Abstract
Expression of the human β amyloid peptide (Aβ) in transgenic Caenorhabditis elegans animals can lead to the formation of intracellular immunoreactive deposits as well as the formation of intracellular amyloid. We have used this model to identify proteins that interact with intracellular Aβ in vivo. Mass spectrometry analysis of proteins that specifically coimmunoprecipitate with Aβ has identified six likely chaperone proteins: two members of the HSP70 family, three αB-crystallin-related small heat shock proteins (HSP-16s), and a putative ortholog of a mammalian small glutamine-rich tetratricopeptide repeat-containing protein proposed to regulate HSP70 function. Quantitative reverse transcription–PCR analysis shows that the small heat shock proteins are also transcriptionally induced by Aβ expression. Immunohistochemistry demonstrates that HSP-16 protein closely colocalizes with intracellular Aβ in this model. Transgenic animals expressing a nonaggregating Aβ variant, a single-chain Aβ dimer, show an altered pattern of coimmunoprecipitating proteins and an altered cellular distribution of HSP-16. Double-stranded RNA inhibition of R05F9.10, the putative C. elegans ortholog of the human small glutamine-rich tetratricopeptide-repeat-containing protein (SGT), results in suppression of toxicity associated with Aβ expression. These results suggest that chaperone function can play a role in modulating intracellular Aβ metabolism and toxicity.
A large body of genetic, transgenic, and cell culture studies has implicated the β amyloid peptide (Aβ), a primary component of the extracellular senile plaques characteristic of Alzheimer's disease (AD), as playing a central role in the pathology of this disease (1). However, there is also substantial evidence from transgenic mouse models that Aβ-dependent toxicity can occur independently of extracellular plaque formation per se. Neuronal pathology preceding plaque formation has been demonstrated in transgenic mice expressing human FAD mutant PS1 (2), FAD mutant APP (3–5), or both mutant proteins (6). These studies suggest that Aβ-dependent toxicity can occur before significant extracellular accumulation, possibly involving intracellular Aβ accumulation. Numerous studies with neuronal cell culture have demonstrated that Aβ can accumulate intracellularly (7–10), after either endogenous Aβ production or uptake of extracellular Aβ. Intracellular Aβ dimers have been detected in primary human neurons and in neuronal cell lines (11), and intraneuronal Aβ42 has also been demonstrated in human brain (12). Immunohistochemical analysis has been used to argue that intraneuronal Aβ contributes to plaque formation after neuronal lysis (13), and that intraneuronal Aβ distribution correlates with expression of the α7 nicotinic acetylcholine receptor (14).
If intracellular Aβ contributes to AD pathology, it would be informative to identify proteins that interact with Aβ intracellularly, as these proteins may be directly involved in Aβ metabolism or toxicity. Although many serum proteins have been identified that bind to Aβ and/or senile plaques, only a few studies have sought to identify candidate intracellular Aβ-binding proteins (15, 16), presumably due to the difficulty of performing coimmunoprecipitation studies against a small nonabundant intracellular peptide. One intracellular protein, HADH II, was initially found to interact with Aβ by yeast two-hybrid studies (17).
We have developed a transgenic Caenorhabditis elegans model that is well suited for identifying intracellular Aβ-interacting proteins. In this model, a strong muscle-specific promoter drives the expression of a chimeric signal peptide/human Aβ1–42 minigene designed to route Aβ into the secretory pathway (18). These animals express high intracellular levels of human Aβ1–42 (19), leading to the formation of intracellular β amyloid (20) and a concomitant progressive paralysis phenotype. We have now used this model to define intracellular binding partners of Aβ that may contribute to, or be a response against, Aβ toxicity.
Materials and Methods
Strains and General Methods.
Construction of transgenic strains CL2006 (dvIs2) and CL3109 (dvIs10) has been previously described (18, 19). Strain CL2179 [smg-1ts(cc546); dvEx179] contains a high-transmittance extrachromosomal array containing Fire lab expression vector L3808 [myo-3/green fluorescent protein (GFP), body wall muscle-specific GFP (see http://ftp.ciwemb.edu/PNF:byName:/FireLabWeb/FireLabInfo/FireLabVectors/) and pRF4 [rol-6(su1006), the dominant Roller morphological marker used in the construction of strains CL2006 and CL3109]. Strains were propagated at 20°C. Large-scale synchronized populations were prepared by alkaline hypochlorite egg purification and propagation of staged animals on 100-mm Petri plates containing nematode growth media supplemented with 2% peptone (21).
Immunoprecipitation.
A coimmunoprecipitation protocol was developed on the basis of previously described procedures (22). Transgenic animals (mixed-stage populations) were resuspended in protease inhibitor, then flash frozen in liquid nitrogen. Frozen pellets were ground with a mortar and pestle, and the resulting slurry was resuspended in a Tris/Triton X-100 immunoprecipitation buffer [50 mM Tris⋅HCl, pH 7.5/0.1% Triton X-100/100 mM NaCl/15 mM EDTA/1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride/1 mM DTT/1 mM phenylmethylsulfonyl fluoride]. After pelleting insoluble material, the lysate was incubated with antibody [0.3 μg/ml of lysate for anti-Aβ monoclonal 4G8 (Signet Laboratories, Dedham, MA) or 0.2 μg/ml for anti-GFP monoclonal antibody (Quantum Biotechnologies, Montreal, Quebec, Canada)] on ice for 90 min, and then the antibody/antigen complexes were recovered by incubation with protein A-Sepharose beads. After pelleting and washing of the beads, recovered proteins were fractionated by SDS/PAGE on 4–20% polyacrylamide Novex gels (Invitrogen).
Mass Spectrometry.
Protein gels were stained with silver nitrate under conditions compatible for in-gel digestion with modified trypsin protease (Promega) and mass spectrometry (23, 24). Peptides were desalted/concentrated by using C18 ZipTips (Millipore) before target-mixing with α-cyano-4-hydroxycinnamic acid matrix (10 mg/ml in 80% acetonitrile) and mass analysis by using a Voyager DE-STR matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (Applied Biosystems) operated in delayed extraction and reflectron mode. Trypsin autolytic peptides (m/z = 515.33, 842.51, 1045.56, 2211.10) were used to internally calibrate each mass spectrum to a mass accuracy within 50 ppm. Mass spectrometry was performed in the Biochemical Mass Spectrometry Facility, School of Pharmacy, University of Colorado Health Sciences Center.
Quantitative Reverse Transcription–PCR (RT-PCR).
Total RNA was prepared from staged worms by using the acid-phenol method (Trizol, GIBCO/BRL). Synthesis of single-stranded cDNA was performed by using an oligo(dT) primer with the SuperScript cDNA synthesis kit (Invitrogen). Gene-specific primers were designed by using primer express software (ABI Prism), and quantitative RT-PCR was performed on the cDNAs by using the SYBR green chemistry on an ABI Prism 7000 (Applied Biosystems). Relative quantitation of mRNA levels was determined by standardizing against a nonvariable control gene (F23B2.13), and data analysis was performed by using methods as described in ABI Prism 7000 user bulletin no. 2. All determinations were replicated at least three times.
RNA Inhibition (RNAi) by Feeding.
To construct Escherichia coli strains expressing specific double-stranded RNAs (dsRNA), genomic sequence encompassing all of a specific C. elegans gene (e.g., R05F9.10) was amplified by using forward and reverse primers containing 5′ extensions encoding a T7 promoter sequence (TGAATTGTAATACGACTCACTATAGGGAGA), and the resulting PCR product was cloned into a TOPO XL vector (Invitrogen). The resulting dsRNA-expressing plasmid was subsequently transformed into E. coli strain HT115, and lawns of induced (dsRNA-expressing) bacteria were prepared on nematode growth media + isopropyl β-D-thiogalactoside + kanamycin plates as previously described (25). Third-larval-stage CL2006 animals were propagated on feeding RNAi plates until they reached adulthood, then transferred to fresh RNAi plates and allowed to lay eggs for 2–4 h. After removal of the adult parental animals, the synchronous progeny populations were allowed to reach adulthood (≈5 days), then scored for paralysis.
Results
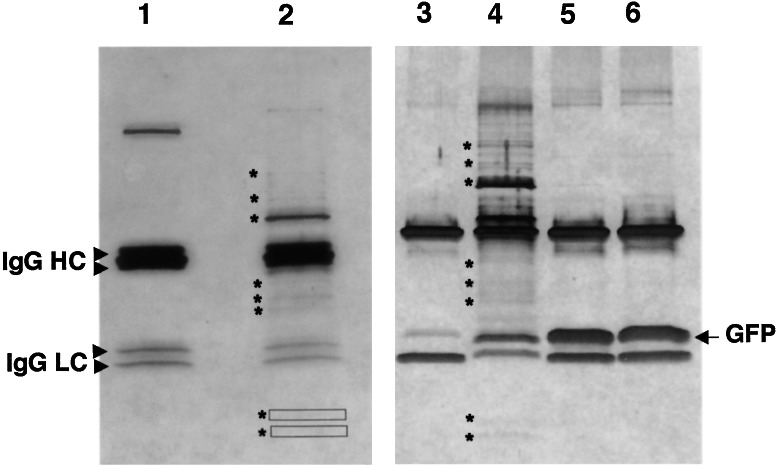
The well characterized anti-Aβ monoclonal antibody 4G8 (26) was used to immunoprecipitate Aβ from mixed-stage populations of transgenic line CL2006 and control wild-type animals. As shown in Fig. 1, fractionation of immunoprecipitates by SDS/PAGE identified >10 bands that specifically coimmunoprecipitated with lysate from Aβ-expressing line CL2006 (Fig. 1, lanes 2 and 4). These bands were not observed in control immunoprecipitations by using a monoclonal antibody against GFP (Fig. 1, lanes 3, 5, 6). All protein bands detectable by silver staining were excised, digested in gel with trypsin, and subjected to matrix-assisted laser desorption ionization/time-of-flight mass spectrometry. Six proteins were successfully identified by comparison of tryptic peptide masses to the set of predicted C. elegans proteins (see Table 1). All of the identified proteins have a likely role in chaperone function. The identified proteins include two HSP70 homologs (C15H9.6 and F26D10.3), three small heat shock proteins with homology to αB-crystallin [T27E4.3 (HSP-16–1), Y46H3A.d (HSP-16–2), and T27E4.3 (HSP-16–48)], and a tetratricopeptide repeat-containing protein (R05F9.10) that is the apparent ortholog of the human small glutamine-rich tetratricopeptide-repeat-containing protein (SGT), which has been reported to bind to hsc7C (27).
Figure 1.
Specific proteins coimmunoprecipitate with intracellular Aβ. Lysates of adult wild-type, CL2006 (Aβ-expressing), or CL2179 (GFP-expressing) animals were incubated with mAb 4G8 or control anti-GFP mAb, and the resulting immunoprecipitates were fractionated on SDS 4–20% polyacrylamide gels and visualized by silver staining. Lane 1, wild-type animals immunoprecipitated with 4G8; lane 2, CL2006 animals immunoprecipitated with 4G8; lane 3, CL2006 animals immunoprecipitated with anti-GFP; lane 4, CL2006 animals immunoprecipitated with 4G8; lane 5, heat-shocked CL2179 animals immunoprecipitated with anti-GFP; lane 6, untreated CL2179 animals immunoprecipitated with anti-GFP. Specific bands reproducibly identified in all CL2006 immunoprecipitation performed to date (nine independent immunoprecipitations) are indicated with asterisks (lanes 2 and 4). All visible bands in lane 2 were subjected to mass spectrometry analysis. The boxes in lane 2 indicate faint bands also recovered for MS analysis. The heavy GFP band in lanes 5 and 6 was confirmed by immunoblot (data not shown). IgG HC, Ig heavy chains; IgG LC, Ig light chains.
Table 1.
Coimmunoprecipitating proteins identified by mass spectrometry and microarray analysis (RNA expression)
| Gene product | Description | Molecular mass,
kDa
|
Molecular weight search score | Z score | Protein coverage, % | Aβ1-42 fold induction | Aβ dimer fold induction | |
|---|---|---|---|---|---|---|---|---|
| Apparent | Predicted | |||||||
| C15H9.6 | HSP70C, BiP/GRP78 ortholog | 71 | 73 | 2.3 × 105 (7.94) | 1.76 | 16 | 0.8 (0.69–0.89) | 0.5 (0.42–0.6) |
| F26D10.3 | HSP70A, cytoplasmic HSP70 | 70 | 69.7 | 4.8 × 106 (23.3) | 2.41 | 31 | 0.55 (0.4–0.76) | 0.86 (0.74–0.98) |
| R05F9.10 | SGT ortholog | 37 | 36.5 | 363 (77) | 1.19 | 20 | 0.86 (0.72–1.05) | 0.6 (0.52–0.69) |
| T27E4.2 | HSP-16-1, αB-crystallin homolog | 16 | 16.4 | 5.9 × 104 (97.3)* | 1.93* | 46 | 9.9 (7.5–11.6) | 4.3 (3.4–5.4) |
| Y46H3A.d | HSP-16-2, αB-crystallin homolog | 16 | 16.4 | 5.5 × 103 (178)*† | 0.45*† | 34 | 9.5 (6.9–13.0) | 6.0 (5.6–6.7) |
| T27E4.3 | HSP-16-48, αB-crystallin homolog | 13 | 16.4 | 407 (148) | 0.97 | 59 | 14 (11.6–20.0) | 6.7 (6.2–8.3) |
Ions derived from Aβ (m/z = 1,325.65 and 1,336.63) included in searches did not match identified proteins but did reduce significance scores.
† Found in a mixture with HSP-16-1. Lower significance scores were due to masses in common between HSP-16-1 and -2. Peptide mass maps were acquired by MALDI-TOF mass spectrometry and used to search the SWISSPROT and NCBInr databases (all species) by using the ms-fit (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm) and ProFound (www.proteometrics.com) algorithms. Significant molecular weight search scores (bold) are at least one order of magnitude greater than the next best match (value in parentheses) that represents a nonhomologous protein. Significant Z scores (bold) are within the 95% confidence interval. Protein coverage (%) reflects the amount of protein sequence accounted for among the tryptic peptides matched in the search. Fold induction determined by assaying transcript levels by quantitative reverse transcription–PCR for transgenic strains CL2006 (Aβ1-42) and CL3109 (Aβ single-chain dimer), then comparing these values to those determined for control transgenic strain CL2179 (GFP). Values are presented as average relative induction and standard deviation-derived range. Genes with significant induction highlighted in bold.
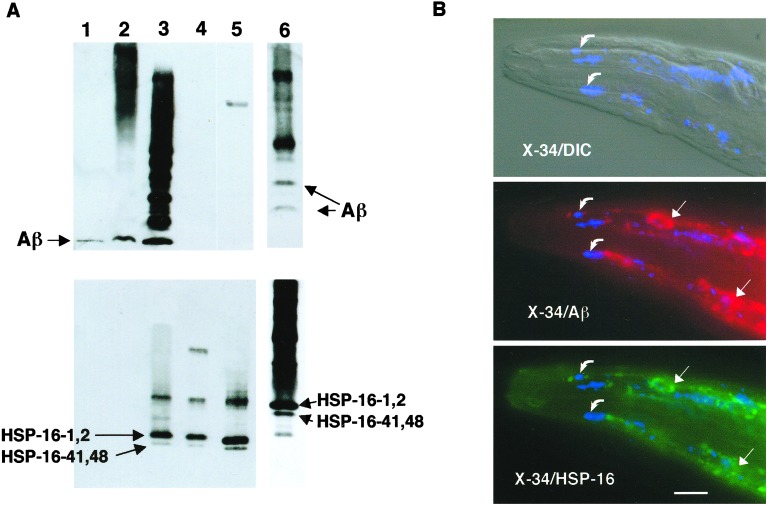
To confirm the identification of HSP-16 proteins in the coimmunoprecipitates, an immunoblot of immunoprecipitated proteins was sequentially probed with mAb 4G8 and a polyclonal antibody raised against an HSP-16–2 peptide (28). This antibody reacts strongly against the closely related HSP-16–1/HSP-16–2 family of small heat shock proteins and weakly against the more distantly related HSP-16–41/HSP-16–48 class. As shown in Fig. 2A, HSP-16 coimmunoprecipitates with Aβ. In complementary experiments immunoprecipitating with anti-HSP-16 antibody, Aβ is also found to coimmunoprecipitate in CL2006 lysates (Fig. 2A, lane 6). Interestingly, the species of Aβ that coimmunoprecipitated with HSP-16 appeared to be predominantly multimeric. To determine whether this Aβ/HSP-16 interaction occurs in vivo, CL2006 transgenic animals were fixed and probed with mAb 4G8, the amyloid-specific dye X-34 (29), and anti-HSP-16 antibody. HSP-16 was found to tightly colocalize with immunoreactive Aβ deposits but not with the fully amyloidic Aβ detected by X-34.
Figure 2.
Association of Aβ with HSP-16. (A) Coimmunoprecipitation of Aβ and HSP-16. Lysates from CL2006 were immunoprecipitated with either mAb 4G8 (lanes 1–5) or anti-HSP-16 antibody (lane 6) and fractionated along with control preparations on a Tris-Bicine/SDS polyacrylamide gel, and then blotted and sequentially probed with 4G8 (Upper) and anti-HSP16 antibody (Lower). Lane 1, synthetic Aβ1–42; lane 2, total CL2006 lysate; lane 3, CL2006 4G8 immunoprecipitate; lane 4, concentrated post-4G8-IP lysate; lane 5, heat-shocked wild-type animals; lane 6, CL2006 anti-HSP-16 immunoprecipitate (run on a separate gel, resulting in different mobilities than lanes 1–5). Note both Aβ and HSP-16 proteins detected in both immunoprecipitates (lanes 3 and 6). (B) Immunohistochemical localization of Aβ, HSP-16, and amyloid dye-reactive deposits in CL2006. CL2006 animals were vitally stained with the amyloid-specific dye X-34 and then fixed, permeabilized, and probed with 4G8 and anti-HSP-16 antibody. (Top) Digitally fused differential contrast (DIC)/shortwave epifluorescence (X-34) image; (Middle) fused X-34/4G8 epifluorescence image; (Bottom) fused X-34/anti-HSP-16 epifluorescence image. (All epifluorescence images are false color.) Note nearly identical patterns of Aβ- and HSP-16-reactive deposits (straight arrows), which do not generally overlap with X-34-reactive amyloid deposits (curved arrows). (Bar = 25 μm.)
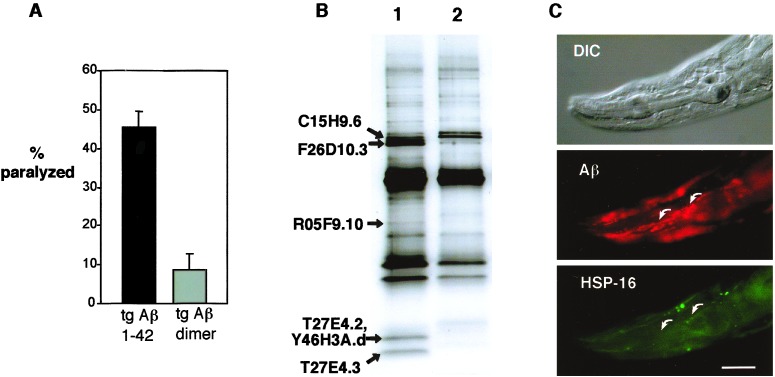
To examine the specificity of this apparent interaction of Aβ with chaperone proteins in this transgenic model, we repeated the coimmunoprecipitations with a transgenic line, CL3109, that expresses a nonamyloidic Aβ variant, the single-chain Aβ dimer. These transgenic animals contain a muscle promoter-Aβ minigene construct similar to that expressed in CL2006 but express a protein consisting of two Aβ1–42 sequences joined by a short linker peptide. Although this strain expresses high levels of this single-chain dimer protein, these animals do not produce detectable amyloid deposits (19) and show significantly reduced levels of paralysis (Fig. 3A). The pattern of proteins immunoprecipitated with mAb 4G8 from CL3109 is significantly different from that of CL2006 (Fig. 3B), with the F26D10.3 HSP70 and HSP-16 bands reduced or absent. The reduction of HSP-16 in the immunoprecipitate was confirmed by immunoblot (data not shown). In addition, CL3109 animals probed with 4G8 and anti-HSP-16 antibody do not show the tight colocalization of Aβ and HSP-16 observed in CL2006, with many (but not all) 4G8-reactive deposits lacking detectable anti-HSP-16 antibody binding (Fig. 3C). These results suggest that in this model, the chaperone response to intracellular Aβ is specifically associated with some conformational or toxic property of Aβ and is not due to a general response to high expression to a foreign protein. We also note that neither HSP-16 nor HSP70 (F26D10.3) bands are found in control experiments in which GFP is immunoprecipitated from heat-shocked CL2179 animals, which have high-level muscle-specific expression of GFP (Fig. 1, lane 5). This result further supports the specific association of these proteins with Aβ.
Figure 3.
Altered phenotype and Aβ/protein interaction in transgenic animals expressing Aβ1–42 single-chain dimer. (A) Synchronous CL2006 and CL3109 (Aβ single-chain dimer-expressing) animals were raised at 20°C and scored for paralysis as young adults. (B) Silver-stained SDS 4–20% polyacrylamide gel of 4G8 immunoprecipitates from CL2006 (lane 1) and CL3109 (lane 2). Note strong reduction of HSP70 (F26D10.3) and HSP-16 (T27E4.2, Y46H3A.d, and T27E4.3) bands in CL3109 immunoprecipitates. (Aβ or Aβ dimer was efficiently recovered in immunoprecipitations as determined by immunoblot; data not shown.) (C) CL3109 animal fixed, permeabilized, and probed with 4G8 (Middle) and anti-HSP-16 (Bottom) (1). Note absence of Aβ/HSP-16 colocalization, arrows (compare with Fig. 2B). (Bar = 25 μm.)
The differential recovery of proteins immunoprecipitated from CL2006 and CL3109 could result from differential association of chaperone proteins with Aβ and the Aβ dimer or from differential induction of the chaperone proteins (or both). We measured transcript levels of the identified chaperone proteins by quantitative PCR in CL2006, CL3109, and control transgenic strain CL2179 (Table 1). We found that the HSP-16 transcripts showed significant up-regulation in both Aβ-expressing strains. Relative to control strain CL2179, CL2006 animals had an ≈10- to 15-fold increase in HSP-16 transcripts, whereas CL3109 showed an ≈4- to 6-fold increase. The reduced recovery of HSP-16 proteins in coimmunoprecipitation experiments with CL3109 may therefore result in part from a weaker induction of these proteins by the Aβ dimer variant. In contrast, transcript levels of the cytoplasmic HSP70 (F26D10.3) were not increased in either CL2006 or CL3109. These results suggest the specific coimmunoprecipitation of cytoplasmic HSP70 with Aβ1–42 results from a specific interaction that does not occur with the nonaggregating Aβ dimer variant. Interestingly, although HSP70 (F26D10.3) and HSP-16 are both transcriptionally induced by heat shock (30, 31), they apparently are not coordinately transcriptionally up-regulated in response to constitutive Aβ expression.
To investigate the role of the coimmunoprecipitating proteins in the pathological phenotype exhibited by the transgenic animals, we attempted to alter the endogenous chaperone response in these animals by inhibiting expression of the identified coimmunoprecipitating proteins by dsRNAi. The most informative of these experiments (see Discussion) resulted from RNAi of R05F9.10, the C. elegans ortholog of the human SGT protein. The SGT protein binds to the C terminus of HSP70 and appears to be a negative regulator of HSP70-dependent chaperone function, based on in vitro luciferase activity reconstitution experiments (32). Thus, inhibition of R05F9.10 would be predicted to enhance HSP70-dependent chaperone function. To inhibit R05F9.10 expression, CL2006 animals were propagated on an E. coli strain expressing dsR05F9.10 RNA (feeding RNAi; ref. 33). (Body wall muscle cells are particularly susceptible to this form of specific gene inhibition, one of the rationales for using this transgenic model.) RNAi of R05F9.10 suppressed the progressive paralysis in CL2006 animals, with the fraction of young adult animals paralyzed reduced from 54% (±6.4 SEM) in animals exposed to control GFP RNAi to 17% (±3.8 SEM) in animals exposed to R05F9.10 RNAi. These suppressed animals showed no detectable change in the amount or distribution of Aβ, HSP-16, or X-34-reactive deposits (data not shown).
Discussion
Previous studies have suggested the involvement of chaperone proteins in AD pathology. Early studies using immunohistochemistry and immunoblotting suggested that expression of both HSP70-class (34, 35) and αB-crystallin-related proteins (36, 37) was increased in AD brains, and both classes of proteins were associated with senile plaques. A more recent study using two-dimensional PAGE coupled with mass spectrometry confirmed the increased expression of αB-crystallin but found that specific HSP70-class proteins may be increased (HSPA4), decreased (HSPA8), or unchanged (HSPA1B, HSPA5) in specific regions of AD brains (38). Interpretation of these analyses of postmortem AD tissue is difficult, because it cannot be determined at what stage in the disease these changes occurred or how directly the chaperone proteins are involved in the pathological process. Our finding that chaperone proteins interact with intracellular Aβ suggests that these proteins may play an early role in Aβ metabolism. We note that, although our studies were designed to identify Aβ-interacting proteins in an unbiased manner, to date the only interacting proteins we have positively identified are likely chaperone proteins.
We find that αB-crystallin-homologous HSP-16 proteins closely colocalize with intracellular Aβ. In vitro, αB-crystallin can physically interact with Aβ1–40 (39) and has been reported to inhibit fibril formation by Aβ1–42 (40). However, Aβ1–40 preparations preincubated with αB-crystallin have been reported to have enhanced toxicity, despite reduced fibril formation (41). We observed HSP-16 protein associated with 4G8-immunoreactive deposits but not with amyloid aggregates detected with the amyloid-specific dye X-34. Thus, HSP-16 is likely to interact with Aβ monomer or some prefibrillar Aβ oligomer. Whether HSP-16 induction by and binding to Aβ are protective in this transgenic C. elegans model is unclear, as we have been unable to completely ablate HSP-16 expression by RNAi (likely due to the family of closely related genes encoding this protein class).
The predominant protein in Aβ coimmunoprecipitates is F26D10.3, which is a cytoplasmic HSP70 whose human homologs are encoded by the HSPA1/2 gene family. This HSP70 apparently has an essential role in C. elegans development, because RNA inhibition of F26D10.3 leads to embryonic lethality or developmental arrest (unpublished observations). The interaction of Aβ1–42 and F26D10.3 appears highly specific, because this protein is not efficiently recovered in immunoprecipitates from transgenic animals expressing an Aβ1–42 single-chain dimer. This observation suggests Aβ1–42 can attain a conformation that the nonamyloidic single-chain dimer cannot. There is strong evidence for the involvement of HSP70 in neurodegenerative diseases (reviewed in ref. 42). HSP70 overexpression can reverse polyglutamine-repeat-dependent toxicity in fly (43), cell culture (44), and mouse (45) models. Expression of a human HSPA1-class HSP70 has also recently been shown to suppress α-synuclein-dependent toxicity in a fly Parkinson's disease model (46). We have used dsRNA inhibition of a putative negative regulator of HSP70 function to endogenously increase HSP70 chaperone activity, and we similarly see protection against Aβ-dependent toxicity. Our results suggest that, as proposed for other age-associated neurodegenerative diseases, chaperone function may play a direct and early role in AD.
Our experiments cannot determine whether the identified chaperone proteins associate with Aβ individually or as part of a multicomponent complex. Interestingly, both a cytoplasmic HSP70 (F26D10.3) and an endoplasmic reticulum-localized HSP70 (C15H9.6, orthologous to GRP78/BiP) are recovered in Aβ immunoprecipitates. We have also performed RNAi experiments against C15H9.6; however, the results of these experiments are difficult to interpret, because RNAi of C15H9.6 leads to the strong up-regulation of another chaperone protein (V.K., unpublished observations). The identification of C15H9.6 as a coimmunoprecipitating protein is consistent with the routing of Aβ to the secretory pathway, as expected with the signal peptide-containing Aβ minigene used in this model. However, Aβ is not effectively secreted from muscle cells in transgenic animals and ultimately appears cytoplasmic, as observed by immuno-electron microscopy (20). We hypothesize that in this transgenic C. elegans model, Aβ is recognized as an abnormal protein and actively rerouted from the secretory pathway to an alternative compartment for refolding or degradation. This metabolism of Aβ may parallel that of prion protein (PrP) expressed in transfected cells, which is apparently continually subjected to endoplasmic reticulum quality control and retrograde transport, and where proteasome inhibitor treatment leads to cytoplasmic accumulation of PrP colocalized with HSP70 (47). It is of particular interest whether this proposed intracellular Aβ metabolism also occurs to some degree in human neurons. We speculate that the ability of some nonsteroidal anti-inflammatory drugs to inhibit Aβ1–42 secretion from cultured cells (48) and to reduce plaque load in transgenic Aβ mice (49) may result from the previously demonstrated ability of these drugs to modulate cellular chaperone functions (50–52).
Acknowledgments
We thank Dr. E. P. Candido (University of British Columbia) for providing the anti-HSP-16 antibody, Dr. A. Fire (Carnegie Institute of Washington) for providing expression vector pPD118.60, and K. Drabikowski (Friedrich Miescher Institute) for advice on the immunoprecipitation protocol. This work was supported by the National Institutes of Health (AG-12423) and a Temple Award from the Alzheimer's Association (to C.D.).
Abbreviations
- Aβ
human β amyloid peptide
- AD
Alzheimer's disease
- GFP
green fluorescent protein
- RNAi
RNA inhibition
- ds
double stranded
References
- 1.Selkoe D J. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 2.Chui D H, Tanahashi H, Ozawa K, Ikeda S, Checler F, Ueda O, Suzuki H, Araki W, Inoue H, Shirotani K, et al. Nat Med. 1999;5:560–564. doi: 10.1038/8438. [DOI] [PubMed] [Google Scholar]
- 3.Hsia A Y, Masliah E, McConlogue L, Yu G Q, Tatsuno G, Hu K, Kholodenko D, Malenka R C, Nicoll R A, Mucke L. Proc Natl Acad Sci USA. 1999;96:3228–3233. doi: 10.1073/pnas.96.6.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Haute C V, Checler F, et al. J Biol Chem. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- 5.Kumar-Singh S, Dewachter I, Moechars D, Lubke U, De Jonghe C, Ceuterick C, Checler F, Naidu A, Cordell B, Cras P, et al. Neurobiol Dis. 2000;7:9–22. doi: 10.1006/nbdi.1999.0272. [DOI] [PubMed] [Google Scholar]
- 6.Wirths O, Multhaup G, Czech C, Blanchard V, Moussaoui S, Tremp G, Pradier L, Beyreuther K, Bayer T A. Neurosci Lett. 2001;306:116–120. doi: 10.1016/s0304-3940(01)01876-6. [DOI] [PubMed] [Google Scholar]
- 7.Wertkin A M, Turner R S, Pleasure S J, Golde T E, Younkin S G, Trojanowski J Q, Lee V M. Proc Natl Acad Sci USA. 1993;90:9513–9517. doi: 10.1073/pnas.90.20.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wild-Bode C, Yamazaki T, Capell A, Leimer U, Steiner H, Ihara Y, Haass C. J Biol Chem. 1997;272:16085–16088. doi: 10.1074/jbc.272.26.16085. [DOI] [PubMed] [Google Scholar]
- 9.Yang A J, Chandswangbhuvana D, Margol L, Glabe C G. J Neurosci Res. 1998;52:691–698. doi: 10.1002/(SICI)1097-4547(19980615)52:6<691::AID-JNR8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 10.Sun X, Cole G M, Chu T, Xia W, Galasko D, Yamaguchi H, Tanemura K, Frautschy S A, Takashima A. Neurobiol Aging. 2002;23:195–203. doi: 10.1016/s0197-4580(01)00265-2. [DOI] [PubMed] [Google Scholar]
- 11.Walsh D M, Hartley D M, Kusumoto Y, Fezoui Y, Condron M M, Lomakin A, Benedek G B, Selkoe D J, Teplow D B. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 12.Gouras G K, Tsai J, Naslund J, Vincent B, Edgar M, Checler F, Greenfield J P, Haroutunian V, Buxbaum J D, Xu H, et al. Am J Pathol. 2000;156:15–20. doi: 10.1016/s0002-9440(10)64700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Andrea M R, Nagele R G, Wang H Y, Peterson P A, Lee D H. Histopathology. 2001;38:120–134. doi: 10.1046/j.1365-2559.2001.01082.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang H Y, D'Andrea M R, Nagele R G. Neurobiol Aging. 2002;23:213–223. doi: 10.1016/s0197-4580(01)00279-2. [DOI] [PubMed] [Google Scholar]
- 15.Ray I, Chauhan A, Wisniewski H M, Wegiel J, Kim K S, Chauhan V P. Neurochem Res. 1998;23:1277–1282. doi: 10.1023/a:1020744216699. [DOI] [PubMed] [Google Scholar]
- 16.Oyama R, Yamamoto H, Titani K. Biochim Biophys Acta. 2000;1479:91–102. doi: 10.1016/s0167-4838(00)00057-1. [DOI] [PubMed] [Google Scholar]
- 17.Yan S D, Fu J, Soto C, Chen X, Zhu H, Al-Mohanna F, Collison K, Zhu A, Stern E, Saido T, et al. Nature (London) 1997;389:689–695. doi: 10.1038/39522. [DOI] [PubMed] [Google Scholar]
- 18.Link C D. Proc Natl Acad Sci USA. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fay D S, Fluet A, Johnson C J, Link C D. J Neurochem. 1998;71:1616–1625. doi: 10.1046/j.1471-4159.1998.71041616.x. [DOI] [PubMed] [Google Scholar]
- 20.Link C D, Johnson C J, Fonte V, Paupard M, Hall D H, Styren S, Mathis C A, Klunk W E. Neurobiol Aging. 2001;22:217–226. doi: 10.1016/s0197-4580(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 21.Lewis J A, Fleming J T. In: Caenorhabditis elegans: Modern Biological Analysis of a Model Organism. Epstein H F, Shakes D C, editors. New York: Academic; 1996. pp. 3–29. [Google Scholar]
- 22.Elion E A. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Deidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1999. pp. 20.5.1–20.5.9. [Google Scholar]
- 23.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 24.Gharahdaghi F, Weinberg C R, Meagher D, Imai B, Mische S. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 25.Kamath R S, Martinez-Campos M, Zipperlen P, Fraser A G, Ahringer J. Genome Biol. 2000;2:1–10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim K S, Wen G Y, Bancher C, Chen C M J, Sapienza V J, Hong H, Wisniewski H M. Neurosci Res Commun. 1990;7:113–122. [Google Scholar]
- 27.Liu F H, Wu S J, Hu S M, Hsiao C D, Wang C. J Biol Chem. 1999;274:34425–34432. doi: 10.1074/jbc.274.48.34425. [DOI] [PubMed] [Google Scholar]
- 28.Hockertz M K, Clark-Lewis I, Candido E P. FEBS Lett. 1991;280:375–378. doi: 10.1016/0014-5793(91)80335-z. [DOI] [PubMed] [Google Scholar]
- 29.Styren S D, Hamilton R L, Styren G C, Klunk W E. J Histochem Cytochem. 2000;48:1223–1232. doi: 10.1177/002215540004800906. [DOI] [PubMed] [Google Scholar]
- 30.Snutch T P, Heschl M F, Baillie D L. Gene. 1988;64:241–255. doi: 10.1016/0378-1119(88)90339-3. [DOI] [PubMed] [Google Scholar]
- 31.Jones D, Russnak R H, Kay R J, Candido E P. J Biol Chem. 1986;261:12006–12015. [PubMed] [Google Scholar]
- 32.Wu S J, Liu F H, Hu S M, Wang C. Biochem J. 2001;359:419–426. doi: 10.1042/0264-6021:3590419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Timmons L, Court D L, Fire A. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 34.Perez N, Sugar J, Charya S, Johnson G, Merril C, Bierer L, Perl D, Haroutunian V, Wallace W. Mol Brain Res. 1991;11:249–254. doi: 10.1016/0169-328x(91)90033-t. [DOI] [PubMed] [Google Scholar]
- 35.Hamos J E, Oblas B, Pulaski-Salo D, Welch W J, Bole D G, Drachman D A. Neurology. 1991;41:345–350. doi: 10.1212/wnl.41.3.345. [DOI] [PubMed] [Google Scholar]
- 36.Shinohara H, Inaguma Y, Goto S, Inagaki T, Kato K. J Neurol Sci. 1993;119:203–208. doi: 10.1016/0022-510x(93)90135-l. [DOI] [PubMed] [Google Scholar]
- 37.Renkawek K, Bosman G J, de Jong W W. Acta Neuropathol (Berlin) 1994;87:511–519. doi: 10.1007/BF00294178. [DOI] [PubMed] [Google Scholar]
- 38.Yoo B C, Kim S H, Cairns N, Fountoulakis M, Lubec G. Biochem Biophys Res Commun. 2001;280:249–258. doi: 10.1006/bbrc.2000.4109. [DOI] [PubMed] [Google Scholar]
- 39.Liang J J. FEBS Lett. 2000;484:98–101. doi: 10.1016/s0014-5793(00)02136-0. [DOI] [PubMed] [Google Scholar]
- 40.Kudva Y C, Hiddinga H J, Butler P C, Mueske C S, Eberhardt N L. FEBS Lett. 1997;416:117–121. doi: 10.1016/s0014-5793(97)01180-0. [DOI] [PubMed] [Google Scholar]
- 41.Stege G J, Renkawek K, Overkamp P S, Verschuure P, van Rijk A F, Reijnen-Aalbers A, Boelens W C, Bosman G J, de Jong W W. Biochem Biophys Res Commun. 1999;262:152–156. doi: 10.1006/bbrc.1999.1167. [DOI] [PubMed] [Google Scholar]
- 42.Ohtsuka K, Suzuki T. Brain Res Bull. 2000;53:141–146. doi: 10.1016/s0361-9230(00)00325-7. [DOI] [PubMed] [Google Scholar]
- 43.Warrick J M, Chan H Y, Gray-Board G L, Chai Y, Paulson H L, Bonini N M. Nat Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 44.Jana N R, Tanaka M, Wang G, Nukina N. Hum Mol Genet. 2000;9:2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 45.Cummings C J, Sun Y, Opal P, Antalffy B, Mestril R, Orr H T, Dillmann W H, Zoghbi H Y. Hum Mol Genet. 2001;10:1511–1518. doi: 10.1093/hmg/10.14.1511. [DOI] [PubMed] [Google Scholar]
- 46.Auluck P K, Chan H Y, Trojanowski J Q, Lee V M, Bonini N M. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 47.Ma J, Lindquist S. Proc Natl Acad Sci USA. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weggen S, Eriksen J L, Das P, Sagi S A, Wang R, Pietrzik C U, Findlay K A, Smith T E, Murphy M P, Bulter T, et al. Nature (London) 2001;414:212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- 49.Lim G P, Yang F, Chu T, Gahtan E, Ubeda O, Beech W, Overmier J B, Hsiao-Ashec K, Frautschy S A, Cole G M. Neurobiol Aging. 2001;22:983–991. doi: 10.1016/s0197-4580(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 50.Lee B S, Chen J, Angelidis C, Jurivich D A, Morimoto R I. Proc Natl Acad Sci USA. 1995;92:7207–7211. doi: 10.1073/pnas.92.16.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ito H, Hasegawa K, Inaguma Y, Kozawa O, Kato K. J Cell Physiol. 1996;166:332–339. doi: 10.1002/(SICI)1097-4652(199602)166:2<332::AID-JCP11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Housby J N, Cahill C M, Chu B, Prevelige R, Bickford K, Stevenson M A, Calderwood S K. Cytokine. 1999;11:347–358. doi: 10.1006/cyto.1998.0437. [DOI] [PubMed] [Google Scholar]