Abstract
The expression of uncoupling protein 2 (UCP2) was reduced in macrophages after stimulation with lipopolysaccharide (LPS). The physiological consequence and the regulatory mechanisms of the UCP2 down-regulation by LPS were investigated in a macrophage cell line, RAW264 cells. UCP2 overexpression in RAW264 cells transfected with eukaryotic expression vector containing ucp2 cDNA markedly reduced the production of intracellular reactive oxygen species. Furthermore, in the UCP2 transfectant, nitric oxide (NO) synthesis, inducible NO synthase (NOS II) protein, NOS II mRNA, and NOS II promoter activity were definitely decreased after LPS stimulation compared with those in parental RAW264 or RAW264 cells transfected with the vector alone. Reporter assays suggested that an enhancer element was located in the region of intron 2 of the UCP2 gene and that the UCP2 expression was down-regulated not by the 7.3-kb promoter region but by the 5′ region of the UCP2 gene containing two introns. Deletion of intron 2 resulted in the low transcriptional activities and abolishment of the LPS-associated negative regulation. In addition, the mRNA expression of transfected UCP2 was suppressed in RAW264 cells transfected with expression vector containing UCP2 genomic DNA, but was markedly increased in cells transfected with the vector containing UCP2 intronless cDNA. These findings suggest that the LPS-stimulated signals suppress UCP2 expression by interrupting the function of intronic enhancer, leading to an up-regulation of intracellular reactive oxygen species, which activate the signal transduction cascade of NOS II expression, probably to ensure rapid and sufficient cellular responses to a microbial attack.
Uncoupling protein 2 (UCP2) is a recently discovered member of the mitochondrial inner membrane carrier family with high homology to the brown adipose tissue-specific proton transporter, UCP1 (1–3). Because the gene ucp2 resides within a region of genetic linkage to obesity (1) and its product UCP2 uncouples respiration (4), a role in energy dissipation has been proposed. Mice lacking Ucp2 after targeted gene disruption, however, are not obese and have a normal response to cold exposure or high-fat diet (5). On the other hand, it has been proposed that UCP2 limits production of reactive oxygen species (ROS) by decreasing the mitochondrial membrane potential (6). Indeed, Ucp2−/− mice are resistant to Toxoplasma gondii infection, and macrophages of the mutant mice have higher levels of ROS (5), which are associated with higher cytolytic activity (7). In addition, unlike UCP1, expression of UCP2 teems in spleen, lung, and isolated macrophages (1, 2, 8). These findings suggest a role for UCP2 in immunity or inflammatory responsiveness.
Recognition of lipopolysaccharide (LPS) is crucial for host
antimicrobial defense reactions (9, 10). Nitric oxide (NO) production
by the inducible isoform of NO synthase (NOS II) after LPS stimulation
plays a pivotal role in numerous and diverse biological functions, in
particular, as a principal mediator of the microbicidal and tumoricidal
actions of macrophages (11). Also, ROS are rapidly produced from
macrophages after LPS stimulation and involved in cellular signaling
for NOS II gene expression (12, 13). Further,
O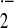 and NO combine to form the potent oxidant
peroxynitrite (ONOO−) which mediates
bactericidal activity (14). Thus, both ROS and NO are important
mediators of cellular immune response. It is well established that
mitochondria are the main source of ROS (15), but the role of UCP2 in
the regulation of the response to LPS has not been elucidated.
and NO combine to form the potent oxidant
peroxynitrite (ONOO−) which mediates
bactericidal activity (14). Thus, both ROS and NO are important
mediators of cellular immune response. It is well established that
mitochondria are the main source of ROS (15), but the role of UCP2 in
the regulation of the response to LPS has not been elucidated.
Recently, it has been reported that UCP2 mRNA levels do not always reflect the expression of the protein itself (16). Although many publications have described variations of UCP2 mRNA expression, only a few studies examined its protein level (17, 18). In the present study, we first demonstrated that both mRNA and protein levels of UCP2 were markedly decreased in macrophage cell line RAW264 cells stimulated with LPS. Then, to examine directly the roles of UCP2 in the response to LPS stimulation, we established macrophage cell line RAW264 cells transfected with ucp2 cDNA (RAWucp2) and analyzed the effects of UCP2 overexpression on ROS and NO productions. It was shown that overexpression of UCP2 reduced intracellular ROS, accompanied by decreases of NOS II expression. The mechanisms of the down-regulation of UCP2 expression by LPS stimulation were also examined.
Materials and Methods
Cell Culture.
The murine macrophage cell line RAW264 (RCB0535) was purchased from RIKEN Cell Bank (Ibaragi, Japan). Cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, 100 units/ml penicillin, and 100 mg/ml streptomycin (Life Technologies, Rockville, MD) at 37°C in a humidified incubator containing 5% CO2 in air.
Measurement of Intracellular Generation of ROS.
Flow cytometric analysis of intracellular generation of ROS was performed by using 2′,7′-dichlorofluorescein-diacetate (DCFH-DA). The oxidation of DCFH-DA by ROS results in the formation of the fluorescent compound DCF (19). Cells were cultured in the presence or absence of 1 μg/ml LPS and treated with 5 μM DCFH-DA at 37°C for final 30 min. The fluorescence intensities of DCF of more than 10,000 viable cells from each sample were analyzed by a FACSCalibur flow cytometer (Becton Dickinson).
Determination of Nitrite Concentration.
Nitrite in cell-culture supernatants was determined with Griess reagent (20) by using a sodium nitrite as a standard.
Preparation of Mitochondrial and Cytoplasmic Proteins.
For isolation of mitochondria, cells were washed with ice-cold PBS three times, suspended in TES buffer (10 mM Tris, pH 7.5/1 mM EDTA/250 mM sucrose) supplemented with protease inhibitors (4 mg/ml aprotinin/0.1 mM phenylmethylsulfonyl fluoride/10 mM DTT), and disrupted in Thomas's potter at low-speed rotation. Unbroken cells and nuclei were removed by centrifuging the homogenate at 750 × g for 10 min. The supernatant was centrifuged at 10,000 × g for 20 min, and the mitochondrial pellet was resuspended in 1 ml of TES buffer. Mitochondria were submitted to another round of 10 min of centrifugation at 750 and 10,000 × g, respectively. The mitochondrial proteins were dissolved in 0.1% SDS and their concentrations were assayed by the Bradford method by using Protein Assay kit (Bio-Rad). Cytoplasmic protein extracts were prepared as described (21).
Western Blotting Analysis.
Mitochondrial proteins (40 μg) or cytoplasmic proteins (10 μg) were boiled in loading buffer (Tris⋅HCl, pH 6.8/2% SDS/5% glycerol/10% 2-mercaptoethanol), separated in an SDS-10% polyacrylamide gel, and transferred to poly(vinylidene difluoride) membrane (Applied Biosystems). Membranes were blocked with 1% nonfat dried milk in Tris-buffered saline (TBS) for 1 h. Goat polyclonal Abs against UCP2 (sc-6525, Santa Cruz Biotechnology) or rabbit polyclonal Abs against NOS II (06–573, Upstate Biotechnology, Lake Placid, NY) were applied for analysis of intrinsic UCP2 and NOS II, respectively. Thereafter, secondary Abs [horseradish peroxidase-conjugated rabbit anti-goat IgG or horseradish peroxidase-conjugated goat anti-rabbit IgG (Dako)] were applied. For analysis of transfected UCP2 expression, horseradish peroxidase-conjugated anti-myc Abs (Invitrogen) were applied. The immunoreactivity was visualized with an enhanced chemiluminescence reagent (ECL, Amersham Pharmacia).
RNA Isolation and Northern Analysis.
Total cellular RNA was prepared from cells by using TRIzol Reagent (Life Technologies). Total RNA (25 μg) was electrophoresed under denaturing conditions through a 1% (wt/vol) agarose-formaldehyde gel and then transferred to Nyrone membranes and UV covalently cross-linked. Reverse transcription (RT)-PCR products were used as UCP2 probe. The membrane was hybridized with alkalyphosphatase-labeled UCP2 probes and UCP2 mRNA bands were detected by a CDP-Star detection reagent (Amersham Pharmacia).
RT-PCR.
The RT reaction with 2 μg of total RNA proceeded at 42°C for 50 min by using ReverScript I (Wako Pure Chemical, Osaka) with oligo(dT)15 primer (Roche Diagnostics). The reaction mixture was used directly in the PCR with the following oligonucleotide primers: UCP2, sense (UCP2f: 5′-GGGACGTAGCAGGAAATCAG-3′), antisense (5′-GGTGCTTGGTATCTCCGAC-3′, 560-bp product); β-actin, sense (5′-AACCGCGAGAAGATGACCCAGATCATGTTT-3′), antisense (5′-AGCAGCCGTGGCCATCTCTTGCTCGAAGTC-3′, 349-bp product); 18S, sense (5′-GAGAAACGGCTACCACATCC-3′), antisense (5′-CCCAAGATCCAACTACGAGC-3′, 230-bp product). The transfected UCP2 was amplified by using UCP2f primer and antisense primer of polyHis region (HIS, 5′-TGGTGATGGTGATGATGACC-3′, 1,089-bp product) of the vector.
Generation of NOS II Luciferase Reporter Constructs.
The full-length murine NOS II promoter fragment (1,739 bp: −1580 to +159 from the transcription initiation site) was cloned into the pGL3-enhancer luciferase reporter gene vector (Promega) (pGL3-NOS II) as described (22).
UCP2 Plasmid Constructs.
The cDNA of murine UCP2 (ucp2), the cDNA of full-length UCP2 mRNA (ucp/m: sequence corresponding to eight exons of UCP2), and UCP2 genomic DNA (ucp/hn: sequence corresponding to premRNA of UCP2) were obtained by PCR with the following primers: ucp2, sense (UCP2–5′:5′-GGGAGGTAGCAGGAAATCAG-3′), antisense (UCP2–3′:5′-GGGCCCTCTAGAGCCGAAAGGTGCCTCCCGAGATT-3′, where the underlined and bold sequences indicate XbaI site and the mutated stop codon (ATG) in the wild type, respectively); ucp/m, sense [5′ end of the first exon (E1–5′): 5′-GCTACTGTCAGTTCCGCCCT-3′)], antisense (UCP2–3′); ucp/hn: sense (−33: 5′-GGTACCCTAGACTTAAGTTCTGTAGGCGCG-3′), antisense (UCP2–3′). The amplified ucp2 and ucp/m fragments were subcloned into a pGEM-T East vector (Promega), digested at HindIII/XbaI sites and NotI/XbaI sites, respectively, and subcloned into pcDNA4/TO/myc-His A vector (Invitrogen) at the corresponding sites. The amplified ucp/hn fragment was digested at AflII/XbaI sites and subcloned into pcDNA4/TO/myc-His A vector at the corresponding sites. The amplified PCR products were subjected to sequencing with an automatic DNA sequencer (Applied Biosystems). All plasmid DNAs used for transfection were prepared by using an EndoFree Plasmid Kit (Qiagen, Hilden, Germany).
UCP2 Promoter and 5′ Region of UCP2 Gene Plasmid Constructs.
To examine the regulation of UCP2 gene expression, the plasmids for the transient expression analysis were constructed by using the pGL3-basic luciferase reporter gene vector (Promega). The following primers were used to amplify the promoter regions and 5′ region upstream from the translation initiation site of UCP2 (23): sense primers, −7325 (5′-CGCGGTACCTCCACCTTAGGGCAAGAACG); −2746 (5′-CGCGGTACCACACACTAGCCTCCAGGACC-3′); −233 (57-GGTACCCGATCCAGAACAGCTGGCCAGTCA-3); −33 (5′-GGTACCCTAGACTTAAGTTCTGTAGGCGCG-3′), where the underlined sequences indicate KpnI sites, and antisense primers, +100 (5′-ATGAACGCGTGAGAACACAGGAGTGCAGAA-3′); +3433 (5′-AATGACGCGTATTTCCTGCTACCTCCCAGA-3′), where the underlined sequences indicate MluI sites. Each amplified fragment was ligated into the KpnI/MluI sites of the pGL3 basic vector.
Mutagenesis and Creation of Constructs Without Intron(s).
Site-directed mutagenesis of the −233/+3433 constructs (designated as 5′UR) was performed by using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The upstream three-initiator ATG codons and one terminator TGA codon were mutated to TTG and to GGA, respectively. All mutations were confirmed by sequencing.
The CpoI site was introduced in the 3′ end of intron 1 of UCP2 by mutagenesis of the 5′UR construct. Then, the 5′UR (del-i1) construct was created by excision of the first intronic region as a 776-bp CpoI fragment (with the CpoI site located in the 5′ end of intron 1). To create 5′UR (del-i2) and 5′UR (del-i12) constructs, the PCR product of genomic DNA with the sense primer E1–5′ and the antisense primer of the 3′ end of the second exon (5′-GGCTAGCACGCGTCTGGAGGTGCTTTGAGGTC-3′; the underlined sequence indicates MluI site) and the PCR product of UCP2 cDNA with sense primer E1–5′ and antisense primer +3433, respectively, were subcloned into the BstXI/MluI sites of the 5′UR construct after digestion with BstXI and MluI. The deletion constructs, 5′UR(del-i2a) and 5′UR(del-i2s), were created by excision of a 1,588-bp AvrII fragment and two 160- and 928-bp SacI fragments within intron 2, respectively.
Stable Transfection of UCP2.
RAW264 cells suspended in RPMI 1640 medium containing 30 μg pcDNA4/TO/myc-HIS A vector alone, pcDNA4/TO/myc-HIS A-ucp2, -ucp2/m, or -ucp2/hn constructs were electroporated with EasyjectT Optima (EquiBio, Boughton Monchelsea, Kent, U.K.) at an electric pulse of 350 V and 1,500 μF. On the second day after electroporation, selection was started in medium containing 500 μg/ml Zeocine (Invitrogen).
Transient Transfection and Luciferase Assay.
RAW264 cells were transfected by using Lipofectamine Reagent (Life Technologies) with constructs containing the luciferase reporter gene, and luciferase activity was determined by using a Duar Luciferase Assay System kit (Promega) as described (22).
Results
Effects of UCP2 Overexpression on Functions of RAW264 Cells.
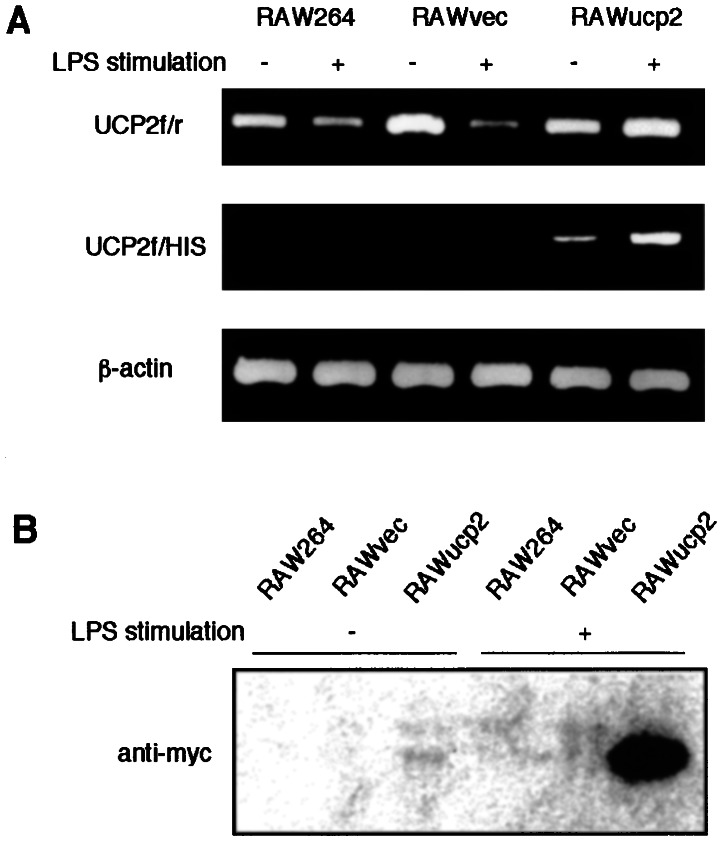
We demonstrated that both levels of mRNA (Fig. 1A) and protein (Fig. 1B) of UCP2 were markedly decreased in RAW264 cells after LPS stimulation. Therefore, to investigate the function of UCP2 in response to LPS, a stable UCP2 transfectant (RAWucp2) and a vector control (RAWvec) were established. Although the expression of UCP2 mRNA was markedly decreased in either RAW264 or RAWvec cells after LPS stimulation, the amplified RT-PCR product of UCP2 mRNA derived from both intrinsic and transfected ucp2 genes was increased in RAWucp2 cells by LPS stimulation (Fig. 2A Top). PCR analysis with UCP2f and HIS primers showed that the mRNA of transfected UCP2 was expressed in RAWucp2 cells and increased markedly by LPS stimulation (Fig. 2A Middle). Also, the transfected UCP2 protein was detected by anti-myc antibodies in RAWucp2 cells and was greatly increased by LPS stimulation (Fig. 2B).
Figure 1.
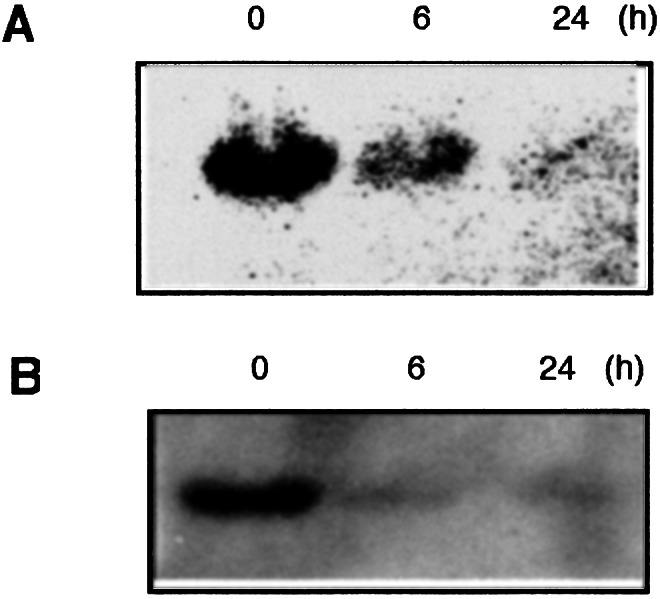
UCP2 expression after LPS stimulation. Total cellular RNA and mitochondrial protein were prepared from RAW264 cells stimulated with LPS for 0, 6, and 24 h. (A) UCP2 mRNA expression was analyzed by Northern blotting analysis. (B) Amount of UCP2 protein in mitochondria was analyzed by Western blotting analysis.
Figure 2.
Intrinsic or transfected UCP2 expression in RAW264 cells and RAW264 cells transfected with ucp2 construct or with vector alone. Total cellular RNA and mitochondrial protein were prepared from these cells after they were cultured with or without LPS for 24 h. (A) The expression of intrinsic and transfected UCP2 mRNA was analyzed by RT-PCR analysis. For normalization, β-actin was used. (B) The expression of transfected ucp2 products was analyzed by Western blotting analysis with anti-myc antibodies.
To examine the effects of UCP2 overexpression on ROS production, the level of intracellular ROS was analyzed by flow cytometry. As shown in Fig. 3, the transfection of ucp2 cDNA resulted in a notable reduction of the intracellular ROS. Further, LPS stimulation increased the production of ROS in RAW264 and RAWvec cells but not in RAWucp2 cells. Then, the effects of UCP2 overexpression on NO production were analyzed. The nitrite concentration in the culture supernatants of RAWucp2 cells stimulated with LPS was considerably smaller than that in the culture supernatants of RAW264 and RAWvec cells (Fig. 4A). After stimulation with LPS for 6 h, a distinct NOS II protein band of 130 kDa was observed in RAW264 and RAWvec cells, but not in RAWucp2 cells (Fig. 4B). Although a NOS II protein band was observed in RAWucp2 cells after stimulation with LPS for 24 h, the expression level was apparently lower than that in RAW264 or RAWvec cells. Similar results were obtained in the RT-PCR analysis of the NOS II mRNA expression (Fig. 4C). In addition, the promoter activities in RAWucp2 cells were considerably lower than those in RAW264 or RAWvec cells and were not enhanced by LPS stimulation. These findings suggest that the overexpression of UCP2 decreases the ROS production, resulting in the abrogation of the NOS II expression after LPS stimulation.
Figure 3.
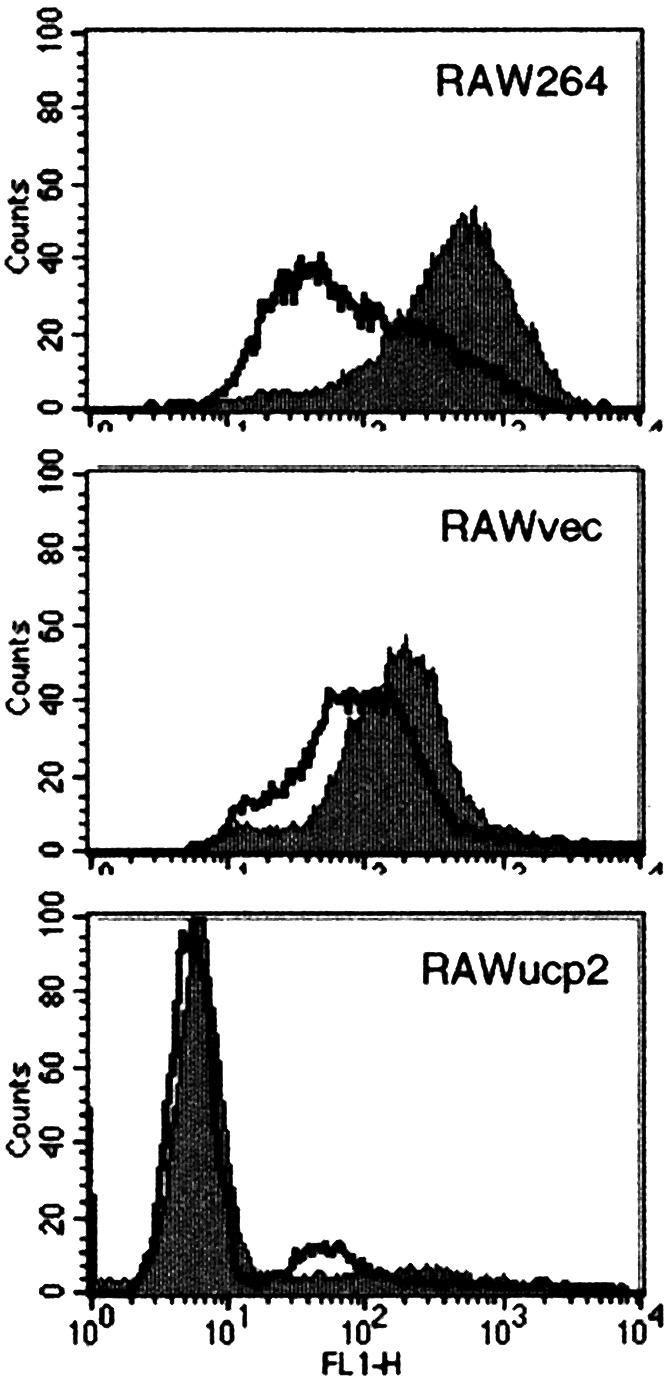
Effects of UCP2 overexpression on intracellular ROS. Cells were cultured with or without LPS for 1 h, and intracellular ROS was analyzed by flow cytometry.
Figure 4.
Effects of UCP2 overexpression on NO production and NOS II expression. (A) Cells were stimulated with LPS for 48 h, and accumulation of nitrite in the supernatants was measured with Griess reagent. (B) Cells were stimulated with LPS for 0, 6, and 24 h. Amount of NOS II protein was analyzed by Western blotting analysis. (C) Cells were stimulated with LPS for 0, 6, and 24 h. NOS II mRNA expression was analyzed by RT-PCR analysis. (D) Cells were transiently transfected with luciferase reporter construct containing NOS II promoter. After LPS stimulation for 24 h, luciferase activities were determined. (A and D) Results are expressed as the mean ± SEM from quadruplicate cultures.
Regulation of UCP2 Expression by the 7.3-kb-Long Promoter and the 3.4-kb-Long 5′ Region of UCP2.
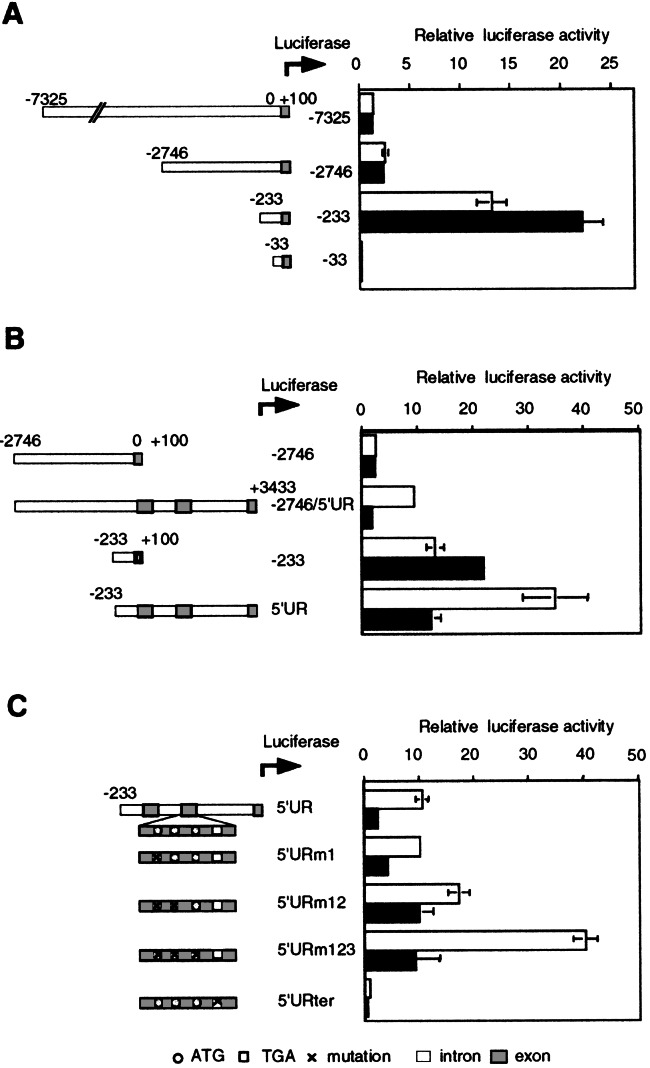
Because it was suggested that the regulation of UCP2 expression plays an important role in the response to LPS, the mechanism of the down-regulation of UCP2 expression was investigated by using various luciferase reporter constructs. We created a series of deletion constructs of the UCP2 promoter and analyzed the transcriptional activities of these constructs. As depicted in Fig. 5A, of four constructs analyzed, the construct −233 (−233/+100) had the highest luciferase activity. The shortest construct −33 (−33/+100) showed negligible luciferase activity, suggesting that strong enhancer elements for basal promoter activity are located in the region from −233 to −34 in murine UCP2 promoter. On the other hand, luciferase activities in the construct −7325 (−7325/+100) and construct −2746 (−2746/+100) were markedly lower than those in the construct −233. Therefore, it seems likely that negative regulatory elements are located within the region from −2746 to −233. Luciferase activities in the construct −233 were significantly enhanced by LPS stimulation.
Figure 5.
Reporter gene assay of promoter and 5′ region of UCP2. RAW 264 cells were transiently transfected with various reporter constructs. Cells were cultured without (□) or with (■) LPS for 24 h, and luciferase activities were determined. Results are expressed as the mean ± SEM from quadruplicate cultures. (A) Reporter constructs containing the UCP2 promoter regions. (B) Reporter constructs containing promoter region and 5′ region upstream from the translation initiation site of UCP2. (C) Reporter constructs containing the UCP2 5′ region with the indicated mutations.
Because an appropriate negative regulation of the ucp2 gene expression by stimulation with LPS could not be found in the promoter region, we examined the regulatory mechanisms in the 5′ region upstream from the translation initiation site of the UCP2 gene. Two constructs (−2746/5′UR:−2746/+3433 and 5′UR:−233/+3433) were created and transiently transfected to RAW264 cells. The 5′ region of the UCP2 gene strikingly enhanced the transcriptional activities of the promoter region (Fig. 5B). In addition, LPS stimulation inhibited the luciferase activities in the constructs with the 5′ region of UCP2 gene. On the other hand, it was reported recently that the upstream open reading flame (uORF) with three ATG codons is located within the second exon and negatively regulates the UCP2 expression in COS cells (16). Thus, we examined whether the uORF would participate in the negative regulation of UCP2 expression by LPS. As shown in Fig. 5C, the mutation of the second and the third ATGs strongly enhanced luciferase activities. In contrast, mutation in the termination codon of the uORF (5′ URter) remarkably reduced luciferase activity. These mutations, however, did not affect the negative regulation by LPS.
Intronic Regulation of the UCP2 Expression.
We next examined whether two intronic regions participate in the negative regulation of UCP2 expression by LPS. The deletion of intron 1 did not affect the negative regulation by LPS. Meanwhile, the deletion of intron 2 not only decreased luciferase activities but also abolished the LPS-associated negative regulation (Fig. 6A). The negative regulation by LPS still existed in the 5′UR construct without the 1,588-bp AvrII fragment or the 1,088-bp SacI fragment in intron 2 (Fig. 6B). Furthermore, the increase of transfected UCP2 mRNA was observed in RAWucp/m cells (Fig. 7A) as well as RAWucp2 cells (Fig. 2A). On the other hand, the expression of transfected UCP2 mRNA was down-regulated in RAWucp/hn cells (Fig. 7B).
Figure 6.
Reporter gene assay of the 5′ region of UCP2 with deletion of intronic region. RAW 264 cells were transiently transfected with various reporter constructs. Cells were cultured without (□) or with (■) LPS for 24 h, and luciferase activities were determined. Results are expressed as the mean ± SEM from quadruplicate cultures. (A) Reporter constructs containing the 5′ region of UCP2 with deletion of intronic region. (B) Reporter constructs containing the 5′ region of UCP2 with deletion of AvrII or SacI fragment.
Figure 7.
Transfected UCP2 expression in RAWucp2/m (A) and RAWucp2/hn (B) cells. Total cellular RNA was prepared from these cells after they were cultured with or without LPS for 0, 1, 3, 6, and 24 h. The expression of transfected UCP2 mRNA was analyzed by RT-PCR analysis. For normalization, 18S was used.
Discussion
In the present study, we first demonstrated that both UCP2 protein and mRNA expression are decreased in macrophage cell line RAW264 cells after LPS stimulation, suggesting a physiological role for UCP2 in macrophages. To investigate the function of UCP2 in the response to LPS directly, we established a macrophage cell line RAWucp2.
The transfected ucp2 product was detected in the UCP2 transfectant and markedly increased after stimulation with LPS. This increase might be attributable to the promoter activity of cytomegalovirus in the pcDNA4 construct, which is known to be potentially responsive to LPS. Recently, Lee et al. (24) reported that macrophages from ob/ob mice, which express less UCP2, generate more ROS than lean mice. Moreover, macrophages from Ucp2−/− mice increased levels of ROS (5). Mice lacking UCP3 also have increased levels of ROS in skeletal muscle (25). These findings agree with the hypothesis (26) that an increase in the mitochondrial membrane potential would slow the transport of electrons through the respiratory chain, increasing the time of interaction between these electrons and molecular oxygen and facilitating the formation of ROS. Thus, we examined whether overexpression of UCP2 inhibits the production of ROS in macrophages on stimulation with LPS. As anticipated, both steady-state and LPS-stimulated ROS productions in RAWucp2 cells were markedly lower than those in RAW264 and RAWvec cells. This finding indicated that UCP2 is a constitutive down-modulator of ROS production, suggesting a role for UCP2 in the regulation of intracellular redox state and macrophage-mediated immunity.
Free radicals were studied within the context of a direct, destructive role in biology. Although this context is true for high radical concentrations, ROS are increasingly recognized to control signal transduction by activation of mitogen-activated protein kinases or being implicated in the regulation of transcription factors such as NF-κB or AP-1 (27). These factors are important components in the transcriptional regulation of NOS II expression (28, 29). We showed that LPS stimulation could not increase the intracellular ROS levels in RAWucp2 cells, accompanied by reduced production of NO. Furthermore, the decreases in the NOS II promoter activity and the NOS II mRNA expression were observed in the RAWucp2 cells, indicating that NOS II expression was transcriptionally down-regulated by UCP2 overexpression. High-output NO synthesis accomplished by NOS II constitutes an important event in the host defense and in the regulation of immune responses. Arsenijevic et al. (5) reported that the ability of Ucp2−/− mice to resist and eliminate an infectious challenge such as T. gondii more efficiently seems to be related to the greater capacity of macrophages to generate ROS, but could not exclude a role for nitric oxide derivatives. It has been demonstrated that the number of toxoplasma tachyzoites and intracellular cysts were increased in the T. gondii-infected mice treated for 2 weeks with aminoguanidine, NOS inhibitor (30). Therefore, it is possible that greater toxoplasmacidal activity of macrophages from Ucp2−/− mice was attributable not only to ROS but also to NO.
On the other hand, the down-regulation of macrophage UCP2 expression by LPS seems to be a cell-specific response, because luciferase activities in lung carcinoma cell line, LLC, transiently transfected with 5′UR construct were significantly increased after LPS stimulation (unpublished observation). Pecqueur et al. (16) have also revealed that UCP2 protein levels increase up to 12 times in lung from mice treated with LPS. Thus, the macrophage-specific regulatory mechanisms of UCP2 expression by LPS stimulation were investigated in RAW264 cells. The reduced level of UCP2 protein was accompanied by the mRNA expression in RAW264 cells stimulated with LPS, suggesting a negative regulation by LPS stimulation at the transcriptional level. Deletion analysis of the UCP2 promoter region revealed that (i) a strong enhancer was located between positions −233 and −33, (ii) a silencer was located between positions −2746 and −233. However, we could not detect any inhibitory activities in these regions after LPS stimulation. Rather, the promoter activity was enhanced by LPS stimulation in the −233 construct.
Evidence is growing that the gene expression is transcriptionally and/or posttranscriptionally regulated by intronic elements (31, 32). In addition, it has been reported that structural diversity in 5′-untranslated region (UTR) sequences may affect gene expression through secondary structural features or the presence of uORFs (33). For example, the 5′-UTR of the mouse ucp2 gene was interrupted by two introns and contains one uORF with three inframe AUG codons located in the second exon (34). These features may be consistent with tight regulation of UCP2 expression. Indeed, 5′UR construct with mutations in the AUG codons definitely increased luciferase activity. On the other hand, 5′UR construct with mutations in the upstream termination codon markedly decreased luciferase activity. It seems likely, thus, that uORF negatively regulates the expression of UCP2 and that this regulation of translation occurs not by leaky scanning but principally by ribosomal reinitiation. However, the inactivation of uORF did not affect the LPS-mediated negative regulation.
Meanwhile, the findings that luciferase activities were markedly decreased by the deletion of intron 2, but not by the deletion of AvrII or SacI fragment within intron 2, suggest the presence of transcriptional enhancer elements in intron 2, probably within its 5′ 43-bp region or 3′ 133-bp region. Simultaneously, LPS-associated negative regulation could be abolished in the construct without intron 2 but not in the construct without AvrII or SacI fragment within intron 2. It seems likely, therefore, that LPS-associated regulatory elements are also located in the same region and negatively regulate the UCP2 transcription by interrupting the function of the enhancer elements. We subcloned the 133-bp fragment corresponding to the 3′ region of intron 2 upstream of pGL3-promoter vector containing the minimal simian virus 40 promoter (Promega). Luciferase activities in these constructs were significantly higher than those in the pGL3-promoter vector and were decreased by LPS stimulation (unpublished observation), thereby suggesting that the 3′ 133-bp region of intron 2 plays an important role for both positive and negative regulation of UCP2 expression in cooperation with the promoter region. The transfected UCP2 expression in RAWucp2/m cells was greatly increased by LPS stimulation, probably because of the activity of the cytomegalovirus promoter. In contrast, the transfected UCP2 expression in RAWucp2/hn cells was substantially decreased even in the presence of cytomegalovirus promoter, supporting the presence of intron-associated tight down-regulatory mechanisms for the UCP2 expression under the LPS stimulation.
The production of reactive nitrogen and ROS by macrophages is critical for host defense. These mediators also cause tissue damage in inflammation. Brüne et al. (35) reported that the balance between reactive nitrogen and ROS regulates macrophage apoptosis. Therefore, the production of these mediators must be rapidly and strictly regulated. In the present study, we demonstrated that the potent negative regulation of UCP2 expression occurs rapidly in response to LPS, probably to ensure rapid and sufficient cellular responses against microbial attack. The present findings may contribute to the understanding of regulatory mechanisms of LPS-stimulated NOS II expression. Further studies on signal transduction cascades that participate in the positive/negative regulation of UCP2 expression would contribute to designing possible drugs that control bacterial infections.
Acknowledgments
This study was supported in part by grants from the Japanese Ministry of Education, Science, Sports, and Culture and from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
Abbreviations
- UCP2
uncoupling protein 2
- LPS
lipopolysaccharide
- ROS
reactive oxygen species
- NOS II
NO synthase II
- uORF
upstream open reading flame
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, Levi-Meyrueis C, Bouillaud F, Seldin M F, Surwit R S, Ricquier D, Warden C H. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 2.Gimeno R E, Dembski M, Weng X, Deng N, Shyjan A W, Gimeno C J, Iris F, Ellis S J, Woolf E A, Tartaglia L A. Diabetes. 1997;46:900–906. doi: 10.2337/diab.46.5.900. [DOI] [PubMed] [Google Scholar]
- 3.Klingenberg M, Huang S G. Biochim Biophys Acta. 1999;1415:271–296. doi: 10.1016/s0005-2736(98)00232-6. [DOI] [PubMed] [Google Scholar]
- 4.Rial E, Gonzalez-Barroso M, Fleury C, Iturrizaga S, Sanchis D, Jimenez-Jimenez J, Ricquier D, Goubern M, Bouillaud F. EMBO J. 1999;18:5827–5833. doi: 10.1093/emboj/18.21.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning B S, Miroux B, Couplan E, Alves-Guerra M C, Goubern M, Surwit R, et al. Nat Genet. 2000;26:435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 6.Negre-Salvayre A, Hirtz C, Carrera G, Cazenave R, Troly M, Salvayre R, Penicaud L, Casteilla L. FASEB J. 1997;11:809–815. [PubMed] [Google Scholar]
- 7.Murray H W, Juangbhanich C W, Nathan C F, Cohn Z A. J Exp Med. 1979;150:950–964. doi: 10.1084/jem.150.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larrouy D, Laharrague P, Carrera G, Viguerie-Bascands N, Levi-Meyrueis C, Fleury C, Pecqueur C, Nibbelink M, Andre M, Casteilla L, Ricquier D. Biochem Biophys Res Commun. 1997;235:760–764. doi: 10.1006/bbrc.1997.6852. [DOI] [PubMed] [Google Scholar]
- 9.Schletter J, Heine H, Ulmer A J, Rietschel E T. Arch Microbiol. 1995;164:383–389. doi: 10.1007/BF02529735. [DOI] [PubMed] [Google Scholar]
- 10.Ulevitch R J, Tobias P S. Curr Opin Immunol. 1999;11:19–22. doi: 10.1016/s0952-7915(99)80004-1. [DOI] [PubMed] [Google Scholar]
- 11.MacMicking J, Xie Q, Nathan C. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 12.Sanlioglu S, Williams C M, Samavati L, Butler N S, Wang G, McCray P B, Jr, Ritchie T C, Hunninghake G W, Zandi E, Engelhardt J F. J Biol Chem. 2001;276:30188–30198. doi: 10.1074/jbc.M102061200. [DOI] [PubMed] [Google Scholar]
- 13.Han Y J, Kwon Y G, Chung H T, Lee S K, Simmons R L, Billiar T R, Kim Y M. Nitric Oxide. 2001;5:504–513. doi: 10.1006/niox.2001.0367. [DOI] [PubMed] [Google Scholar]
- 14.Beckman J S, Beckman T W, Chen J, Marshall P A, Freeman B A. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wallace D C. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 16.Pecqueur C, Alves-Guerra M C, Gelly C, Levi-Meyrueis C, Couplan E, Collins S, Ricquier D, Bouillaud F, Miroux B. J Biol Chem. 2001;276:8705–8712. doi: 10.1074/jbc.M006938200. [DOI] [PubMed] [Google Scholar]
- 17.Ricquier D, Bouillaud F. Biochem J. 2000;345:161–179. [PMC free article] [PubMed] [Google Scholar]
- 18.Boss O, Hagen T, Lowell B B. Diabetes. 2000;49:143–156. doi: 10.2337/diabetes.49.2.143. [DOI] [PubMed] [Google Scholar]
- 19.Rothe G, Valet G. J Leukocyte Biol. 1990;47:440–448. [PubMed] [Google Scholar]
- 20.Ding H A, Nathan C F, Stuehr D J. J Immunol. 1991;141:2407–2412. [PubMed] [Google Scholar]
- 21.Kizaki T, Ookawara T, Iwabuchi K, Onoé K, Day N K, Good R A, Maruyama N, Haga S, Matsuura N, Ohira Y, Ohno H. J Leukocyte Biol. 2000;68:21–30. [PubMed] [Google Scholar]
- 22.Kizaki T, Suzuki K, Hitomi Y, Iwabuchi K, Onoé K, Haga S, Ishida H, Ookawara T, Suzuki K, Ohno H. Biochem Biophys Res Commun. 2001;289:1031–1038. doi: 10.1006/bbrc.2001.6123. [DOI] [PubMed] [Google Scholar]
- 23.Yoshitomi H, Yamazaki K, Tanaka I. Biochem J. 1999;340:397–404. [PMC free article] [PubMed] [Google Scholar]
- 24.Lee F Y, Li Y, Yang E K, Yang S Q, Lin H Z, Trush M A, Dannenberg A J, Diehl A M. Am J Physiol. 1999;276:C386–C394. doi: 10.1152/ajpcell.1999.276.2.C386. [DOI] [PubMed] [Google Scholar]
- 25.Vidal-Puig A J, Grujic D, Zhang C Y, Hagen T, Boss O, Ido Y, Szczepanik A, Wade J, Mootha V, Cortright R, et al. J Biol Chem. 2000;275:16258–16266. doi: 10.1074/jbc.M910179199. [DOI] [PubMed] [Google Scholar]
- 26.Korshunov S S, Skulachev V P, Starkov A A. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 27.Schulze-Osthoff K, Los M, Baeuerle P A. Biochem Pharmacol. 1995;50:735–741. doi: 10.1016/0006-2952(95)02011-z. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 29.Marks-Konczalik J, Chu S C, Moss J. J Biol Chem. 1998;273:22201–22208. doi: 10.1074/jbc.273.35.22201. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi S, Chan C C, Gazzinelli R, Roberge F G. J Immunol. 1996;156:1476–1481. [PubMed] [Google Scholar]
- 31.Schjerven H, Brandtzaeg P, Johansen F E. J Immunol. 2001;167:6412–6420. doi: 10.4049/jimmunol.167.11.6412. [DOI] [PubMed] [Google Scholar]
- 32.Morimura T, Miyatani S, Kitamura D, Goitsuka R. J Immunol. 2001;166:3277–3283. doi: 10.4049/jimmunol.166.5.3277. [DOI] [PubMed] [Google Scholar]
- 33.Griffin E, Re A, Hamel N, Fu C, Bush H, McCaffrey T, Asch A S. Nat Med. 2001;7:840–846. doi: 10.1038/89969. [DOI] [PubMed] [Google Scholar]
- 34.Pecqueur C, Cassard-Doulcier A M, Raimbault S, Miroux B, Fleury C, Gelly C, Bouillaud F, Ricquier D. Biochem Biophys Res Commun. 1999;255:40–46. doi: 10.1006/bbrc.1998.0146. [DOI] [PubMed] [Google Scholar]
- 35.Brune B, Gotz C, Messmer U K, Sandau K, Hirvonen M R, Lapetina E G. J Biol Chem. 1997;272:7253–7258. doi: 10.1074/jbc.272.11.7253. [DOI] [PubMed] [Google Scholar]