table 1).
Discrepancy Between Functional Avidity and Tetramer Staining of Tumor Ag-Reactive T Cell Clones.
Initial experiments were performed to analyze the capacity of H2307 and H23018 to lyse the IGR-Heu autologous tumor cell line. As depicted in Fig. 1, both clones lysed IGR-Heu tumor cells much less efficiently than Heu127 and Heu171 TIL. The former cytotoxicity was enhanced when target cells were pulsed with the mutated α-actinin-4 peptide but not with the wild-type peptide (data not shown).
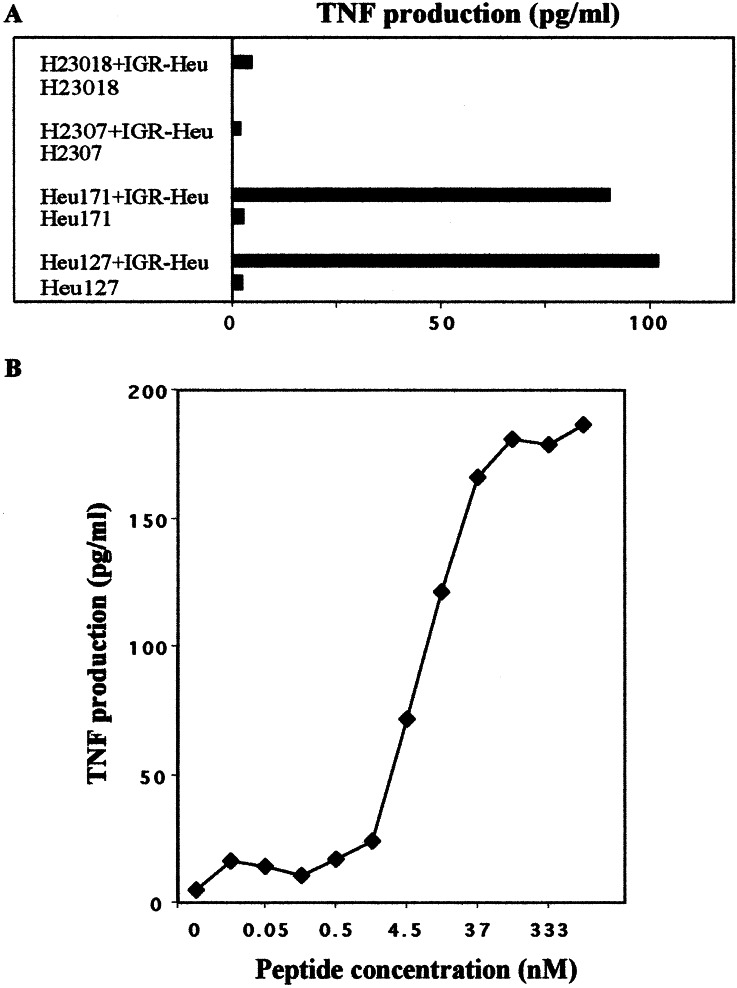
Further experiments were performed to investigate cytokine secretion by TIL and PBL-derived T cell clones after stimulation with the autologous tumor cell line. Fig. 3A shows that H23018 released much less TNFα than Heu127 and Heu171 T cell clones. This secretion was highly increased when IGR-Heu target cells were incubated with the relevant peptide (Fig. 3B). To ascertain whether weak functional activities of the H23018 T cell clone correlated with its TCR avidity for the HLA-A2/mutated α-actinin-4 peptide complex, specific TNFα secretion was performed in the presence of the allogeneic HLA-A2-transfected melanoma cell line loaded with different concentrations of the antigenic peptide. Titration curves over a wide range of mutated peptide concentrations (0.1 nM to 1 μM) were generated, and clone avidity was defined as a concentration of peptide required to obtain 50% of maximal TNFα secretion in a cytokine release assay (Fig. 4). All T cell clones specifically secreted TNFα when incubated with peptide-pulsed HLA-A2-transfected melanoma cells, but they demonstrated an avidity varying from 4.5 to 50 nM. Indeed, as depicted in Fig. 4, Heu127 and Heu171 displayed respectively high (4.5 nM) and intermediate (20 nM) avidities for the HLA-A2/peptide complex, whereas H2307 and H23018 T cell clones displayed low avidity (50 nM).
Figure 3.
(A) Recognition of IGR-Heu tumor cell line by autologous T cell clones. IGR-Heu cells (30,000 cells/well) were incubated with Heu127, Heu171, H2307, or H23018 T cells (3,000 cells/well). After 24 h, the amount of TNFα produced by the clones was measured by testing the killing activity of the culture supernatant on the TNF-sensitive WEHI-164c13 cells. (B) Recognition of the peptide-pulsed IGR-Heu cell line by the autologous H23018 clone. IGR-Heu cells were incubated for 20 min at RT with the indicated concentrations of mutated α-actinin-4 peptide before addition of H23018 lymphocytes. After 24 h, the amount of TNFα was measured as in A.
Figure 4.
TNFα secretion and functional avidity determination of mutated peptide-reactive T cell clones. Allogeneic HLA-A2 melanoma cells were incubated for 20 min at RT with the indicated concentrations of α-actinin-4 peptide before addition of the clones. TNFα production in the culture supernatant was measured after 24 h as in Fig. 3.
To determine whether T cell clones displaying distinct avidity for α-actinin-4 mutated peptide may be stained with similar intensity with HLA tetramers, fluorescence analyses were performed under different time and temperature conditions and with serial concentrations of multimers. Efficient labeling with HLA tetramers of all T cell clones, particularly H23018, was observed on staining during 1 h at RT and was slightly decreased after 4 h. Labeling of the clones at 4°C resulted in slightly less intense staining after 4 h as compared with 1 h but was comparable to that obtained after 4 h at RT. Furthermore, the three clones were stained with similar intensities with tetramers used at different concentrations (Fig. 5). These results indicate that T cell clones with distinct avidity can display similar ability to be stained with HLA multimers. An even slightly more intense staining was observed with H23018 displaying the lowest avidity.
Figure 5.
Binding of HLA tetramers to mutated α-actinin-4-specific T cell clones. Clones were stained with serial concentrations of HLA-A2/α-actinin-4 multimers starting from 2 × 10−2 to 2 × 102 nM under the indicated time and temperature conditions. The cells were washed, fixed with paraformaldehyde, and analyzed by flow cytometry. Data are shown as relative mean fluorescence.
Selective Intratumoral Expansion of High-Avidity CTL Clones.
An initial study demonstrated a selective expansion of tetramers+ Vβ22-Jβ1.4 and Vβ8-Jβ1.5 expressing cells at the tumor site as compared with autologous peripheral blood collected at the same time as cancer biopsy (Fig. 6A). Undetectable (Vβ22) and low (Vβ8) frequencies of specific T cell clones at patient PBL were confirmed by using clonotypic primers (see Materials and Methods) corresponding to TCRVβ CDR3 regions (Fig. 6B). To determine whether Vβ13.2-Jβ2.7+ CTLs were present and potentially overexpressed at the tumor site, we analyzed the corresponding transcript after PCR amplification in the fresh tumor and autologous PBMCs. CDR3-size distribution analysis was performed by using Vβ13.2 oligonucleotide in combination with Cβ or Jβ2.7 primers. A monoclonal peak of 391 nucleotides corresponding to Vβ13.2-Jβ2.7 combination was observed in the H23018 clone. Interestingly, as opposed to Vβ22-Jβ1.4 and Vβ8-Jβ1.5, oligoclonal expansion of H23018 TCR transcript was observed in PBMCs and not at all in the fresh tumor (Fig. 6A). Indeed, a dominant peak corresponding to the H23018 Vβ13.2-Jβ2.7 rearrangement was detected in unstimulated PBMCs as well as in a PBL T cell line stimulated with autologous tumor cells but not in a TIL cell line and tumor biopsy. In contrast, Vβ22-Jβ1.4-expressing cells were not at all detectable in PBLs stimulated with autologous tumor, and Vβ8-Jβ1.5+ T cells were represented in both TIL and PBL cell lines (Fig. 6A). These results indicate a selective in situ clonal expansion of CTL clones with high and intermediate avidities exhibiting strong cytotoxic activity toward tumor cells.
Figure 6.
(A) CDR3 size distribution patterns of Vβ22, Vβ8, and Vβ13.2 transcripts in T cell clones, uncultured tumor and autologous PBMCs, and in in vitro autologous tumor cell-stimulated TIL PBL cell lines analyzed with fluorescent Cβ and indicated Jβ primers. Total RNA from the different samples was extracted, reverse transcribed, and amplified by PCR by using Vβ and Cβ primers. Amplified cDNA was copied by a fluorescent Cβ or Jβ primer in a run-off reaction and subjected to electrophoresis on an automated sequencer. The patterns obtained show the size in nucleotides (x axis) and fluorescence intensity (y axis) of different amplified products. (B) Quantitation of mutated α-actinin-4-specific T cell clones by using clonotypic primers. T cell clones were derived either from TILs (Heu127, Vβ22+ and Heu171, Vβ8+) or PBLs (H23018, Vβ13.2+). Their TCRVβ chain was sequenced, and their presence in fresh tumor and peripheral blood samples was assessed by using clonotypic primers corresponding to CDR3 regions (see Material and Methods for details). The high-avidity Vβ22+-mutated α-actinin-4-specific T cell clone was found only at the tumor site, whereas the low-avidity Vβ13.2+ anti-mutated α-actinin-4 T cell clone was found only in patient peripheral blood. Rates (%) beside the profiles indicate the proportion of the respective recurrent sequence among the total corresponding TCRVβ population. The numbers in parentheses beside the profiles indicate fluorescence intensity of the different amplified products.
Quantitative Assessment of α-Actinin-4-Specific TCR Clonotypes.
To estimate the frequency of TIL-derived Heu127 and Heu171 and PBL-derived H23018 CTL clones in tumor biopsy and patient peripheral blood, clonotypic primers were used to quantitate the corresponding clonotypes, as described in Materials and Methods. The Heu127 T cell clone represented ≈16% of the total TCRVβ22 mRNA at the tumor site, whereas it was undetectable in PBLs (Fig. 6B). With respect to the Heu171 CTL clone, ≈25% and ≈1% of the total TCRVβ8 mRNA were detected at the tumor site and PBLs, respectively. In contrast, the H23018 clone represented ≈4% of the total TCRVβ13.2 mRNA in PBLs, whereas it was undetectable at the tumor site (Fig. 6B). These results confirm a selective in situ accumulation of high-avidity tumor-specific CTL, whereas low-avidity T cell clones were expanded at the periphery.
Discussion
It has become clear that the use of tetramers to selectively sort high-avidity tumor-specific CTLs from the peripheral blood of tumor patients has opened new strategies for the therapy of cancers. Although HLA multimers have already been used to enumerate and characterize tumor-specific CD8+ T lymphocytes in primary and advanced melanomas, no data are available concerning similar studies in patients bearing other solid tumors, including lung carcinoma. A unique study has been performed in the blood of a squamous cell carcinomas patient by using tetramers of soluble HLA-A2 molecules loaded with the antigenic peptide encoded by a mutated malic enzyme gene (12). The present study represents the first direct assessment, to our knowledge, of functional avidity and tetramer staining of tumor-specific mutated Ag-reactive T cell clones isolated from TILs and PBLs of a cancer patient and Ag-specific TCRVβ repertoire usage in both cell compartments. We have previously identified a tumor Ag-derived epitope presented by HLA-A2 molecules and encoded by a mutated α-actinin-4 (ACTN4) gene on a LCC cell line generated from a patient with long survival (19). Tumor Ag-specific T cell clones were isolated from TILs and PBLs by using tetramers of soluble HLA-A2/mutated α-actinin-4 peptide.
TCR analysis indicated that mutated Ag-reactive TIL clones used two different receptors encoded either by Vβ22-Jβ1.4/Vα2.1-Jα24 or Vβ8-Jβ1.5/Vα12-Jα8 rearranged gene segments. In contrast, all tetramer+ clones isolated from patient PBL stimulated with the antigenic peptide expressed a unique Vβ13.2-Jβ2.7/Vα16-Jα17.11 chain receptor. This result may be because of in vitro selection after stimulation with the mutated peptide. However, the high frequency of Vβ13.2+ T lymphocytes was also induced after stimulation of PBL with the autologous tumor. One explanation for the dominance of one tumor Ag-specific T cell clone in PBLs is that it may result from the proliferation of an antitumor precursor that was possibly stimulated with the primary tumor. Alternatively, we may have selected in vitro tetramer+ cells with high proliferative potential. Diverse in vitro expansion potential of tumor specific tetramer-selected T lymphocytes has been reported in melanoma (27).
Our data indicate that T cell clones directed against a truly tumor-specific Ag isolated from TILs and PBLs show similar tetramer staining intensity but distinct avidity of Ag recognition. It has been proposed that the avidity of T cells for Ag recognition is largely because of the affinity of their TCR for peptide–MHC ligand and that this affinity phenotype correlates with the intensity of staining by the specific peptide/HLA tetramers by flow cytometry (28, 29). Comparison of the levels of tetramer fluorescence intensities of antimutated α-actinin-4 T cell clones indicated that no correlation between functional avidity and tetramer staining intensity could be made. Indeed, high-avidity TIL clones exhibited similar, even slightly lower, tetramer fluorescence intensity than low-avidity T cell clones. Concordant results were obtained in melanoma Ag-specific T cells (27, 30, 31), and MHC–tetramer staining was reported to rather reflect structural avidity (32). Furthermore, it has been reported that functional avidity correlates with stability of MHC/peptide multimer binding to TCR (31).
TCR β chain usage by T lymphocytes directed against a tumor-specific mutated Ag in TILs and PBLs indicated distinct but overlapping T cell repertoires. Indeed, a CDR3 size distribution study clearly showed that, whereas high-avidity Vβ22+Ag-specific T lymphocytes were exclusively expanded at the tumor site, low-avidity Vβ13.2+ T cells were amplified in patient PBL only. With respect to a Vβ8+ T cell clone displaying intermediate avidity, although it is expanded in situ, it is slightly represented in peripheral blood. These results indicate that T cell clones exhibiting high avidity for Ag recognition and mediating strong cytotoxic activity toward autologous tumor are selectively expanded at the tumor site. In contrast, the low-avidity T cell clone is not detected in tumor biopsy.
Our data suggest a selective in situ expansion of tumor-specific T cell clones exhibiting high functional avidity. This observation favors a strategy based on HLA tetramer selection of T cell clones from patient TIL rather than from PBL for their use in immunotherapy protocols. The combination of tetramer technique with TCR gene usage at the tumor site and PBL corresponds to a powerful method to select tumor Ag able to induce strong CTL response as potential candidates for cancer vaccine trials. This method will also be useful in following changes in the repertoire of tumor-specific CTLs during vaccination and disease progression. Indeed, a recent study performed after vaccination of melanoma patients with melan-A peptide indicated an increase in functional avidity of Ag recognition and in tumor reactivity (33).
Acknowledgments
We thank Dr. P. Coulie for helpful discussion and Drs. D. Colau and V. Karanikas for help with tetramer production. We also thank Yann Lecluse for FACS analyses and D. Grunenwald from the Thoracic Department Surgery of Institut Mutualiste Montsouris (Paris). This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Institut Gustave Roussy, the Association pour la Recherche sur le Cancer (Grants 9307, 5253, 2129), the Fondation de France, and Groupement des Entreprises Françaises dans la Lutte Contre le Cancer (GEFLUC). H.E. was supported by a fellowship from the Fondation de France (Comité Tumeurs Solides).
Abbreviations
- TIL
tumor-infiltrating lymphocyte
- CDR3
complementarity-determining region 3
- CTL
cytotoxic T lymphocyte
- TCR
T cell receptor
- PBL
peripheral blood lymphocyte
- PBMC
peripheral blood mononuclear cell
- Ag
antigen
- LCC
large-cell carcinoma
- EBV
Epstein–Barr virus
- TNF
tumor necrosis factor
- RT
room temperature
References
- 1.Van den Eynde B J, van der Bruggen P. Curr Opin Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- 2.Marchand M, van Baren N, Weynants P, Brichard V, Dreno B, Tessier M H, Rankin E, Parmiani G, Arienti F, Humblet Y, et al. Int J Cancer. 1999;80:219–230. doi: 10.1002/(sici)1097-0215(19990118)80:2<219::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 3.Thurner B, Haendle I, Roder C, Dieckmann D, Keikavoussi P, Jonuleit H, Bender A, Maczek C, Schreiner D, von den Driesch P, et al. J Exp Med. 1999;190:1669–1678. doi: 10.1084/jem.190.11.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Echchakir H, Vergnon I, Dorothee G, Grunenwald D, Chouaib S, Mami-Chouaib F. Int Immunol. 2000;12:537–546. doi: 10.1093/intimm/12.4.537. [DOI] [PubMed] [Google Scholar]
- 5.Baurain J F, Colau D, van Baren N, Landry C, Martelange V, Vikkula M, Boon T, Coulie P G. J Immunol. 2000;164:6057–6066. doi: 10.4049/jimmunol.164.11.6057. [DOI] [PubMed] [Google Scholar]
- 6.Weynants P, Thonnard J, Marchand M, Delos M, Boon T, Coulie P G. Am J Respir Crit Care Med. 1999;159:55–62. doi: 10.1164/ajrccm.159.1.9805073. [DOI] [PubMed] [Google Scholar]
- 7.Slingluff C L, Cox A L, Stover J M, Moore M M, Hunt D F, Engelhard V H. Cancer Res. 1994;54:2731–2737. [PubMed] [Google Scholar]
- 8.Yoshino I, Goedegebuure P S, Peoples G E, Parikh A S, DiMaio J M, Lyerly H K, Gazdar A F, Eberlein T J. Cancer Res. 1994;54:3387–3390. [PubMed] [Google Scholar]
- 9.Hogan K T, Eisinger D P, Cupp S B, Lekstrom K J, Deacon D D, Shabanowitz J, Hunt D F, Engelhard V H, Slingluff C L, Ross M M. Cancer Res. 1998;58:5144–5150. [PubMed] [Google Scholar]
- 10.Gomi S, Nakao M, Niiya F, Imamura Y, Kawano K, Nishizaka S, Hayashi A, Sobao Y, Oizumi K, Itoh K. J Immunol. 1999;163:4994–5004. [PubMed] [Google Scholar]
- 11.Kawano K, Gomi S, Tanaka K, Tsuda N, Kamura T, Itoh K, Yamada A. Cancer Res. 2000;60:3550–3558. [PubMed] [Google Scholar]
- 12.Karanikas V, Colau D, Baurain J F, Chiari R, Thonnard J, Gutierrez-Roelens I, Goffinet C, Van Schaftingen E V, Weynants P, Boon T, et al. Cancer Res. 2001;61:3718–3724. [PubMed] [Google Scholar]
- 13.Altman J D, Moss P A, Goulder P J, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 14.Dunbar P R, Ogg G S, Chen J, Rust N, van der Bruggen P, Cerundolo V. Curr Biol. 1998;8:413–416. doi: 10.1016/s0960-9822(98)70161-7. [DOI] [PubMed] [Google Scholar]
- 15.Lee P P, Yee C, Savage P A, Fong L, Brockstedt D, Weber J S, Johnson D, Swetter S, Thompson J, Greenberg P D, et al. Nat Med. 1999;5:677–685. doi: 10.1038/9525. [DOI] [PubMed] [Google Scholar]
- 16.Romero P, Dunbar P R, Valmori D, Pittet M, Ogg G S, Rimoldi D, Chen J L, Lienard D, Cerottini J C, Cerundolo V. J Exp Med. 1998;188:1641–1650. doi: 10.1084/jem.188.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valmori D, Dutoit V, Lienard D, Lejeune F, Speiser D, Rimoldi D, Cerundolo V, Dietrich P Y, Cerottini J C, Romero P. J Immunol. 2000;165:533–538. doi: 10.4049/jimmunol.165.1.533. [DOI] [PubMed] [Google Scholar]
- 18.Dietrich P Y, Walker P R, Quiquerez A L, Perrin G, Dutoit V, Liénard D, Guillaume P, Cerottini J C, Romero P, Valmori D. Cancer Res. 2001;61:2047–2054. [PubMed] [Google Scholar]
- 19.Echchakir H, Mami-Chouaib F, Vergnon I, Baurain J F, Karanikas V, Chouaib S, Coulie P G. Cancer Res. 2001;61:4078–4083. [PubMed] [Google Scholar]
- 20.Nikolopoulos S N, Spengler B A, Kisselbach K, Evans A E, Biedler J L, Ross R A. Oncogene. 2000;19:380–386. doi: 10.1038/sj.onc.1203310. [DOI] [PubMed] [Google Scholar]
- 21.Asselin-Paturel C, Echchakir H, Carayol G, Gay F, Opolon P, Grunenwald D, Chouaib S, Mami-Chouaib F. Int J Cancer. 1998;77:7–12. doi: 10.1002/(sici)1097-0215(19980703)77:1<7::aid-ijc2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 22.Espevik T, Nissen-Meyer J. J Immunol Methods. 1986;95:99–105. doi: 10.1016/0022-1759(86)90322-4. [DOI] [PubMed] [Google Scholar]
- 23.Hansen M B, Nielsen S E, Berg K. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 24.Genevee C, Diu A, Nierat J, Caignard A, Dietrich P Y, Ferradini L, Roman-Roman S, Triebel F, Hercend T. Eur J Immunol. 1992;22:1261–1269. doi: 10.1002/eji.1830220522. [DOI] [PubMed] [Google Scholar]
- 25.Pannetier C, Even J, Kourilsky P. Immunol Today. 1995;16:176–181. doi: 10.1016/0167-5699(95)80117-0. [DOI] [PubMed] [Google Scholar]
- 26.Puisieux I, Even J, Pannetier C, Jotereau F, Favrot M, Kourilsky P. J Immunol. 1994;153:2807–2818. [PubMed] [Google Scholar]
- 27.Palermo B, Campanelli R, Mantovani S, Lantelme E, Manganoni A M, Carella G, Da Prada G, Zzdella Cuna G R, Romagne F, Gauthier L, et al. Eur J Immunol. 2001;31:412–420. doi: 10.1002/1521-4141(200102)31:2<412::aid-immu412>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 28.Yee C, Savage P A, Lee P P, Davis M M, Greenberg P D. J Immunol. 1999;162:2227–2234. [PubMed] [Google Scholar]
- 29.Dutoit V, Rubio-Godoy V, Dietrich P Y, Quiqueres A L, Schnuriger V, Rimoldi D, Liénard D, Speiser D, Guillaume P, Batard P, et al. Cancer Res. 2001;61:5850–5856. [PubMed] [Google Scholar]
- 30.Rubio-Godoy V, Dutoit V, Rimoldi D, Liénard D, Lejeune F, Speiser D, Guillaume P, Cerottini J C, Romero P, Valmori D. Proc Natl Acad Sci USA. 2001;98:10302–10307. doi: 10.1073/pnas.181348898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutoit V, Rubio-Godoy V, Doucey M A, Batard P, Liénard D, Rimoldi D, Speiser D, Guillaume P, Cerottini J C, Romero P, et al. J Immunol. 2002;168:1167–1171. doi: 10.4049/jimmunol.168.3.1167. [DOI] [PubMed] [Google Scholar]
- 32.Bullock T N J, Mullins D W, Colella T A, Engelhard V H. J Immunol. 2001;167:5824–5831. doi: 10.4049/jimmunol.167.10.5824. [DOI] [PubMed] [Google Scholar]
- 33.Valmori D, Dutoit V, Schnuriger V, Quiquerez A L, Pittet M J, Guillaume T, Rubio-Godoy V, Walker P R, Rimoldi D, Liénard D, et al. J Immunol. 2002;168:4231–4240. doi: 10.4049/jimmunol.168.8.4231. [DOI] [PubMed] [Google Scholar]